Targeted Metabolic Profiling of Urine Highlights a Potential Biomarker Panel for the Diagnosis of Alzheimer’s Disease and Mild Cognitive Impairment: A Pilot Study
Abstract
1. Introduction
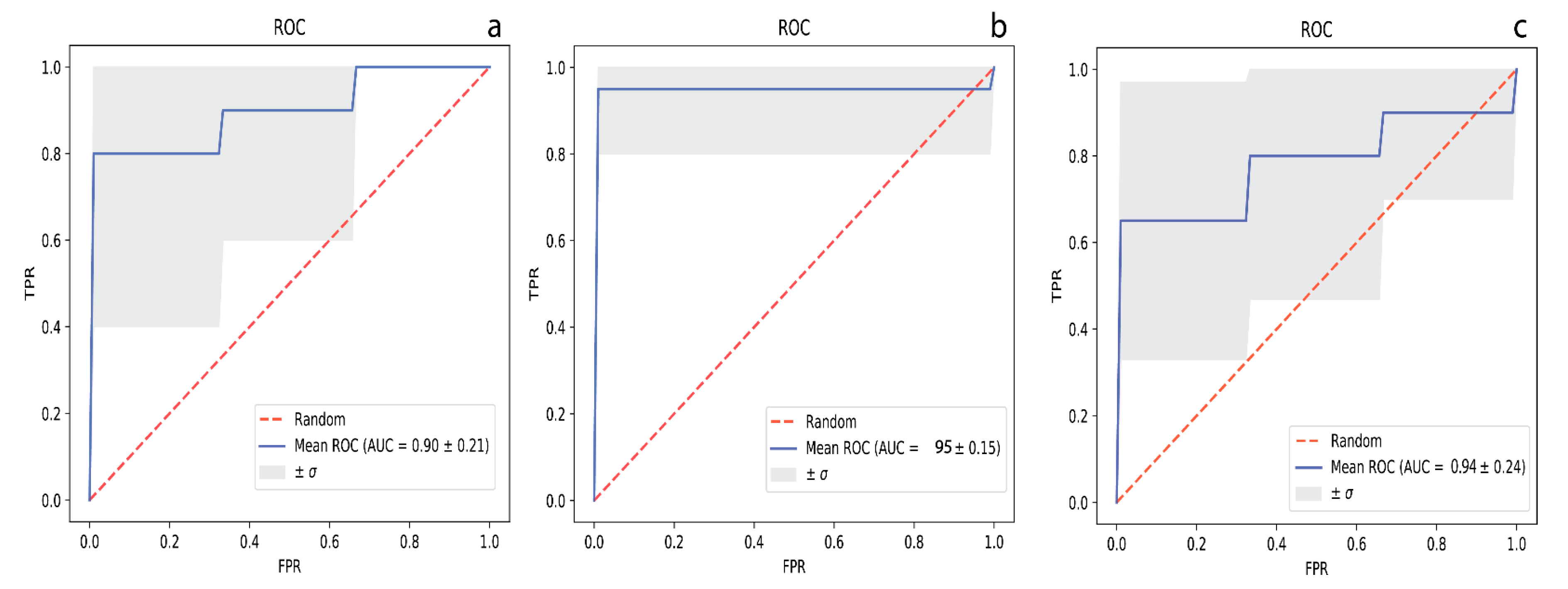
2. Results
3. Discussion
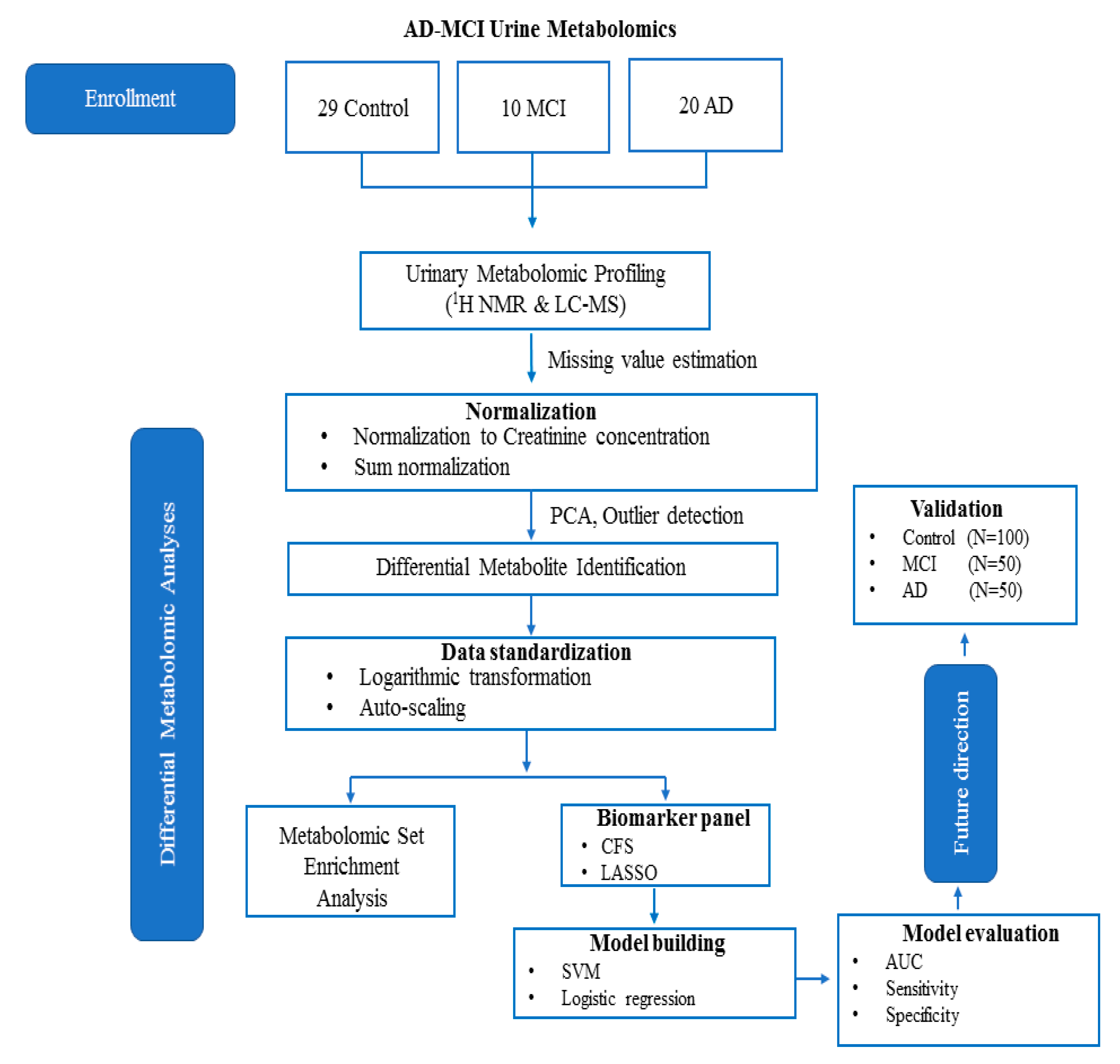
4. Materials and Methods
4.1. Urine Samples
4.2. 1H NMR Analysis
4.2.1. Sample Preparation and Acquisition
4.2.2. Metabolite Identification and Quantification
4.3. DI/LC-MS/MS Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Guo, R.; Fan, G.; Zhang, J.; Wu, C.; Du, Y.; Ye, H.; Li, Z.; Wang, L.; Zhang, Z.; Zhang, L.; et al. A 9-microRNA Signature in Serum Serves as a Noninvasive Biomarker in Early Diagnosis of Alzheimer’s Disease. J. Alzheimer Dis. JAD 2017, 60, 1365–1377. [Google Scholar] [CrossRef] [PubMed]
- Olsson, B.; Lautner, R.; Andreasson, U.; Öhrfelt, A.; Portelius, E.; Bjerke, M.; Hölttä, M.; Rosén, C.; Olsson, C.; Strobel, G.; et al. Csf and blood biomarkers for the diagnosis of alzheimer’s disease: A systematic review and meta-analysis. Lancet. Neurol. 2016, 15, 673–684. [Google Scholar] [CrossRef]
- Graham, S.F.; Chevallier, O.P.; Roberts, D.; Hölscher, C.; Elliott, C.T.; Green, B.D. Investigation of the Human Brain Metabolome to Identify Potential Markers for Early Diagnosis and Therapeutic Targets of Alzheimer’s Disease. Anal. Chem. 2013, 85, 1803–1811. [Google Scholar] [CrossRef] [PubMed]
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef]
- Khoury, R.; Patel, K.; Gold, J.; Hinds, S.; Grossberg, G.T. Recent Progress in the Pharmacotherapy of Alzheimer’s Disease. Drugs Aging 2017, 34, 811–820. [Google Scholar] [CrossRef]
- Geda, Y.E. Mild cognitive impairment in older adults. Curr. Psychiatry Rep. 2012, 14, 320–327. [Google Scholar] [CrossRef]
- Roberts, R.; Knopman, D.S. Classification and epidemiology of MCI. Clin. Geriatr. Med. 2013, 29, 753–772. [Google Scholar] [CrossRef]
- Yesavage, J.A.; O’Hara, R.; Kraemer, H.; Noda, A.; Taylor, J.L.; Ferris, S.; Gély-Nargeot, M.C.; Rosen, A.; Friedman, L.; Sheikh, J.; et al. Modeling the prevalence and incidence of Alzheimer’s disease and mild cognitive impairment. J. Psychiatr. Res. 2002, 36, 281–286. [Google Scholar] [CrossRef]
