Hypoglycemic, Antioxidant Activities, and Probiotic Characteristics of Lacticaseibacillus rhamnosus LBUX2302 Isolated from Stool Samples of Neonates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Isolation
2.2. Molecular Identification
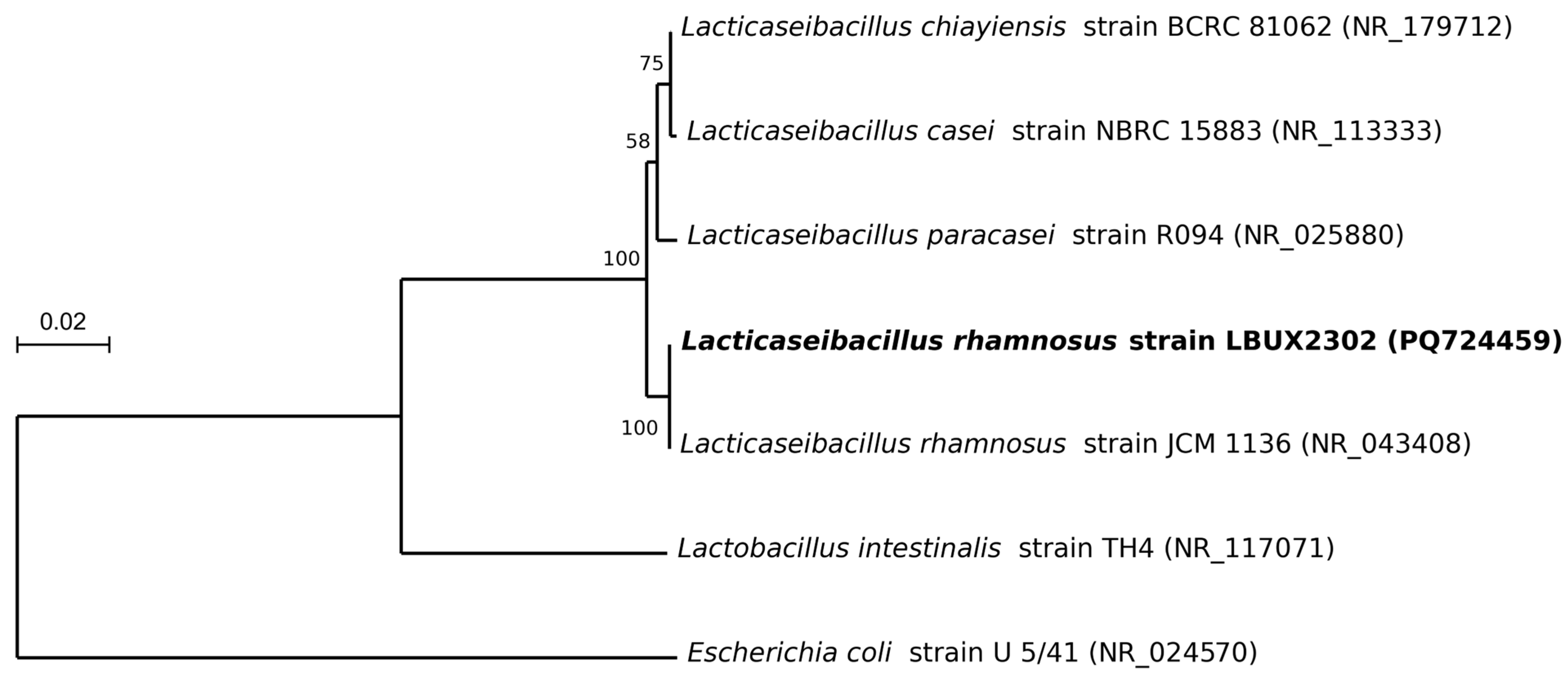
2.3. Phylogeny Analysis
2.4. Catalase Test
2.5. Hemolysis Test
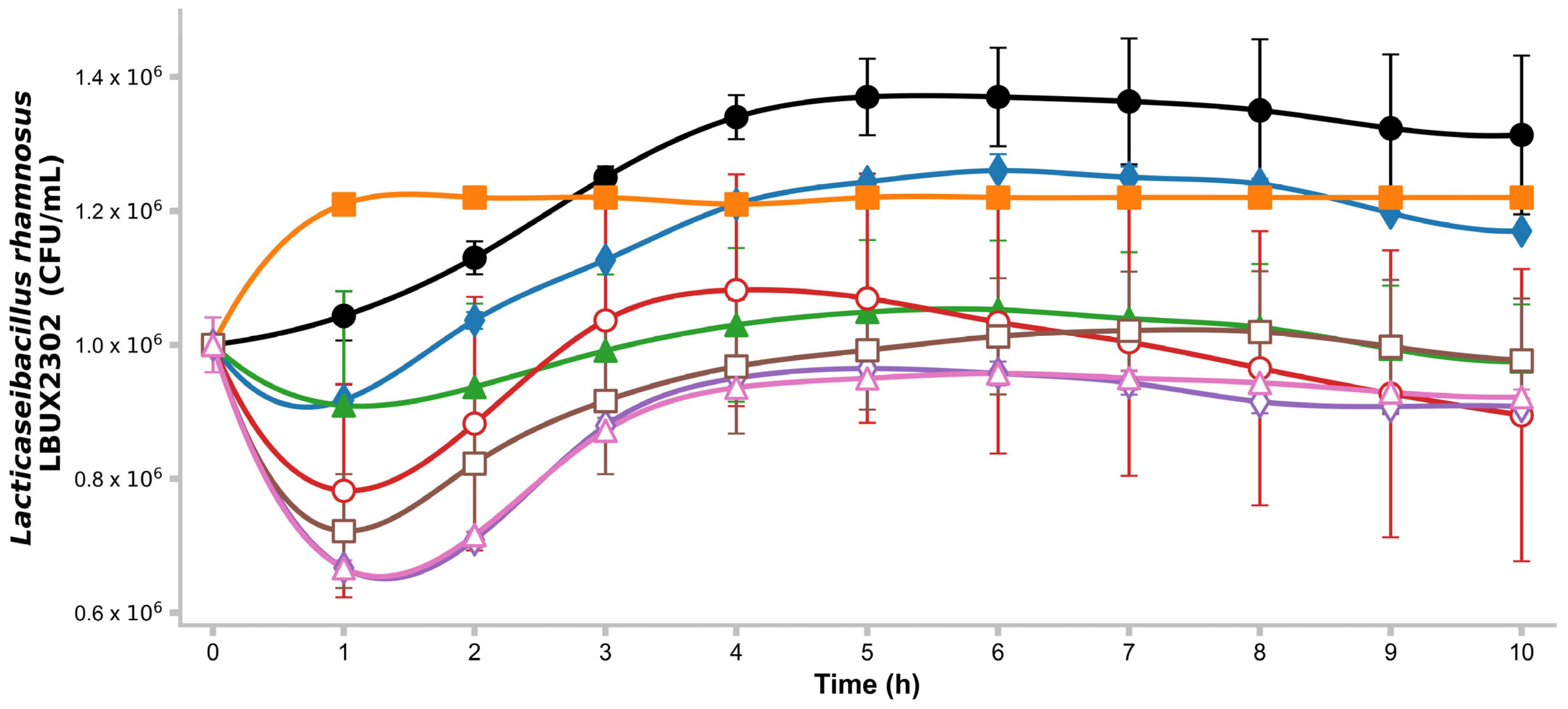
2.6. Growth Kinetics on Different Carbon Sources
2.7. Antimicrobial Activity
2.8. Antibiotic Resistance
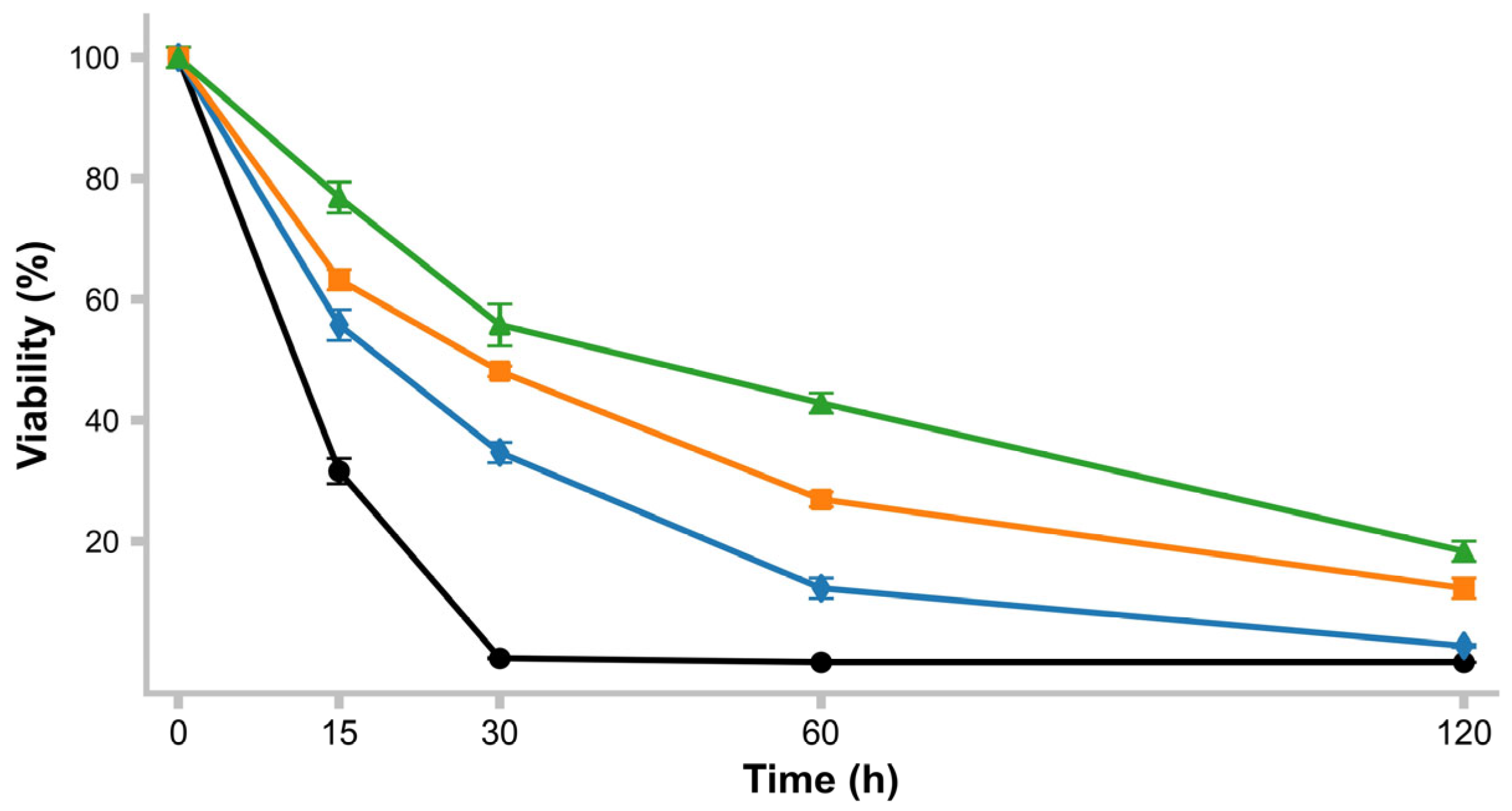
2.9. pH Resistance
2.10. Hydrophobicity
2.11. Autoaggregation
2.12. Coaggregation
2.13. Qualitative Assay of Bile Salt Tolerance
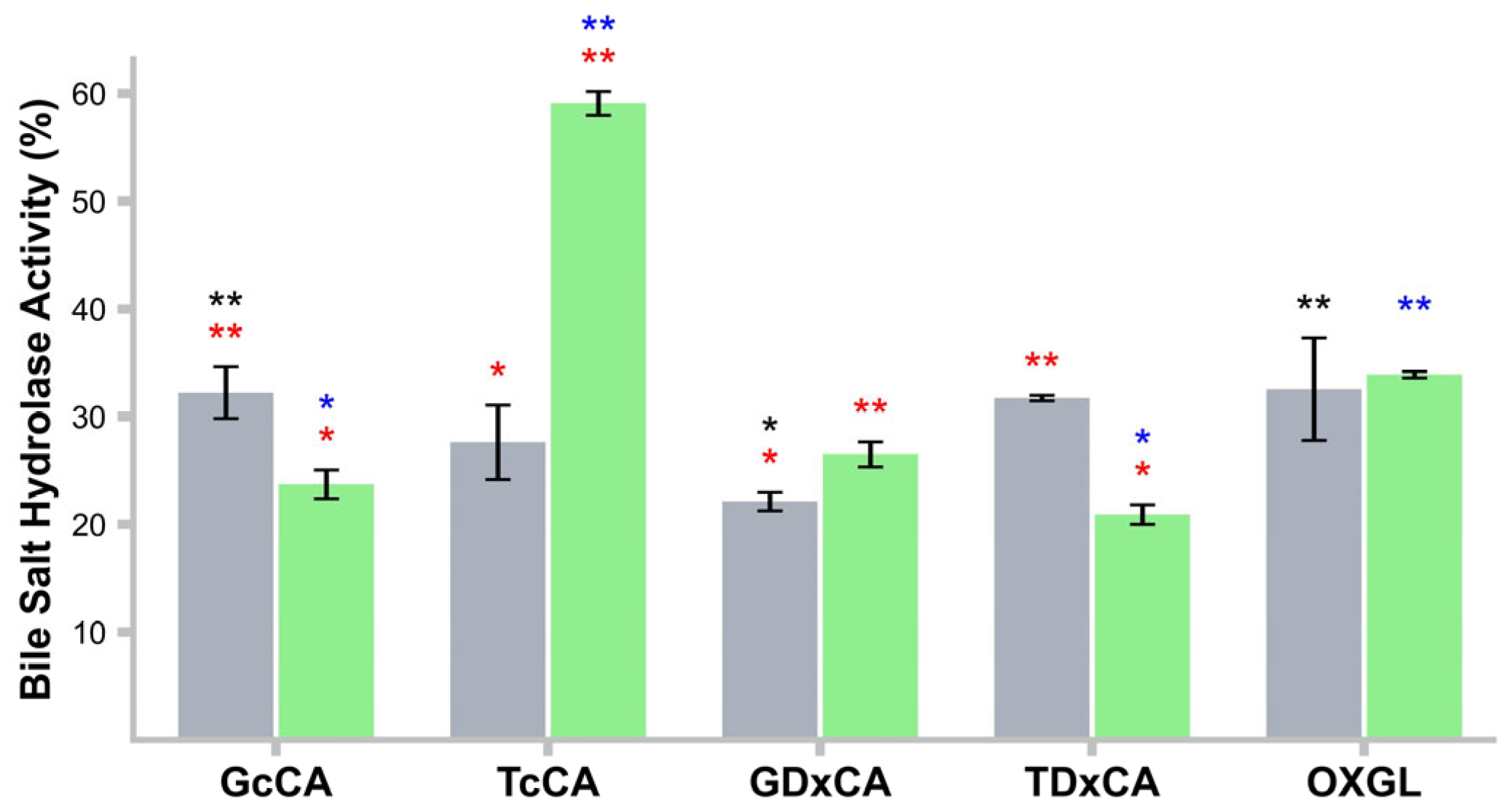
2.14. Bile Salt Hydrolase (BSH) Activity
2.15. 2,2-Diphenyl-1 Picrylhydrazyl (DPPH) Radical Inhibitory Activity
2.16. Hydroxyl Radical Inhibitory Activity
2.17. Superoxide Anion Radical Inhibitory Activity
2.18. Qualitative Ferulic Acid Activity (EFA)
2.19. Inhibition of α-Amylase
2.20. Inhibition of α-Glucosidase
2.21. Adhesion to Cancer Cell Lines
2.22. Statistical Analysis
3. Results
3.1. Identification and Phylogeny
3.2. Growth Kinetics on Different Carbon Sources
3.3. Catalase and Hemolysis
3.4. Antimicrobial Activity
3.5. Antibiotic Resistance
3.6. Bile Salt Tolerance
3.7. Hydrophobicity and Autoaggregation
3.8. Coaggregation
3.9. Ferulic Acid Activity
3.10. BSH Activity in CFs
3.11. pH Survival
3.12. Antioxidant Activity
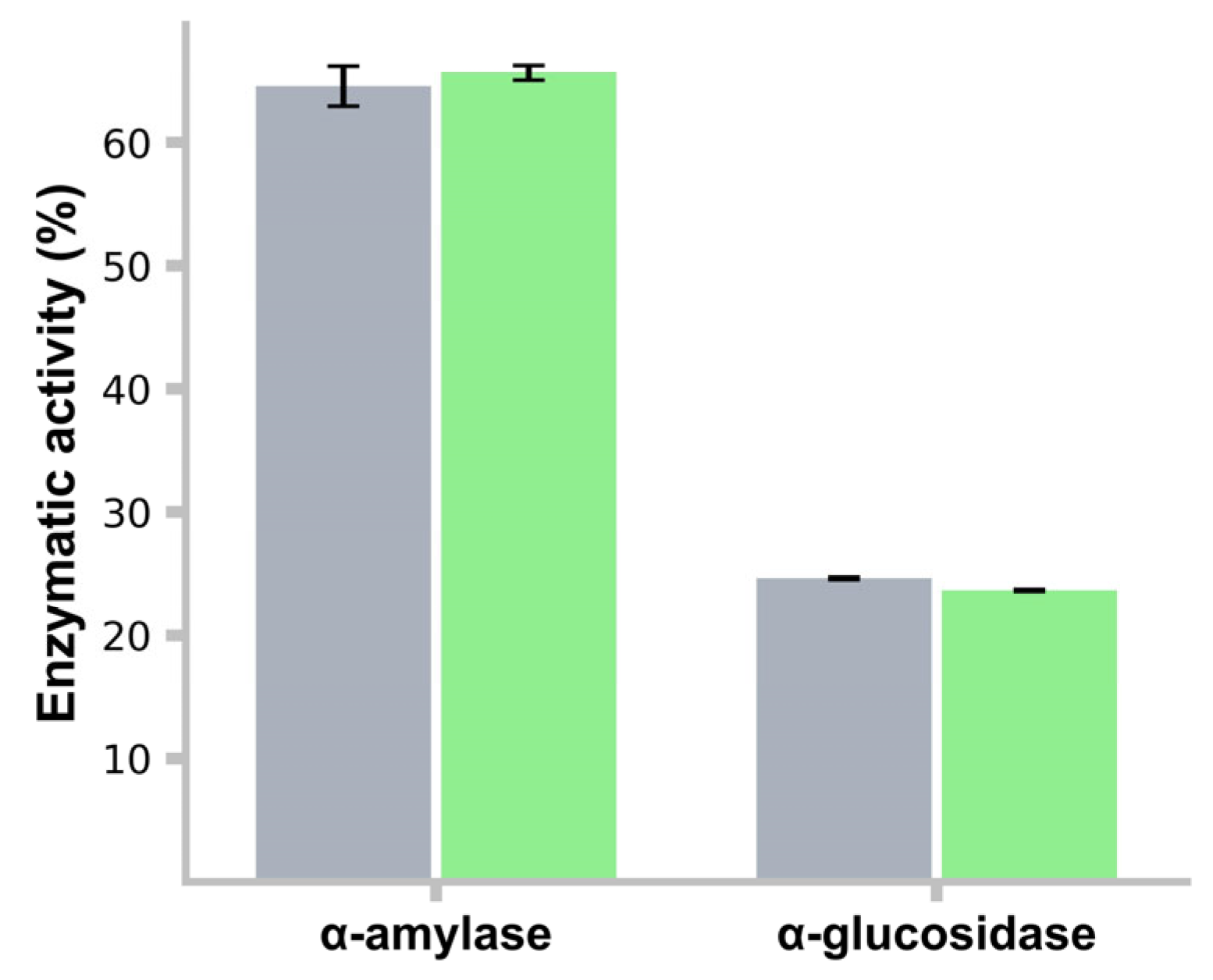
3.13. α-Amylase and α-Glucosidase Activity Inhibition
3.14. Adhesion to Cancer Cell Lines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Mora-Villalobos, J.A.; Montero-Zamora, J.; Barboza, N.; Rojas-Garbanzo, C.; Usaga, J.; Redondo-Solano, M.; Schroedter, L.; Olszewska-Widdrat, A.; López-Gómez, J.P. Multi-product lactic acid bacteria fermentations: A review. Fermentation 2020, 6, 23. [Google Scholar] [CrossRef]
- Rossi, F. Special Issue “Functional Characterization of Lactic Acid Bacteria”: Editorial. Microorganisms 2023, 11, 1190. [Google Scholar] [CrossRef]
- Hotel, A.C.P.; Cordoba, A. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Prevention 2001, 5, 1–10. [Google Scholar]
- Latif, A.; Shehzad, A.; Niazi, S.; Zahid, A.; Ashraf, W.; Iqbal, M.W.; Rehman, A.; Riaz, T.; Aadil, R.M.; Khan, I.M.; et al. Probiotics: Mechanism of action, health benefits and their application in food industries. Front. Microbiol. 2023, 14, 1216674. [Google Scholar] [CrossRef]
- Saarela, M.; Mogensen, G.; Fonden, R.; Matto, J.; Mattila-Sandholm, T. Probiotic bacteria: Safety, functional and technological properties. J. Biotechnol. 2000, 84, 197–215. [Google Scholar] [CrossRef]
- Poimenidou, S.V.; Skarveli, A.; Saxami, G.; Mitsou, E.K.; Kotsou, M.; Kyriacou, A. Inhibition of Listeria monocytogenes Growth, Adherence and Invasion in Caco-2 Cells by Potential Probiotic Lactic Acid Bacteria Isolated from Fecal Samples of Healthy Neonates. Microorganisms 2023, 11, 363. [Google Scholar] [CrossRef]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Gao, J.; Li, X.; Zhang, G.; Sadiq, F.A.; Simal-Gandara, J.; Xiao, J.; Sang, Y. Probiotics in the dairy industry—Advances and opportunities. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3937–3982. [Google Scholar] [CrossRef]
