Physiological and Molecular Timing of the Glucose to Acetate Transition in Escherichia coli
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Macrophenotype Parameters of the Glucose/Acetate Transition in a Model Condition
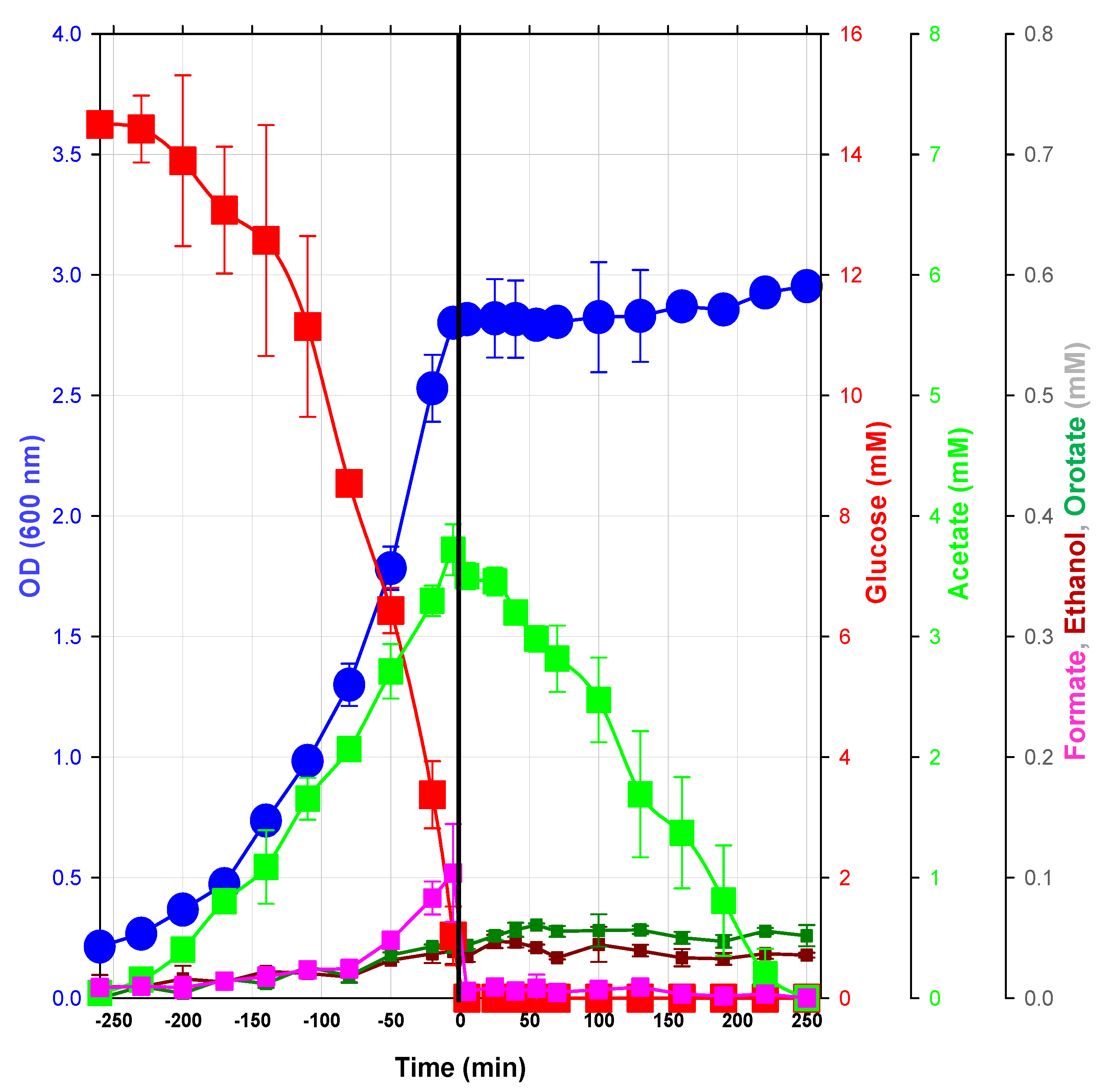
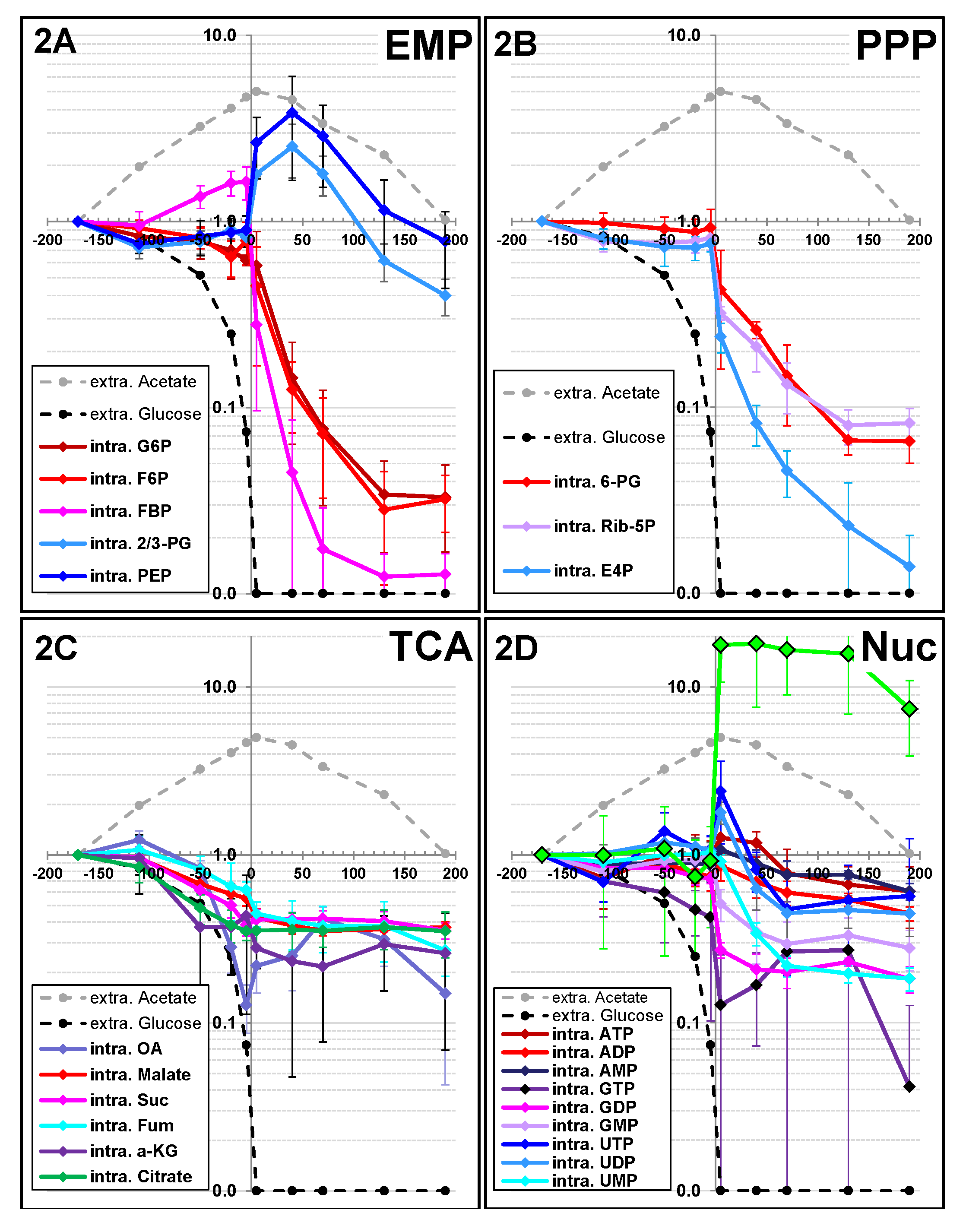
2.1.2. Changes in Metabolite Profiles
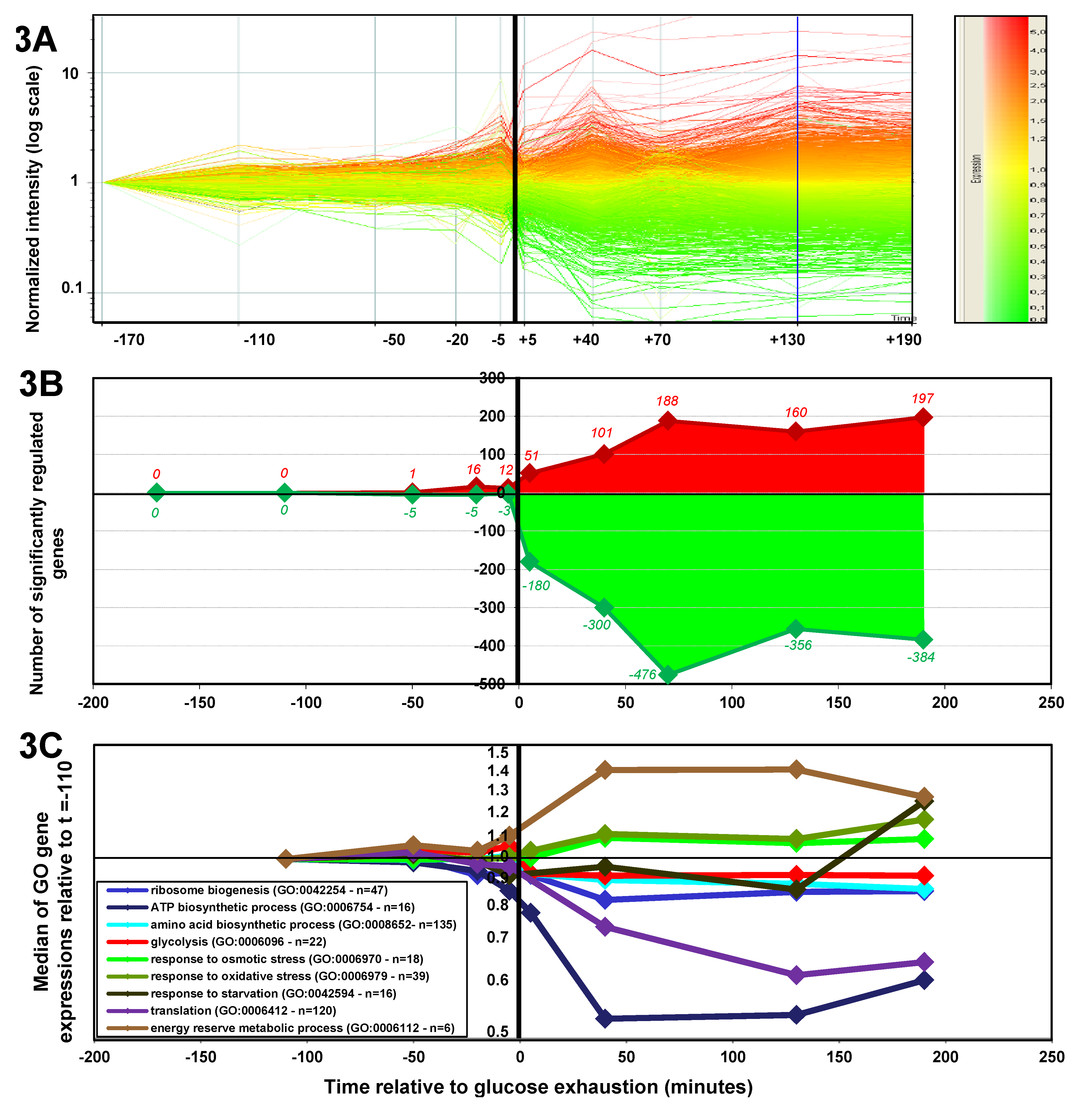
2.1.3. Changes in Gene Expression during the Glucose-Acetate Transition
2.1.4. Expression of Key Metabolic Genes during the Glucose-Acetate Transition
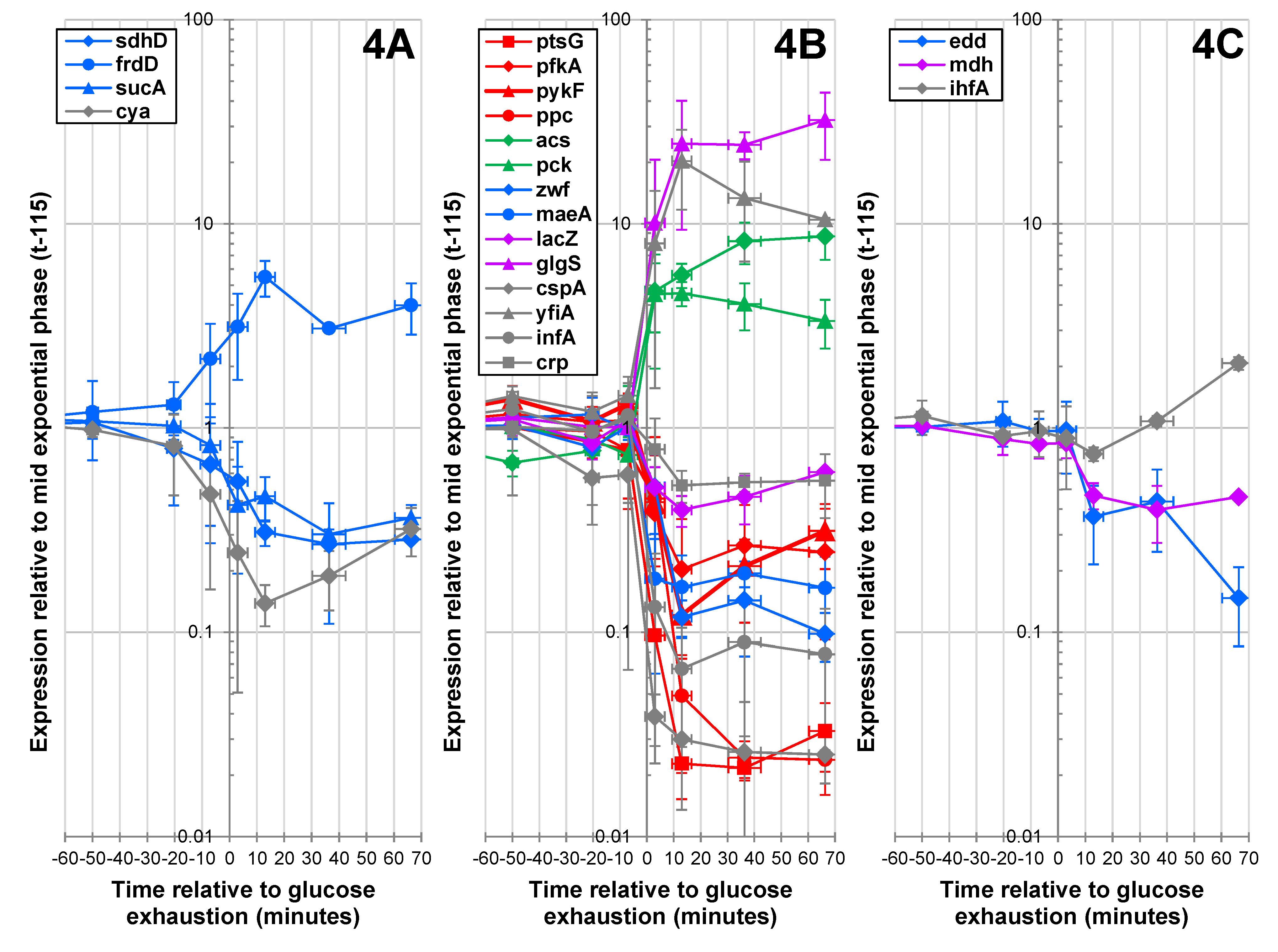
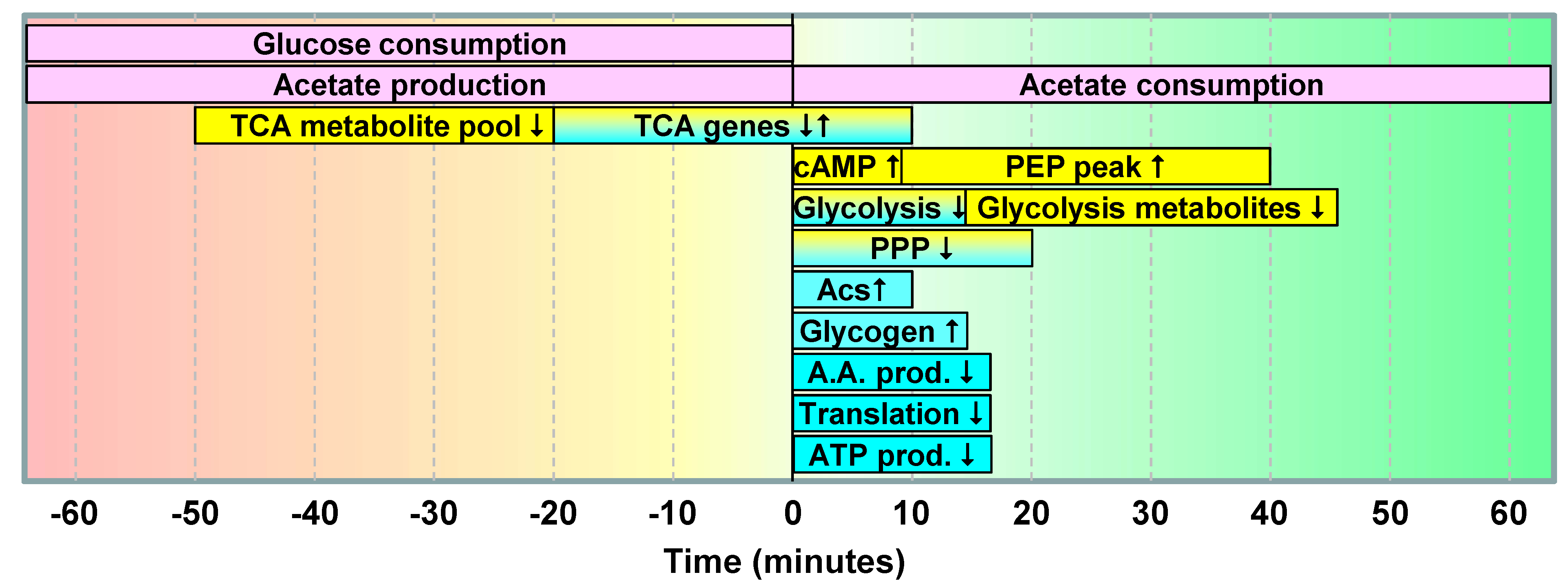
2.1.5. Data Integration

2.2. Discussion
3. Experimental Section
3.1. Strain, Media and Growth Conditions
3.2. Metabolome Analysis
3.3. Transcriptome Analysis
3.4. RT-PCR Analysis
| Name | Sequence (5' to 3') | Name | Sequence (5' to 3') |
|---|---|---|---|
| Q-16S-3' | ATCCGGACTACGACGCACTT | Q-lacZ3' | GCATAACCACCACGCTCATCG |
| Q-16S-5' | ACGACCAGGGCTACACACG | Q-lacZ5' | ACCTACGGGTAACAGTTTCTTTATG |
| Q-AceA-3' | AACCAGCAGGGTTGGAACG | Q-maeA3' | ATTGCGCGGGAGACTTTCTG |
| Q-AceA-5' | ACATGGGCGGCAAAGTTTTA | Q-maeA5' | AAGTGAAACGCTGGCGCAGT |
| Q-aceB-3' | TCAGGCCATAAATCGGCACA | Q-mdh3' | TGGCCCATAGACAGGGTTGC |
| Q-aceB-5' | GGTGAACGCACCGAAGAAGG | Q-mdh5' | CCGAGCAGGAAGTGGCTGAT |
| Q-acs-3' | GGATCTTCGGCGTTCATCTC | Q-pck3' | GTGTCTACGCCCGGCAGTTC |
| Q-acs-5' | GGGAAAATTGACTGGCAGGA | Q-pck5' | GACGCCATCCTCAACGGTTC |
| Q-crp3' | ACAGGCCCAGTTCGCCAATA | Q-pfkA3' | CACCCATGTAGGAACCGTCA |
| Q-crp5' | AGGCTCTGTGGCAGTGCTGA | Q-pfkA5' | AATTCCGCGACGAGAACATC |
| Q-cya3' | CCCGGCGGCACATAAATAAA | Q-ppc3' | CAGGCGAGAACGCAGGTTTT |
| Q-cya5' | GCCGCGTTTGAAGCATTACC | Q-ppc5' | ATGGTTGAAGCGACCCCTGA |