- Caraci, F.; Castellano, S.; Salomone, S.; Drago, F.; Bosco, P.; Di Nuovo, S. Searching for disease-modifying drugs in AD: Can we combine neuropsychological tools with biological markers? CNS Neurol. Disord. Drug Targets 2014, 13, 173–186. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- Yilmaz, A.; Geddes, T.; Han, B.; Bahado-Singh, R.O.; Wilson, G.D.; Imam, K.; Maddens, M.; Graham, S.F. Diagnostic Biomarkers of Alzheimer’s Disease as Identified in Saliva using 1H NMR-Based Metabolomics. J. Alzheimer Dis. JAD 2017, 58, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Nasaruddin, M.B.; Elliott, C.T.; McGuinness, B.; Passmore, A.P.; Kehoe, P.G.; Hölscher, C.; McClean, P.L.; Graham, S.F.; Green, B.D. Alzheimer’s disease-like pathology has transient effects on the brain and blood metabolome. Neurobiol. Aging 2016, 38, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.F.; Rey, N.L.; Yilmaz, A. Biochemical Profiling of the Brain and Blood Metabolome in a Mouse Model of Prodromal Parkinson’s Disease Reveals Distinct Metabolic Profiles. J. Proteome Res. 2018, 17, 2460–2469. [Google Scholar] [CrossRef] [PubMed]
- Alpay Savasan, Z.; Yilmaz, A.; Ugur, Z.; Aydas, B.; Bahado-Singh, R.O.; Graham, S.F. Metabolomic Profiling of Cerebral Palsy Brain Tissue Reveals Novel Central Biomarkers and Biochemical Pathways Associated with the Disease: A Pilot Study. Metabolites 2019, 9, 27. [Google Scholar] [CrossRef]
- Koç, E.R.; Ilhan, A.; Zübeyde, A.; Acar, B.; Gürler, M.; Altuntaş, A.; Karapirli, M.; Bodur, A.S. A comparison of hair and serum trace elements in patients with Alzheimer disease and healthy participants. Turk. J. Med. Sci. 2015, 45, 1034–1039. [Google Scholar] [CrossRef]
- Trushina, E.; Dutta, T.; Persson, X.M.; Mielke, M.M.; Petersen, R.C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE 2013, 8, e63644. [Google Scholar] [CrossRef]
- González-Domínguez, R.; García-Barrera, T.; Gómez-Ariza, J.L. Using direct infusion mass spectrometry for serum metabolomics in Alzheimer’s disease. Anal. Bioanal. Chem. 2014, 406, 7137–7148. [Google Scholar] [CrossRef]