- Reyes-Castillo, P.A.; González-Vázquez, R.; Torres-Maravilla, E.; Bautista-Hernández, J.I.; Zúñiga-León, E.; Leyte-Lugo, M.; Mateos-Sánchez, L.; Mendoza-Pérez, F.; Gutiérrez-Nava, M.A.; Reyes-Pavón, D.; et al. Bifidobacterium longum LBUX23 Isolated from Feces of a Newborn; Potential Probiotic Properties and Genomic Characterization. Microorganisms 2023, 11, 1648. [Google Scholar] [CrossRef]
- Wang, H.; Li, L. Comprehensive Evaluation of Probiotic Property, Hypoglycemic Ability and Antioxidant Activity of Lactic Acid Bacteria. Foods 2022, 11, 1363. [Google Scholar] [CrossRef] [PubMed]
- Afshari, A.; Hashemi, M.; Tavassoli, M.; Eraghi, V.; Noori, S.M.A. Probiotic bacteria from 10 different traditional Iranian cheeses: Isolation, characterization, and investigation of probiotic potential. Food Sci. Nutr. 2022, 10, 2009–2020. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Na, R.; Niu, X.; Xiao, B.; Yang, H. Lactobacillus rhamnosus and Lactobacillus casei affect various stages of Gardnerella species biofilm formation. Front. Cell. Infect. Microbiol. 2021, 11, 568178. [Google Scholar] [CrossRef]
- Galkiewicz, J.P.; Kellogg, C.A. Cross-kingdom amplification using bacteria-specific primers: Complications for studies of coral microbial ecology. Appl. Environ. Microbiol. 2008, 74, 7828–7831. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Makarova, O.; Johnston, P.; Walther, B.; Rolff, J.; Roesler, U. Complete Genome Sequence of the Disinfectant Susceptibility Testing Reference Strain Staphylococcus aureus subsp. aureus ATCC 6538. Genome Announc. 2017, 5, e00293-17. [Google Scholar] [CrossRef]
- Gonzalez-Vazquez, R.; Azaola-Espinosa, A.; Mayorga-Reyes, L.; Reyes-Nava, L.A.; Shah, N.P.; Rivera-Espinoza, Y. Isolation, Identification and Partial Characterization of a Lactobacillus casei Strain with Bile Salt Hydrolase Activity from Pulque. Probiotics Antimicrob. Proteins 2015, 7, 242–248. [Google Scholar] [CrossRef]
- Minogue, T.D.; Daligault, H.A.; Davenport, K.W.; Bishop-Lilly, K.A.; Broomall, S.M.; Bruce, D.C.; Chain, P.S.; Chertkov, O.; Coyne, S.R.; Freitas, T. Complete genome assembly of Escherichia coli ATCC 25922, a serotype O6 reference strain. Genome Announc. 2014, 2, 10-1128. [Google Scholar] [CrossRef]
- Hilborn, E.D.; Mshar, P.A.; Fiorentino, T.R.; Dembek, Z.F.; Barrett, T.J.; Howard, R.T.; Cartter, M.L. An outbreak of Escherichia coli O157 [ratio] H7 infections and haemolytic uraemic syndrome associated with consumption of unpasteurized apple cider. Epidemiol. Infect. 2000, 124, 31–36. [Google Scholar] [CrossRef]
- Agbankpe, A.J.; Dougnon, T.V.; Balarabe, R.; Deguenon, E.; Baba-Moussa, L. In vitro assessment of antibacterial activity from Lactobacillus spp. strains against virulent Salmonella species isolated from slaughter animals in Benin. Vet. World 2019, 12, 1951. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, T.; Li, Y.-Y.; Li, J.; Zhang, Y.; Wang, Y.; Wang, H.; Li, H. Antioxidant properties of wine lactic acid bacteria: Oenococcus oeni. Appl. Microbiol. Biotechnol. 2015, 99, 5189–5202. [Google Scholar] [CrossRef] [PubMed]
- Vinderola, C.G.; Reinheimer, J.A. Lactic acid starter and probiotic bacteria: A comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Zuo, F.; Yu, R.; Feng, X.; Chen, L.; Zeng, Z.; Khaskheli, G.B.; Ma, H.; Chen, S. Characterization and in vitro properties of potential probiotic Bifidobacterium strains isolated from breast-fed infant feces. Ann. Microbiol. 2016, 66, 1027–1037. [Google Scholar] [CrossRef]