| Q-edd3' | ACCAAGTGGCGGCATGAGTT | Q-pps3' | CTGGCTCGTAACGCTCACCA |
| Q-edd5' | GTTTGCTGGACCGCGATTGT | Q-pps5' | GTGCCGCGTTTTATCCGAAG |
| Q-frdD3' | CCGCAGGTACGTGGATTTTC | Q-ptsG3' | GGAATGTCGCCGTGGAAAAC |
| Q-frdD5' | TGGTCGCGTATTCCTGTTCC | Q-ptsG5' | CCGTTTGGTCTGCACCACAT |
| Q-glgS3' | GCTCTCTTGCCTGCATCATCTG | Q-pykF3' | GCAACCATGATGCCGTCAGA |
| Q-glgS5' | CGGTCGATATTCTGGCCGTTA | Q-pykF5' | CGGCGAAAACATCCACATCA |
| Q-Icd-3' | TTCGTCACCGATGTTTGCAC | Q-sdhD3' | ACACACCCCACACCACAACG |
| Q-Icd-5' | CGCCTGTATGAACCTGAACG | Q-sdhD5' | CGTTAAACCGCTGGCTTTGC |
| Q-ihfA3' | TACTCGTCTTTGGGCGAAGC | Q-sucA3' | GGTGTCAGGGTCGGAGATCG |
| Q-ihfA5' | GCGAGGATATTCCCATTACAGC | Q-sucA5' | ACGGGAGTCAAACCGGATCA |
| Q-ihfB-3' | CAAAGAGAAACTGCCGAAACC | Q-yfiA3' | TGTGGCGTCAGCAACAAACC |
| Q-ihfB-5' | GCCAAGACGGTTGAAGATGC | Q-yfiA5' | ACCGTCTCGCCAAACTGGAA |
| Q-infA3' | ACAATGCGGCCTTTGCTCAG | Q-zwf3' | GCCCTTCGATCCCCACTTCT |
| Q-infA5' | GCACACATCTCCGGTAAAATGC | Q-zwf5' | GGCGCTGCGTTTTGCTAACT |
4. Conclusions
Abbreviations
| 1,3DPG | 1,3-bisphospho-D-glycerate |
| 2/3PG | 2-/3-phosphoglycerate |
| 6PG | 6-phospho D-gluconate |
| 6-PG | 6-phosphogluconate |
| 6PGL | 6-phospho D-glucono-1,5-lactone |
| a-KG | 2-oxoglutarate |
| AC | acetate |
| acea | isocitrate lyase |
| aceb | malate synthase |
| aceef | pyruvate dehydrogenase |
| ack | acetate kinase |
| acn | aconitate hydratase |
| ACoA | acetyl-CoA |
| ACP | acetyl phosphate |
| acs | acetyl-CoA synthetase |
| AMP | adenosine-monophosphate |
| ADP | adenosine-diphosphate |
| ATP | adenosine-triphosphate |
| CACN | cis-aconitate |
| CIT | citrate |
| crp | CRP transcriptional dual regulator |
| cya | adenylate cyclase |
| DHAP | dihydroxyacetone phosphate |
| E4P | erythrose-4-phosphate |