- Botosoa, E.P.; Zhu, M.; Marbeuf-Gueye, C.; Triba, M.N.; Dutheil, F.; Duyckäerts, C.; Beaune, P.; Loriot, M.A.; Le Moyec, L. NMR metabolomic of frontal cortex extracts: First study comparing two neurodegenerative diseases, Alzheimer disease and amyotrophic lateral sclerosis. IRBM 2012, 33, 281–286. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, H.; Zhang, T.; Jiang, Y.; Xing, H.; Zhang, A.-H. Metabolomics-based screening of salivary biomarkers for early diagnosis of Alzheimer’s disease. RSC Adv. 2015, 5, 96074–96079. [Google Scholar] [CrossRef]
- Koal, T.; Klavins, K.; Seppi, D.; Kemmler, G.; Humpel, C. Sphingomyelin SM(d18:1/18:0) is significantly enhanced in cerebrospinal fluid samples dichotomized by pathological amyloid-β42, tau, and phospho-tau-181 levels. J. Alzheimer Dis. JAD 2015, 44, 1193–1201. [Google Scholar] [CrossRef]
- Nho, K.; Kueider-Paisley, A.; MahmoudianDehkordi, S.; Arnold, M.; Risacher, S.L.; Louie, G.; Blach, C.; Baillie, R.; Han, X.; Kastenmüller, G.; et al. Altered bile acid profile in mild cognitive impairment and Alzheimer’s disease: Relationship to neuroimaging and CSF biomarkers. Alzheimer Dement. J. Alzheimer Assoc. 2019, 15, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wei, M.; Yue, K.; Hu, M.; Li, S.; Men, L.; Pi, Z.; Liu, Z.; Liu, Z. Study on Urine Metabolic Profile of Aβ25-35-Induced Alzheimer’s Disease Using UHPLC-Q-TOF-MS. Neuroscience 2018, 394, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Echeverry, G.; Hortin, G.L.; Rai, A.J. Introduction to urinalysis: Historical perspectives and clinical application. Methods Mol. Biol. 2010, 641, 1–12. [Google Scholar] [CrossRef]
- Garrod, A.E. The incidence of alkaptonuria: A study in chemical individuality. 1902. Mol. Med. 1996, 2, 274–282. [Google Scholar] [CrossRef]
- Issaq, H.J.; Nativ, O.; Waybright, T.; Luke, B.; Veenstra, T.D.; Issaq, E.J.; Kravstov, A.; Mullerad, M. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J. Urol. 2008, 179, 2422–2426. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Aronov, P.; Zakharkin, S.O.; Anderson, D.; Perroud, B.; Thompson, I.M.; Weiss, R.H. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol. Cell. Proteom. MCP 2009, 8, 558–570. [Google Scholar] [CrossRef]
- Pasikanti, K.K.; Esuvaranathan, K.; Ho, P.C.; Mahendran, R.; Kamaraj, R.; Wu, Q.H.; Chiong, E.; Chan, E.C. Noninvasive urinary metabonomic diagnosis of human bladder cancer. J. Proteome Res. 2010, 9, 2988–2995. [Google Scholar] [CrossRef]