- Yan, F.; Li, N.; Yue, Y.; Wang, C.; Zhao, L.; Evivie, S.E.; Li, B.; Huo, G. Screening for Potential Novel Probiotics With Dipeptidyl Peptidase IV-Inhibiting Activity for Type 2 Diabetes Attenuation in vitro and in vivo. Front. Microbiol. 2020, 10, 2855. [Google Scholar] [CrossRef]
- Tomaro-Duchesneau, C.; Saha, S.; Malhotra, M.; Coussa-Charley, M.; Al-Salami, H.; Jones, M.; Labbé, A.; Prakash, S. Lactobacillus fermentum NCIMB 5221 has a greater ferulic acid production compared to other ferulic acid esterase producing Lactobacilli. Int. J. Probiotics Prebiotics 2012, 7, 23–32. [Google Scholar]
- Won, G.; Choi, S.I.; Park, N.; Kim, J.E.; Kang, C.H.; Kim, G.H. In Vitro Antidiabetic, Antioxidant Activity, and Probiotic Activities of Lactiplantibacillus plantarum and Lacticaseibacillus paracasei Strains. Curr. Microbiol. 2021, 78, 3181–3191. [Google Scholar] [CrossRef]
- González-Vázquez, R.; Zúñiga-León, E.; Torres-Maravilla, E.; Leyte-Lugo, M.; Mendoza-Pérez, F.; Hernández-Delgado, N.C.; Pérez-Pastén-Borja, R.; Azaola-Espinosa, A.; Mayorga-Reyes, L. Genomic and biochemical characterization of bifidobacterium pseudocatenulatum JCLA3 isolated from human intestine. Microorganisms 2022, 10, 2100. [Google Scholar] [CrossRef]
- Punthakee, Z.; Goldenberg, R.; Katz, P. Definition, Classification and Diagnosis of Diabetes, Prediabetes and Metabolic Syndrome. Can. J. Diabetes 2018, 42 (Suppl. S1), S10–S15. [Google Scholar] [CrossRef]
- Salis, S.; Virmani, A.; Priyambada, L.; Mohan, M.; Hansda, K.; Beaufort, C. ‘Old Is Gold’: How Traditional Indian Dietary Practices Can Support Pediatric Diabetes Management. Nutrients 2021, 13, 4427. [Google Scholar] [CrossRef]
- Kumari, V.B.C.; Huligere, S.S.; Alotaibi, G.; Al Mouslem, A.K.; Bahauddin, A.A.; Shivanandappa, T.B.; Ramu, R. Antidiabetic Activity of Potential Probiotics Limosilactobacillus spp., Levilactobacillus spp., and Lacticaseibacillus spp. Isolated from Fermented Sugarcane Juice: A Comprehensive In Vitro and In Silico Study. Nutrients 2023, 15, 1882. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, N.; Yin, B.; Fang, D.; Jiang, T.; Fang, S.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Effects of Lactobacillus plantarum CCFM0236 on hyperglycaemia and insulin resistance in high-fat and streptozotocin-induced type 2 diabetic mice. J. Appl. Microbiol. 2016, 121, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Muganga, L.; Liu, X.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Screening for lactic acid bacteria based on antihyperglycaemic and probiotic potential and application in synbiotic set yoghurt. J. Funct. Foods 2015, 16, 125–136. [Google Scholar] [CrossRef]
- Huligere, S.S.; Chandana Kumari, V.B.; Alqadi, T.; Kumar, S.; Cull, C.A.; Amachawadi, R.G.; Ramu, R. Isolation and characterization of lactic acid bacteria with potential probiotic activity and further investigation of their activity by α-amylase and α-glucosidase inhibitions of fermented batters. Front. Microbiol. 2023, 13, 1042263. [Google Scholar] [CrossRef]