| ED | Entner-Doudoroff |
| eno | enolase |
| F6P | fructose-6-phosphate |
| fba | fructose bisphosphate aldolase |
| FBP | fructose-bisphosphate |
| Fbp | fructose-1,6-bisphosphatase |
| FOR | formate |
| frd | fumarate reductase |
| fum | fumarase |
| FUM | fumarate |
| G6P | glucose-6-phosphate |
| gap | glyceraldehyde 3-phosphate dehydrogenase |
| GHAP | D-glyceraldehyde 3-phosphate |
| GLC | glucose |
| GLX | glyoxylate |
| GLYC | glycolysis |
| gltA | citrate synthase |
| GMP | guanosine-monophosphate |
| GDP | guanosine-diphosphate |
| GTP | guanosine-triphosphate |
| gnd | 6-phosphogluconate dehydrogenase |
| gpm | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase |
| IC | ionic chromatography |
| icd | isocitrate dehydrogenase |
| ICIT | iso-citrate |
| mae | malate dehydrogenase |
| MAL | malate |
| mdh | malate dehydrogenase |
| MS | mass spectrometry |
| NAD+ | beta-nicotinamide adenine dinucleotide |
| NMR | nuclear magnetic resonance |
| OA | oxaloacetate |
| pck | phosphoenolpyruvate carboxykinase |
| PEP | phosphoenolpyruvate |
| Pfk | 6-phosphofructokinase |
| pfl | pyruvate formate-lyase |
| pgi | phosphoglucose isomerase |
| pgk | phosphoglycerate kinase |
| pgl | 6-phosphogluconolactonase |
| PP | pentose phosphate |
| ppc | phosphoenolpyruvate carboxylase |
| PPP | pentose phosphate pathway |
| pps | phosphoenolpyruvate synthetase |
| pta | phosphate acetyltransferase |
| PTS | phosphotransferase system |
| pyk | pyruvate kinase |
| PYR | pyruvate |
| R5P | D-ribose 5-phosphate |
| Rib-5P | ribose-5-phosphate |
| rpe | ribulose-5-phosphate 3-epimerase |
| rpi | ribose-5-phosphate isomerase |
| RT-PCR | reverse transcription polymerase chain reaction |