- Yu, J.; Kong, L.; Zhang, A.; Han, Y.; Liu, Z.; Sun, H.; Liu, L.; Wang, X. High-Throughput Metabolomics for Discovering Potential Metabolite Biomarkers and Metabolic Mechanism from the APPswe/PS1dE9 Transgenic Model of Alzheimer’s Disease. J. Proteome Res. 2017, 16, 3219–3228. [Google Scholar] [CrossRef]
- Fukuhara, K.; Ohno, A.; Ota, Y.; Senoo, Y.; Maekawa, K.; Okuda, H.; Kurihara, M.; Okuno, A.; Niida, S.; Saito, Y.; et al. NMR-based metabolomics of urine in a mouse model of Alzheimer’s disease: Identification of oxidative stress biomarkers. J. Clin. Biochem. Nutr. 2013, 52, 133–138. [Google Scholar] [CrossRef]
- Peng, J.; Guo, K.; Xia, J.; Zhou, J.; Yang, J.; Westaway, D.; Wishart, D.S.; Li, L. Development of isotope labeling liquid chromatography mass spectrometry for mouse urine metabolomics: Quantitative metabolomic study of transgenic mice related to Alzheimer’s disease. J. Proteome Res. 2014, 13, 4457–4469. [Google Scholar] [CrossRef]
- Castor, K.J.; Shenoi, S.; Edminster, S.P.; Tran, T.; King, K.S.; Chui, H.; Pogoda, J.M.; Fonteh, A.N.; Harrington, M.G. Urine dicarboxylic acids change in pre-symptomatic Alzheimer’s disease and reflect loss of energy capacity and hippocampal volume. PLoS ONE 2020, 15, e0231765. [Google Scholar] [CrossRef]
- Boksa, P. A way forward for research on biomarkers for psychiatric disorders. J. Psychiatry Neurosci. JPN 2013, 38, 75–77. [Google Scholar] [CrossRef]
- Mapstone, M.; Cheema, A.K.; Fiandaca, M.S. Plasma phospholipids identify antecedent memory impairment in older adults. Nat. Med. 2014, 20, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Vapnik, V.N. An overview of statistical learning theory. IEEE Trans. Neural Netw. 1999, 10, 988–999. [Google Scholar] [CrossRef]
- Xi, B.; Gu, H.; Baniasadi, H.; Raftery, D. Statistical analysis and modeling of mass spectrometry-based metabolomics data. Methods Mol. Biol. 2014, 1198, 333–353. [Google Scholar] [CrossRef]