- Łepecka, A.; Szymański, P.; Okoń, A.; Zielińska, D. Antioxidant activity of environmental lactic acid bacteria strains isolated from organic raw fermented meat products. Lwt 2023, 174, 114440. [Google Scholar] [CrossRef]
- Wang, G.; Chen, Y.; Xia, Y.; Song, X.; Ai, L. Characteristics of probiotic preparations and their applications. Foods 2022, 11, 2472. [Google Scholar] [CrossRef]
- Ardeshirlarijani, E.; Tabatabaei-Malazy, O.; Mohseni, S.; Qorbani, M.; Larijani, B.; Baradar Jalili, R. Effect of probiotics supplementation on glucose and oxidative stress in type 2 diabetes mellitus: A meta-analysis of randomized trials. DARU J. Pharm. Sci. 2019, 27, 827–837. [Google Scholar] [CrossRef]
- Kaprasob, R.; Sarkar, D.; Kerdchoechuen, O.; Laohakunjit, N.; Khanongnuch, C.; Shetty, K. Beneficial lactic acid bacteria based bioprocessing of cashew apple juice for targeting antioxidant nutraceutical inhibitors as relevant antidotes to type 2 diabetes. Process Biochem. 2019, 82, 40–50. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.Y.; Jeong, Y.; Kang, C.-H. Antioxidant Activity and Probiotic Properties of Lactic Acid Bacteria. Fermentation 2022, 8, 29. [Google Scholar] [CrossRef]
- Sathiyaseelan, A.; Saravanakumar, K.; Han, K.; Naveen, K.V.; Wang, M.-H. Antioxidant and Antibacterial Effects of Potential Probiotics Isolated from Korean Fermented Foods. Int. J. Mol. Sci. 2022, 23, 10062. [Google Scholar] [CrossRef]
- Kebouchi, M.; Galia, W.; Genay, M.; Soligot, C.; Lecomte, X.; Awussi, A.A.; Perrin, C.; Roux, E.; Dary-Mourot, A.; Le Roux, Y. Implication of sortase-dependent proteins of Streptococcus thermophilus in adhesion to human intestinal epithelial cell lines and bile salt tolerance. Appl. Microbiol. Biotechnol. 2016, 100, 3667–3679. [Google Scholar] [CrossRef] [PubMed]
- Bustos, A.Y.; Font de Valdez, G.; Fadda, S.; Taranto, M.P. New insights into bacterial bile resistance mechanisms: The role of bile salt hydrolase and its impact on human health. Food Res. Int. 2018, 112, 250–262. [Google Scholar] [CrossRef]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In vitro bile salt hydrolase (BSH) activity screening of different probiotic microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef]
- Dong, Z.; Lee, B.H. Bile salt hydrolases: Structure and function, substrate preference, and inhibitor development. Protein Sci. 2018, 27, 1742–1754. [Google Scholar] [CrossRef]
- Kaya, Y.; Kök, M.Ş.; Öztürk, M. Molecular cloning, expression and characterization of bile salt hydrolase from Lactobacillus rhamnosus E9 strain. Food Biotechnol. 2017, 31, 128–140. [Google Scholar] [CrossRef]
- Miyazaki, T.; Ueda, H.; Ikegami, T.; Honda, A. Upregulation of Taurine Biosynthesis and Bile Acid Conjugation with Taurine through FXR in a Mouse Model with Human-like Bile Acid Composition. Metabolites 2023, 13, 824. [Google Scholar] [CrossRef]
- Lee, M.; Park, J.; Kim, O.K.; Kim, D.; Han, M.J.; Kim, S.H.; Kim, T.H.; Lee, J. Lactobacillus reuteri NCIMB 30242 (LRC) Inhibits Cholesterol Synthesis and Stimulates Cholesterol Excretion in Animal and Cell Models. J. Med. Food 2023, 26, 529–539. [Google Scholar] [CrossRef]
- Guan, C.; Chen, X.; Jiang, X.; Zhao, R.; Yuan, Y.; Chen, D.; Zhang, C.; Lu, M.; Lu, Z.; Gu, R. In vitro studies of adhesion properties of six lactic acid bacteria isolated from the longevous population of China. RSC Adv. 2020, 10, 24234–24240. [Google Scholar] [CrossRef]