| RU5P | D-ribulose 5-phosphate |
| S7P | D-sedoheptulose 7-phosphate |
| sdh | succinate dehydrogenase |
| suc | 2-oxoglutarate decarboxylase |
| SUC | succinate |
| Succ | succinyl-CoA synthetase |
| SUCCoA | succinyl-CoA |
| tal | transaldolase |
| TCA | Tricarboxylic acid |
| tkt | transketolase |
| tpi | triose phosphate isomerase |
| UMP | uridine-monophosphate |
| UDP | uridine-diphosphate |
| UTP | uridine-triphosphate |
| XUP | D-xylulose 5-phosphate |
| zwf | glucose 6-phosphate-1-dehydrogenase |
Acknowledgments
Conflicts of Interest
References
- Koch, A.L. The adaptive responses of Escherichia coli to a feast and famine existence. Adv. Microb. Physiol. 1971, 6, 147–217. [Google Scholar] [CrossRef]
- Chang, D.E.; Smalley, D.J.; Tucker, D.L.; Leatham, M.P.; Norris, E.; Stevenson, S.J.; Anderson, B.; Grissom, J.E.; Laux, D.C.; Cohen, P.S.; et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 2004, 101, 7427–7432. [Google Scholar] [CrossRef]
- Miranda, R.L.; Conway, T.; Leatham, M.P.; Chang, D.E.; Norris, W.E.; Allen, J.H.; Stevenson, S.J.; Laux, D.C.; Cohen, P.S. Glycolytic and gluconeogenic growth of Escherichia coli O156:H7 EDL933 and E. coli K-12 MG1655 in the mouse intestine. Infec. Immun. 2004, 723, 1666–1676. [Google Scholar]
- Fabich, A.J.; Jones, S.A.; Chowdhury, F.Z.; Cernosek, A.; Anderson, A.; Smalley, D.; McHargue, J.W.; Hightower, G.A.; Smith, J.T.; Autieri, S.M.; et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 2008, 76, 1143–1152. [Google Scholar] [CrossRef]
- Papagianni, M. Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb. Cell. Fact. 2012. [Google Scholar] [CrossRef]
- Wolfe, A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef]