- Cunnane, S.; Nugent, S.; Roy, M.; Courchesne-Loyer, A.; Croteau, E.; Tremblay, S.; Castellano, A.; Pifferi, F.; Bocti, C.; Paquet, N.; et al. Brain fuel metabolism, aging, and Alzheimer’s disease. Nutrition 2011, 27, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Kan, H.E.; Renema, W.K.J.; Isbrandt, D.; Heerschap, A. Phosphorylated guanidinoacetate partly compensates for the lack of phosphocreatine in skeletal muscle of mice lacking guanidinoacetate methyltransferase. J. Physiol. 2004, 560, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Parra, M.; Stahl, S.; Hellmann, H. Vitamin B6 and Its Role in Cell Metabolism and Physiology. Cells 2018, 7, 84. [Google Scholar] [CrossRef]
- Morris, M.C.; Schneider, J.A.; Tangney, C.C. Thoughts on B-vitamins and dementia. J. Alzheimer Dis. JAD 2006, 9, 429–433. [Google Scholar] [CrossRef]
- Albrahim, T. The potential role of nutritional components in improving brain function among patients with Alzheimers disease: A meta-analysis of RCT studies. Neurosciences 2020, 25, 4–17. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Soliman, O.S.; Rassem, A. Correlation Based Feature Selection Using Quantum Bio Inspired Estimation of Distribution Algorithm; Springer: Berlin/Heidelberg, Germany, 2012; pp. 318–329. [Google Scholar]
- Nova, D.; Estévez, P.A. A review of learning vector quantization classifiers. Neural Comput. Appl. 2014, 25, 511–524. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ahn, J.K.; Cheong, Y.E.; Lee, S.-J.; Cha, H.-S.; Kim, K.H. Systematic re-evaluation of the long-used standard protocol of urease-dependent metabolome sample preparation. PLoS ONE 2020, 15, e0230072. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. 2019, 58, 968–994. [Google Scholar] [CrossRef]
- Ravanbakhsh, S.; Liu, P.; Bjorndahl, T.C.; Mandal, R.; Grant, J.R.; Wilson, M.; Eisner, R.; Sinelnikov, I.; Hu, X.; Luchinat, C.; et al. Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS ONE 2015, 10, e0124219. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
| Controls | MCI | AD | P-Value | |
|---|---|---|---|---|
| n | 29 | 10 | 20 | |
| Age, Mean (SD) | 79.12 (6.28) | 76.57 (9.37) | 79.92 (9.11) | 0.43 a |
| Gender | ||||
| Male | 13 | 5 | 9 | 0.56 b |
| Female | 16 | 5 | 11 |
| Name | Mean (SD) of HC | Mean (SD) of MCI | Mean (SD) of AD | P-Value HC vs MCI | P-Value MCI vs AD | P-Value HC vs AD |
|---|---|---|---|---|---|---|
| 2-Hydroxybutyric acid | 2.774 (1.379) | 2.431 (1.244) | 4.271 (2.599) | 0.4368 (W) | 0.01762 (W) | 0.0423 (W) |
| 2-Hydroxyisovaleric acid | 0.948 (0.352) | 0.891 (0.351) | 0.013 (0.014) | 0.2522 | 0.03331 (W) | 0.0461 (W) |