- Rajab, S.; Tabandeh, F.; Shahraky, M.K.; Alahyaribeik, S. The effect of lactobacillus cell size on its probiotic characteristics. Anaerobe 2020, 62, 102103. [Google Scholar] [CrossRef]
- Wang, S.; Li, L.; Yu, L.; Tian, F.; Zhao, J.; Zhai, Q.; Chen, W. Natural aggregation of Lactobacillus: Mechanisms and influencing factors. Food Biosci. 2024, 62, 105007. [Google Scholar] [CrossRef]
- Cozzolino, A.; Vergalito, F.; Tremonte, P.; Iorizzo, M.; Lombardi, S.J.; Sorrentino, E.; Luongo, D.; Coppola, R.; Di Marco, R.; Succi, M. Preliminary Evaluation of the Safety and Probiotic Potential of Akkermansia muciniphila DSM 22959 in Comparison with Lactobacillus rhamnosus GG. Microorganisms 2020, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef] [PubMed]
- Isenring, J.; Geirnaert, A.; Lacroix, C.; Stevens, M.J.A. Bistable auto-aggregation phenotype in Lactiplantibacillus plantarum emerges after cultivation in in vitro colonic microbiota. BMC Microbiol. 2021, 21, 268. [Google Scholar] [CrossRef]
- Grigoryan, S.; Bazukyan, I.; Trchounian, A. Aggregation and Adhesion Activity of Lactobacilli Isolated from Fermented Products In Vitro and In Vivo: A Potential Probiotic Strain. Probiotics Antimicrob. Proteins 2018, 10, 269–276. [Google Scholar] [CrossRef]
- Zawistowska-Rojek, A.; Kośmider, A.; Stępień, K.; Tyski, S. Adhesion and aggregation properties of Lactobacillaceae strains as protection ways against enteropathogenic bacteria. Arch. Microbiol. 2022, 204, 285. [Google Scholar] [CrossRef]
- Sophatha, B.; Piwat, S.; Teanpaisan, R. Adhesion, anti-adhesion and aggregation properties relating to surface charges of selected Lactobacillus strains: Study in Caco-2 and H357 cells. Arch. Microbiol. 2020, 202, 1349–1357. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Thananimit, S.; Pahumunto, N.; Teanpaisan, R. Characterization of short chain fatty acids produced by selected potential probiotic lactobacillus strains. Biomolecules 2022, 12, 1829. [Google Scholar] [CrossRef]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy. Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef]
- Inturri, R.; Trovato, L.; Volti, G.L.; Oliveri, S.; Blandino, G. In vitro inhibitory activity of Bifidobacterium longum BB536 and Lactobacillus rhamnosus HN001 alone or in combination against bacterial and Candida reference strains and clinical isolates. Heliyon 2019, 5, e02891. [Google Scholar] [CrossRef]
- Davoodabadi, A.; Soltan Dallal, M.M.; Lashani, E.; Tajabadi Ebrahimi, M. Antimicrobial Activity of Lactobacillus spp. Isolated From Fecal Flora of Healthy Breast-Fed Infants Against Diarrheagenic Escherichia coli. Jundishapur J. Microbiol. 2015, 8, e27852. [Google Scholar] [CrossRef] [PubMed]
- Johnson-Henry, K.C.; Donato, K.A.; Shen-Tu, G.; Gordanpour, M.; Sherman, P.M. Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157:H7-induced changes in epithelial barrier function. Infect. Immun. 2008, 76, 1340–1348. [Google Scholar] [CrossRef]