- Kremling, A.; Bettenbrock, K.; Gilles, E.D. Analysis of global control of Escherichia coli carbohydrate uptake. BMC Syst. Biol. 2007. [Google Scholar] [CrossRef]
- Kotte, O.; Zaugg, J.B.; Heinemann, M. Bacterial adaptation through distributed sensing of metabolic fluxes. Mol. Syst. Biol. 2010. [Google Scholar] [CrossRef]
- Peskov, K.; Mogilevskaya, E.; Demin, O. Kinetic modelling of central carbon metabolism in Escherichia coli. FEBS J. 2012, 279, 3374–3385. [Google Scholar] [CrossRef]
- El-Mansi, M.; Cozzone, A.J.; Shiloach, J.; Eikmanns, B.J. Control of carbon flux through enzymes of central and intermediary metabolism during growth of Escherichia coli on acetate. Curr. Opin. Microbiol. 2006, 9, 173–179. [Google Scholar]
- Oh, M.K.; Rohlin, L.; Kao, K.C.; Liao, J.C. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 2002, 277, 13175–13183. [Google Scholar]
- Kao, K.C.; Yang, Y.L.; Boscolo, R.; Sabatti, C.; Roychowdhury, V.; Liao, J.C. Transcriptome-based determination of multiple transcription regulator activities in Escherichia coli by using network component analysis. Proc. Natl. Acad. Sci. USA 2004, 101, 641–646. [Google Scholar] [CrossRef]
- Kao, K.C.; Tran, L.M.; Liao, J.C. A global regulatory role of gluconeogenic genes in Escherichia coli revealed by transcriptome network analysis. J. Biol. Chem. 2005, 280, 36079–36087. [Google Scholar] [CrossRef]
- Sunya, S.; Delvigne, F.; Uribelarrea, J.L.; Molina-Jouve, C.; Gorret, N. Comparison of the transient responses of Escherichia coli to a glucose pulse of various intensities. Appl. Microbiol. Biotechnol. 2012, 95, 1021–1034. [Google Scholar] [CrossRef]