| 3-Hydroxybutyric acid | 3.938 (5.098) | 2.559 (2.904) | 3.295 (2.450) | 0.0495 (W) | 0.0795 (W) | 0.0832(W) |
| 3-Hydroxyisovaleric acid | 3.861 (1.909) | 2.833 (0.929) | 3.393 (1.350) | 0.0048 | 0.0347 (W) | 0.1528 (W) |
| 5-Aminopentanoic acid | 3.955 (4.715) | 2.882 (2.831) | 3.445 (3.628) | 0.0308 (W) | 0.0257 (W) | 0.7668 (W) |
| Alpha-ketoisovaleric acid | 3.402 (1.864) | 2.564 (1.548) | 1.015 (0.904) | 0.0463 (W) | 0.0411 (W) | 0.0307 (W) |
| C6:1 | 0.009 (0.007) | 0.014 (0.012) | 0.008 (0.008) | 0.1643 (W) | 0.0166 (W) | 0.2476 (W) |
| Cytosine | 6.279 (7.311) | 11.623 (15.351) | 19.403 (8.093) | 0.0487 (W) | 0.0386 (W) | 0.06467 (W) |
| D-Glucose | 14.035 (8.563) | 9.128 (3.037) | 13.541 (6.780) | 0.0336 (W) | 0.01232 | 0.0204 |
| Dimethylsulfone | 9.238 (7.167) | 5.028 (3.839) | 4.173 (5.835) | 0.9646 | 0.0190 (W) | 0.0820 (W) |
| Guanidoacetic acid | 15.031 (9.884) | 9.077 (3.838) | 16.389 (7.515) | 0.0103 | 0.0038 (W) | 0.4371 (W) |
| Hippuric acid | 55.489 (7.874) | 40.655 (6.302) | 57.376 (6.537) | 0.3908 (W) | 0.0111 (W) | 0.9945 |
| Mannitol | 13.260 (4.916) | 17.071 (6.889) | 7.808 (6.440) | 0.4368 (W) | 0.0429 (W) | 0.1414 (W) |
| Methanol | 52.958 (6.169) | 59.581 (5.870) | 47.690 (3.050) | 0.0266 (W) | 0.0021 | 0.0552 (W) |
| PC aa C32:0 | 0.019 (0.430) | 0.02 (0.001) | 0.02 (0.003) | 0.1850 (W) | 0.0403 (W) | 0.4136 (W) |
| Trimethylamine | 0.958 (2.582) | 3.073 (5.811) | 1.197 (3.040) | 0.0121 (W) | 0.0412 (W) | 0.0439 (W) |
| Tryptophan | 22.649 (22.057) | 20.443 (11.526) | 17.337 (9.148) | 0.4646 | 0.0114 (W) | 0.8012 (W) |
| Alanine | 7.553 (7.690) | 6.386 (3.828) | 7.401 (3.007) | 0.8868 (W) | 0.0439 (W) | 0.7395 (W) |
| Proline | 4.727 (2.369) | 5.641 (3.053) | 6.804 (3.828) | 0.4954 (W) | 0.3735 (W) | 0.0394 |
| Pyridoxine | 0.976 (1.215) | 0.477 (0.375) | 0.390 (0.373) | 0.2720 (W) | 0.5884 (W) | 0.0249 (W) |
| Isoleucine | 1.563 (0.917) | 1.283 (0.740) | 0.968 (0.416) | 0.7158 (W | 0.02364 | 0.9438 (W) |
| Myo-inositol | 18.945 (6.379) | 15.869 (8.629) | 16.034 (5.995) | 0.0331 (W) | 0.0134 | 0.3440 (W) |
| Trimethylamine n-oxide | 10.229 (7.735) | 19.907 (10.822) | 18.864 (11.571) | 0.0425 | 0.7488 | 0.0134 |