- Peng, H.; Zhou, G.; Yang, X.M.; Chen, G.J.; Chen, H.B.; Liao, Z.L.; Zhong, Q.P.; Wang, L.; Fang, X.; Wang, J. Transcriptomic Analysis Revealed Antimicrobial Mechanisms of Lactobacillus rhamnosus SCB0119 against Escherichia coli and Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 15159. [Google Scholar] [CrossRef]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T.; et al. In vitro Probiotic Potential and Safety Evaluation (Hemolytic, Cytotoxic Activity) of Bifidobacterium Strains Isolated from Raw Camel Milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef]
- Kim, M.J.; Ku, S.; Kim, S.Y.; Lee, H.H.; Jin, H.; Kang, S.; Li, R.; Johnston, T.V.; Park, M.S.; Ji, G.E. Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. Int. J. Mol. Sci. 2018, 19, 1422. [Google Scholar] [CrossRef]
- Žuntar, I.; Petric, Z.; Bursać Kovačević, D.; Putnik, P. Safety of Probiotics: Functional Fruit Beverages and Nutraceuticals. Foods 2020, 9, 947. [Google Scholar] [CrossRef]
- Gunzburg, W.H.; Aung, M.M.; Toa, P.; Ng, S.; Read, E.; Tan, W.J.; Brandtner, E.M.; Dangerfield, J.; Salmons, B. Efficient protection of microorganisms for delivery to the intestinal tract by cellulose sulphate encapsulation. Microb. Cell Fact. 2020, 19, 216. [Google Scholar] [CrossRef]
- Ayyash, M.M.; Abdalla, A.K.; AlKalbani, N.S.; Baig, M.A.; Turner, M.S.; Liu, S.-Q.; Shah, N.P. Invited review: Characterization of new probiotics from dairy and nondairy products—Insights into acid tolerance, bile metabolism and tolerance, and adhesion capability. J. Dairy. Sci. 2021, 104, 8363–8379. [Google Scholar] [CrossRef]
- Lund, P.A.; De Biase, D.; Liran, O.; Scheler, O.; Mira, N.P.; Cetecioglu, Z.; Fernández, E.N.; Bover-Cid, S.; Hall, R.; Sauer, M.; et al. Understanding How Microorganisms Respond to Acid pH Is Central to Their Control and Successful Exploitation. Front. Microbiol. 2020, 11, 556140. [Google Scholar] [CrossRef]
- Laulund, S.; Wind, A.; Derkx, P.M.F.; Zuliani, V. Regulatory and Safety Requirements for Food Cultures. Microorganisms 2017, 5, 28. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Lv, H.; Zhang, H.; Liu, Y.; Zhang, M.; Wang, Y.; Tan, Z. Probiotic Potential and Wide-spectrum Antimicrobial Activity of Lactic Acid Bacteria Isolated from Infant Feces. Probiotics Antimicrob. Proteins 2021, 13, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Pino, A.; Vaccalluzzo, A.; Caggia, C.; Balzaretti, S.; Vanella, L.; Sorrenti, V.; Ronkainen, A.; Satokari, R.; Randazzo, C.L. Lacticaseibacillus rhamnosus CA15 (DSM 33960) as a Candidate Probiotic Strain for Human Health. Nutrients 2022, 14, 4902. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Díaz, J.A.; Hernández García, J.E.; Sebastián Frizzo, L.; Fernández León, K.J.; Sánchez, L.; Solenzal Valdivia, Y. Caracterización in vitro de propiedades probióticas de Lactobacillus ssp. aislados del tracto digestivo de abejas. Rev. Salud Anim. 2021, 43. [Google Scholar]
- Serwecińska, L. Antimicrobials and antibiotic-resistant bacteria: A risk to the environment and to public health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Kromann, S.; Chen, M.; Shi, L.; Meng, H. Antibiotic resistance of Lactobacillus spp. and Streptococcus thermophilus isolated from Chinese fermented milk products. Foodborne Pathog. Dis. 2019, 16, 221–228. [Google Scholar] [CrossRef]
- Capurso, L. Thirty Years of Lactobacillus rhamnosus GG: A Review. J. Clin. Gastroenterol. 2019, 53, S1–S41. [Google Scholar] [CrossRef]