- Schuetz, R.; Zamboni, N.; Zampieri, M.; Heinemann, M.; Sauer, U. Multidimensional optimality of microbial metabolism. Science 2012, 336, 601–604. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd Ed. ed; Cold Spring Harbor Lab. Press: Plainview, NY, USA, 2011. [Google Scholar]
- Womack, J.E.; O’Donovan, G.A. Orotic acid excretion in some wild-type strains of Escherichia coli K-12. J. Bacteriol. 1978, 136, 825–827. [Google Scholar]
- Bolten, C.J.; Kiefer, P.; Letisse, F.; Portais, J.C.; Wittmann, C. Sampling for metabolome analysis of microorganisms. Anal. Chem. 2007, 79, 3843–3849. [Google Scholar] [CrossRef]
- Kiefer, P.; Nicolas, C.; Letisse, F.; Portais, J.C. Determination of carbon labelling distribution of intracellular metabolites from single fragment ions by ion chromatography tandem mass spectrometry. Anal. Biochem. 2007, 360, 182–188. [Google Scholar] [CrossRef]
- Wu, L.; Mashego, M.R.; van Dam, J.C.; Proll, A.M.; Winke, J.L.; et al. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 2005, 336, 164–171. [Google Scholar] [CrossRef]
- Chassagnole, C.; Noisommit-Rizzi, N.; Schmid, J.W.; Mauch, K.; Reuss, M. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng. 2002, 79, 53–73. [Google Scholar] [CrossRef]
- Keseler, I.M.; Collado-Vides, J.; Gama-Castro, S.; Ingraham, J.; Paley, S.; Paulsen, I.T.; Peralta-Gil, M.; Karp, P.D. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005, 33, 334–337. [Google Scholar]
- Wang, Y.; Barbacioru, C.; Hyland, F.; Xiao, W.; Hunkapiller, K.L.; Blake, J.; Chan, F.; Gonzalez, C.; Zhang, L.; Samaha, R.R. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics 2006. [Google Scholar] [CrossRef]
- Yang, J.K.; Epstein, W. Purification and characterization of adenylate cyclase from Escherichia coli K12. J. Biol. Chem. 1983, 258, 3750–3758. [Google Scholar]