| Glycolic acid | 12.043 (7.354) | 15.671 (9.141) | 8.274 (4.972) | 0.9370 (W) | 0.3735 (W) | 0.0518 |
| Acetic acid | 6.136 (1.867) | 14.663 (2.450) | 9.336 (2.758) | 0.0485 (W) | 0.0103 | 0.7548 (W) |
| Acetone | 0.884 (0.802) | 1.442 (1.767) | 1.068 (0.907) | 0.7856 (W) | 0.0446 | 1.0000 (W) |
| PC ae C36:4 | 0.002 (0.001) | 0.002 (0.003) | 0.019 (0.034) | 0.0134 (W) | 0.0495 (W) | 0.2720 (W) |
| SM C26:0 | 0.674 (0.974) | 0.350 (0.876) | 0.674 (0.974) | 0.0475 (W) | 0.0457 (W) | 0.1643 (W) |
| PC ae C36:0 | 2.376 (0.769) | 1.622 (3.323) | 2.878 (1.428) | 0.02241 | 0.0403 (W) | 0.3934 (W) |
| Caffeine | 2.934 (1.724) | 1.962 (2.014) | 2.274 (1.375) | 0.0491 (W) | 0.3115 (W) | 0.0691 (W) |
| Isobutyric acid | 1.237 (0.840) | 1.698 (1.201) | 2.776 (1.724) | 0.0406 (W) | 0.0646 (W) | 0.0628 (W) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yilmaz, A.; Ugur, Z.; Bisgin, H.; Akyol, S.; Bahado-Singh, R.; Wilson, G.; Imam, K.; Maddens, M.E.; Graham, S.F. Targeted Metabolic Profiling of Urine Highlights a Potential Biomarker Panel for the Diagnosis of Alzheimer’s Disease and Mild Cognitive Impairment: A Pilot Study. Metabolites 2020, 10, 357. https://doi.org/10.3390/metabo10090357
Yilmaz A, Ugur Z, Bisgin H, Akyol S, Bahado-Singh R, Wilson G, Imam K, Maddens ME, Graham SF. Targeted Metabolic Profiling of Urine Highlights a Potential Biomarker Panel for the Diagnosis of Alzheimer’s Disease and Mild Cognitive Impairment: A Pilot Study. Metabolites. 2020; 10(9):357. https://doi.org/10.3390/metabo10090357
Chicago/Turabian StyleYilmaz, Ali, Zafer Ugur, Halil Bisgin, Sumeyya Akyol, Ray Bahado-Singh, George Wilson, Khaled Imam, Michael E. Maddens, and Stewart F. Graham. 2020. "Targeted Metabolic Profiling of Urine Highlights a Potential Biomarker Panel for the Diagnosis of Alzheimer’s Disease and Mild Cognitive Impairment: A Pilot Study" Metabolites 10, no. 9: 357. https://doi.org/10.3390/metabo10090357
APA StyleYilmaz, A., Ugur, Z., Bisgin, H., Akyol, S., Bahado-Singh, R., Wilson, G., Imam, K., Maddens, M. E., & Graham, S. F. (2020). Targeted Metabolic Profiling of Urine Highlights a Potential Biomarker Panel for the Diagnosis of Alzheimer’s Disease and Mild Cognitive Impairment: A Pilot Study. Metabolites, 10(9), 357. https://doi.org/10.3390/metabo10090357