- Shahali, A.; Soltani, R.; Akbari, V. Probiotic Lactobacillus and the potential risk of spreading antibiotic resistance: A systematic review. Res. Pharm. Sci. 2023, 18, 468–477. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-Wide Assessment of Antibiotic Resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef]
- Hasan, M.; Arif, A.; Hasnain, A.; Abbas, T. Antibiotic susceptibility and antibacterial activity of neutralized cell-free supernatant of Lactobacillus rhamnosus MT539286 against Foodborne and Clinical pathogens. Int. J. Endorsing Health Sci. Res. (IJEHSR) 2020, 9, 4–9. [Google Scholar] [CrossRef]
- Anisimova, E.; Gorokhova, I.; Karimullina, G.; Yarullina, D. Alarming Antibiotic Resistance of Lactobacilli Isolated from Probiotic Preparations and Dietary Supplements. Antibiotics 2022, 11, 1557. [Google Scholar] [CrossRef]
- Chamberlain, M.; O’Flaherty, S.; Cobián, N.; Barrangou, R. Metabolomic Analysis of Lactobacillus acidophilus, L. gasseri, L. crispatus, and Lacticaseibacillus rhamnosus Strains in the Presence of Pomegranate Extract. Front. Microbiol. 2022, 13, 863228. [Google Scholar] [CrossRef] [PubMed]
- Indira, M.; Venkateswarulu, T.C.; Abraham Peele, K.; Nazneen Bobby, M.; Krupanidhi, S. Bioactive molecules of probiotic bacteria and their mechanism of action: A review. 3 Biotech. 2019, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Mora-Flores, L.P.; Moreno-Terrazas Casildo, R.; Fuentes-Cabrera, J.; Pérez-Vicente, H.A.; de Anda-Jáuregui, G.; Neri-Torres, E.E. The Role of Carbohydrate Intake on the Gut Microbiome: A Weight of Evidence Systematic Review. Microorganisms 2023, 11, 1728. [Google Scholar] [CrossRef]
- Ceapa, C.; Lambert, J.; van Limpt, K.; Wels, M.; Smokvina, T.; Knol, J.; Kleerebezem, M. Correlation of Lactobacillus rhamnosus Genotypes and Carbohydrate Utilization Signatures Determined by Phenotype Profiling. Appl. Environ. Microbiol. 2015, 81, 5458–5470. [Google Scholar] [CrossRef]
- Li, Q.-Q.; Zeng, S.-P.; Liang, M.-H.; Yousaf, M.; Wu, Y.-P.; Tang, J.; Xiong, J.; Liu, D.-M. Safety and metabolism characteristics of Lacticaseibacillus rhamnosus LR-ZB1107-01 based on complete genome and corresponding phenotype. LWT 2024, 204, 116443. [Google Scholar] [CrossRef]
- Jeckelmann, J.-M.; Erni, B. Carbohydrate transport by group translocation: The bacterial phosphoenolpyruvate: Sugar phosphotransferase system. Bact. Cell Walls Membr. 2019, 92, 223–274. [Google Scholar]
- Kaewarsar, E.; Chaiyasut, C.; Lailerd, N.; Makhamrueang, N.; Peerajan, S.; Sirilun, S. Optimization of mixed inulin, fructooligosaccharides, and galactooligosaccharides as prebiotics for stimulation of probiotics growth and function. Foods 2023, 12, 1591. [Google Scholar] [CrossRef]
- Niu, Z.; Zou, M.; Bei, T.; Zhang, N.; Li, D.; Wang, M.; Li, C.; Tian, H. Effect of fructooligosaccharides on the colonization of Lactobacillus rhamnosus AS 1.2466T in the gut of mice. Food Sci. Human. Wellness 2023, 12, 607–613. [Google Scholar] [CrossRef]
- Zunga, M.; Yebra, M.J.; Monedero, V. Complex Oligosaccharide Utilization Pathways in Lactobacillus. Curr. Issues Mol. Biol. 2021, 40, 49–80. [Google Scholar] [CrossRef]



| (A) Catalase | Hemolysis | |||||||||||||||||||||
| L. casei | − | − | ||||||||||||||||||||
| L. rhamnosus LBUX2302 | − | − | ||||||||||||||||||||
| S. aureus | + | + | ||||||||||||||||||||