- Botsford, J.L.; Harman, J.G. Cyclic AMP in procaryotes. Microbiol. Rev. 1992, 56, 100–122. [Google Scholar]
- Enjalbert, B.; Jourdan, F.; Portais, J.C. Intuitive visualization and analysis of multi-omics data and application to Escherichia coli carbon metabolism. PLoS One 2011, 6, e21318. [Google Scholar] [CrossRef]
- Mitchell, A.; Romano, G.H.; Groisman, B.; Yona, A.; Dekel, E.; Kupiec, M.; Dahan, O.; Pilpel, Y. Adaptive prediction of environmental changes by microorganisms. Nature 2009, 460, 220–224. [Google Scholar] [CrossRef]
- Berthoumieux, S.; de Jong, H.; Baptist, G.; Pinel, C.; Ranquet, C.; Ropers, D.; Geiselmann, J. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Mol. Syst. Biol. 2013. [Google Scholar] [CrossRef]
- Kochanowski, K.; Sauer, U.; Chubukov, V. Somewhat in control-the role of transcription in regulating microbial metabolic fluxes. Curr. Opin. Biotechnol. 2013. [Google Scholar] [CrossRef]
- Ferenci, T. “Growth of bacterial cultures” 50 years on: towards an uncertainty principle instead of constants in bacterial growth kinetics. Res. Microbiol. 1999, 150, 431–438. [Google Scholar] [CrossRef]
- Traxler, M.F.; Chang, D.E.; Conway, T. Guanosine 3',5'-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl. Acad. Sci. USA 2006, 103, 2374–2379. [Google Scholar] [CrossRef]
- Ferenci, T. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv. Microb. Physiol. 2008, 53, 169–229. [Google Scholar] [CrossRef]
- Valgepea, K.; Aamberg, K.; Nahku1, R.; Lahtvee, P.J.; Arike, L.; Vilu, R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Sys Biol. 2010, 4, 166–179. [Google Scholar] [CrossRef]
- Arrayexpress Database. Available online: http://www.ebi.ac.uk/arrayexpress/ (accessed on 28 August 2013).