| (B) Antimicrobial Activity | ||||||||||||||||||||||
| E. coli ATCC25922 | E. coli O157:H7 | S. typhi ATCC14028 | S. aureus ATCC6538 | |||||||||||||||||||
| L. casei | + | + | + | + | ||||||||||||||||||
| L. rhamnosus LBUX2302 | ++ | + | ++ | + | ||||||||||||||||||
| (C) Antibiotic Resistance | ||||||||||||||||||||||
| Va | Am | Stx | Ge | Dc | Cf | Clm | E | Pe | Te | Cfx | Cpf | |||||||||||
| L. casei | r | s | s | r | r | s | s | s | r | s | s | s | ||||||||||
| L. rhamnosus LBUX2302 | s | s | s | r | r | s | s | s | r | s | s | s | ||||||||||
| (D) Bile Salt Tolerance | ||||||||||||||||||||||
| GcCA | TcCA | GDxCA | TDxCA | OXGL | ||||||||||||||||||
| 0.1 | 0.3 | 0.5 | 0.1 | 0.3 | 0.5 | 0.1 | 0.3 | 0.5 | 0.1 | 0.3 | 0.5 | 0.1 | 0.3 | 0.5 | ||||||||
| L. casei | r | r | r | r | r | r | r | r | r | r | r | r | r | r | r | |||||||
| L. rhamnosus LBUX2302 | r | r | r | r | r | r | r | r | r | r | r | r | r | r | r | |||||||
| (E) % Hydrophobicity | %Autoaggregation | |||||||||||||||||||||
| L. casei | 0 | 94 | ||||||||||||||||||||
| L. rhamnosus LBUX2302 | 61 | 54 | ||||||||||||||||||||
| (F) % Coaggregation | ||||||||||||||||||||||
| E. coli ATCC25922 | S. aureus ATCC6538 | S. typhi ATCC14028 | ||||||||||||||||||||
| L. casei | 7.3 | 25 | 10 | |||||||||||||||||||
| L. rhamnosus LBUX2302 | 74 | 77 | 69 | |||||||||||||||||||
| (G) Ferulic Acid Activity | ||||||||||||||||||||||
| L. casei | + | |||||||||||||||||||||
| L. rhamnosus LBUX2302 | + | |||||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes-Castillo, P.A.; Esquivel-Campos, A.L.; Torres-Maravilla, E.; Zúñiga-León, E.; Mendoza-Pérez, F.; González-Vázquez, R.; Córdova-Espinoza, M.G.; Gutiérrez-Nava, M.A.; González-Vázquez, R.; Mayorga-Reyes, L. Hypoglycemic, Antioxidant Activities, and Probiotic Characteristics of Lacticaseibacillus rhamnosus LBUX2302 Isolated from Stool Samples of Neonates. Life 2025, 15, 804. https://doi.org/10.3390/life15050804
Reyes-Castillo PA, Esquivel-Campos AL, Torres-Maravilla E, Zúñiga-León E, Mendoza-Pérez F, González-Vázquez R, Córdova-Espinoza MG, Gutiérrez-Nava MA, González-Vázquez R, Mayorga-Reyes L. Hypoglycemic, Antioxidant Activities, and Probiotic Characteristics of Lacticaseibacillus rhamnosus LBUX2302 Isolated from Stool Samples of Neonates. Life. 2025; 15(5):804. https://doi.org/10.3390/life15050804
Chicago/Turabian StyleReyes-Castillo, Pedro A., Ana Laura Esquivel-Campos, Edgar Torres-Maravilla, Eduardo Zúñiga-León, Felipe Mendoza-Pérez, Rosa González-Vázquez, María Guadalupe Córdova-Espinoza, María Angélica Gutiérrez-Nava, Raquel González-Vázquez, and Lino Mayorga-Reyes. 2025. "Hypoglycemic, Antioxidant Activities, and Probiotic Characteristics of Lacticaseibacillus rhamnosus LBUX2302 Isolated from Stool Samples of Neonates" Life 15, no. 5: 804. https://doi.org/10.3390/life15050804
APA StyleReyes-Castillo, P. A., Esquivel-Campos, A. L., Torres-Maravilla, E., Zúñiga-León, E., Mendoza-Pérez, F., González-Vázquez, R., Córdova-Espinoza, M. G., Gutiérrez-Nava, M. A., González-Vázquez, R., & Mayorga-Reyes, L. (2025). Hypoglycemic, Antioxidant Activities, and Probiotic Characteristics of Lacticaseibacillus rhamnosus LBUX2302 Isolated from Stool Samples of Neonates. Life, 15(5), 804. https://doi.org/10.3390/life15050804