- Nicolas, C.; Kiefer, P.; Letisse, F.; Krömer, J.; Massou, S.; Soucaille, P.; Wittmann, C.; Lindley, N.D.; Portais, J.C. Response of the central metabolism of Escherichia coli to modified expression of the gene encoding the glucose-6-phosphate dehydrogenase. FEBS Lett. 2007, 581, 3771–3776. [Google Scholar] [CrossRef]
- Primer3 Input. Available online: http://frodo.wi.mit.edu/ (accessed on 28 August 2013).
- Covert, M.W.; Palsson, B.Ø. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J. Biol. Chem. 2002, 277, 28058–28064. [Google Scholar] [CrossRef]
- Cho, B.K.; Charusanti, P.; Herrgård, M.J.; Palsson, B.Ø. Microbial regulatory and metabolic networks. Curr. Opin. Biotechnol. 2007, 18, 360–364. [Google Scholar] [CrossRef]
- Metatoul home page. Available online: http://www.metatoul.fr/ (accessed on 28 August 2013).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Enjalbert, B.; Letisse, F.; Portais, J.-C. Physiological and Molecular Timing of the Glucose to Acetate Transition in Escherichia coli. Metabolites 2013, 3, 820-837. https://doi.org/10.3390/metabo3030820
Enjalbert B, Letisse F, Portais J-C. Physiological and Molecular Timing of the Glucose to Acetate Transition in Escherichia coli. Metabolites. 2013; 3(3):820-837. https://doi.org/10.3390/metabo3030820
Chicago/Turabian StyleEnjalbert, Brice, Fabien Letisse, and Jean-Charles Portais. 2013. "Physiological and Molecular Timing of the Glucose to Acetate Transition in Escherichia coli" Metabolites 3, no. 3: 820-837. https://doi.org/10.3390/metabo3030820
APA StyleEnjalbert, B., Letisse, F., & Portais, J.-C. (2013). Physiological and Molecular Timing of the Glucose to Acetate Transition in Escherichia coli. Metabolites, 3(3), 820-837. https://doi.org/10.3390/metabo3030820