Effects of Several Bile Acids on the Production of Virulence Factors by Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Medium
2.2. Cell Line
2.3. Chemical Products
2.4. Effects of Bile Acids on P. aeruginosa PAO1 Growth
2.5. Study of the Virulence of P. aeruginosa PAO1 Treated and Untreated with Bile Acids on HT29 Cells
2.5.1. Measurement of the Adhesion Index
2.5.2. Measurement of the Toxicity of PAO1 Culture Supernatants
2.5.3. Measurement of the Virulence of Bacteria Treated with Bile Acids on HT29 Cells
2.6. Effects of Bile Acids on the Production of Virulence Factors
2.6.1. Recovery of Supernatants and Protein Assay
2.6.2. Measurement of Protease Activities by Hydrolysis of Azocasein
2.6.3. Measurement of Pyocyanin Production
2.7. Extraction of RNA
2.8. RT-PCR
| DNA Synthesis: Primers Used in This Study | |||
| Gene | Amplicon Size | Fw Primer Sequence | Rev Primer Sequence |
| lasB | 261 bp | GCCGCAAGACCGAGAATGA | GTCGTTGACGATCAGCGGAC |
| aprA | 488 bp | TACGGCTTCAACTCCAACAC | TCGACGTATTGCAGCACCA |
| nan2 | 893 bp | CCTTGCAAACTCAACGACCA | AGGCGGGAGGACAGGATTTT |
| rDNA16S | 202 bp | AAGCAACGCGAAGAACCTTA | AAGCAACGCGAAGAACCTTA |
2.9. Mathematical Analysis
3. Results and Discussion
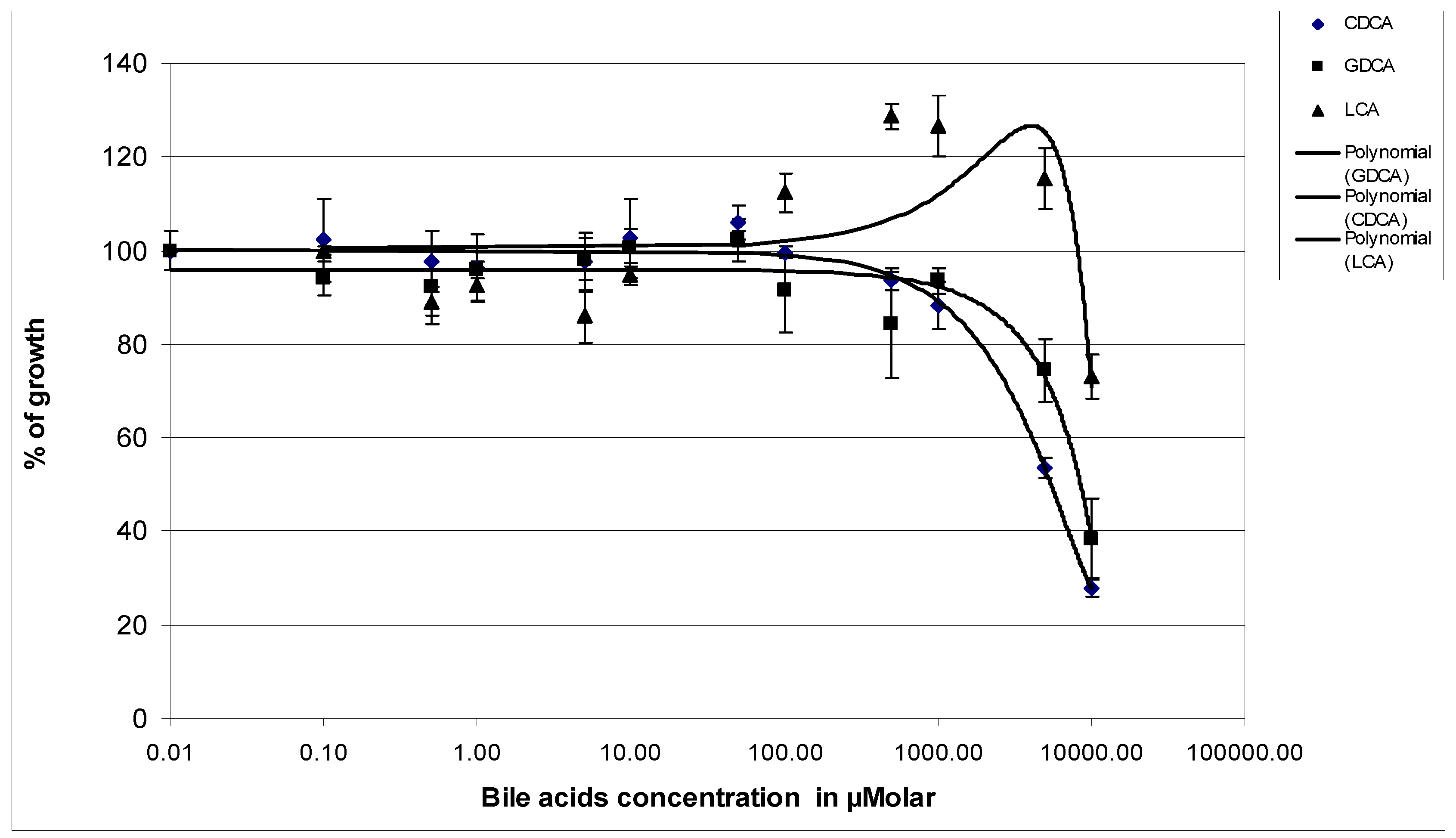
3.1. Effects of Bile Acids on the Growth of P. aeruginosa PAO1
3.2. Effects of Exposure of P. aeruginosa PAO1 to 1 mM Bile Acids on the Virulence of the Bacteria on HT29 Cell Line
3.2.1. Adhesion Index Measurement
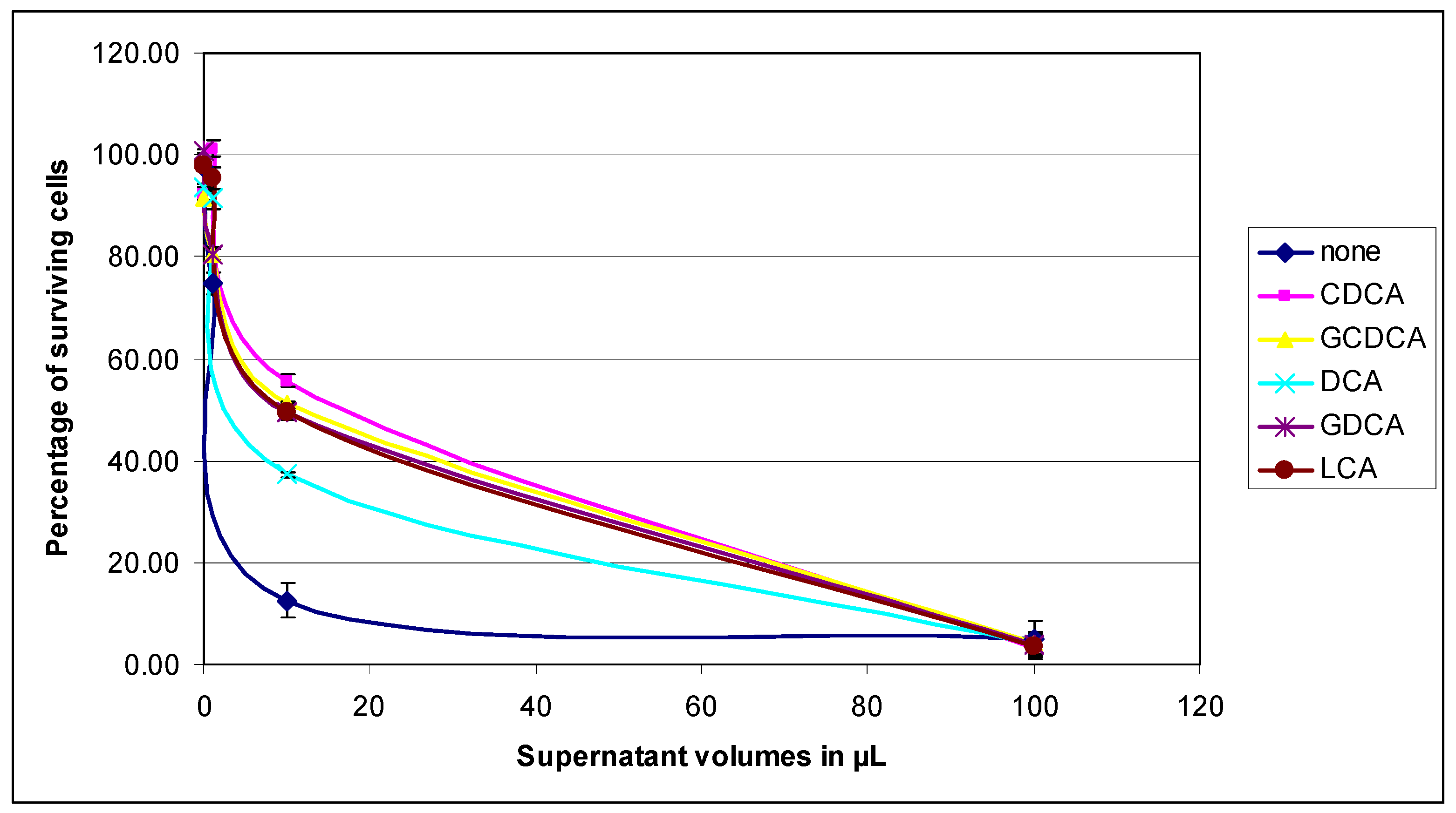
3.2.2. Virulence of the P. aeruginosa Growth Supernatants on HT29 Cells
3.2.3. Virulence of the PAO1 Strain on the HT29 Cells
3.3. Effects of Bile Acids on the Production of Virulence Factors of P. aeruginosa PAO1
3.3.1. Protease Activities in PAO1 Culture Supernatants
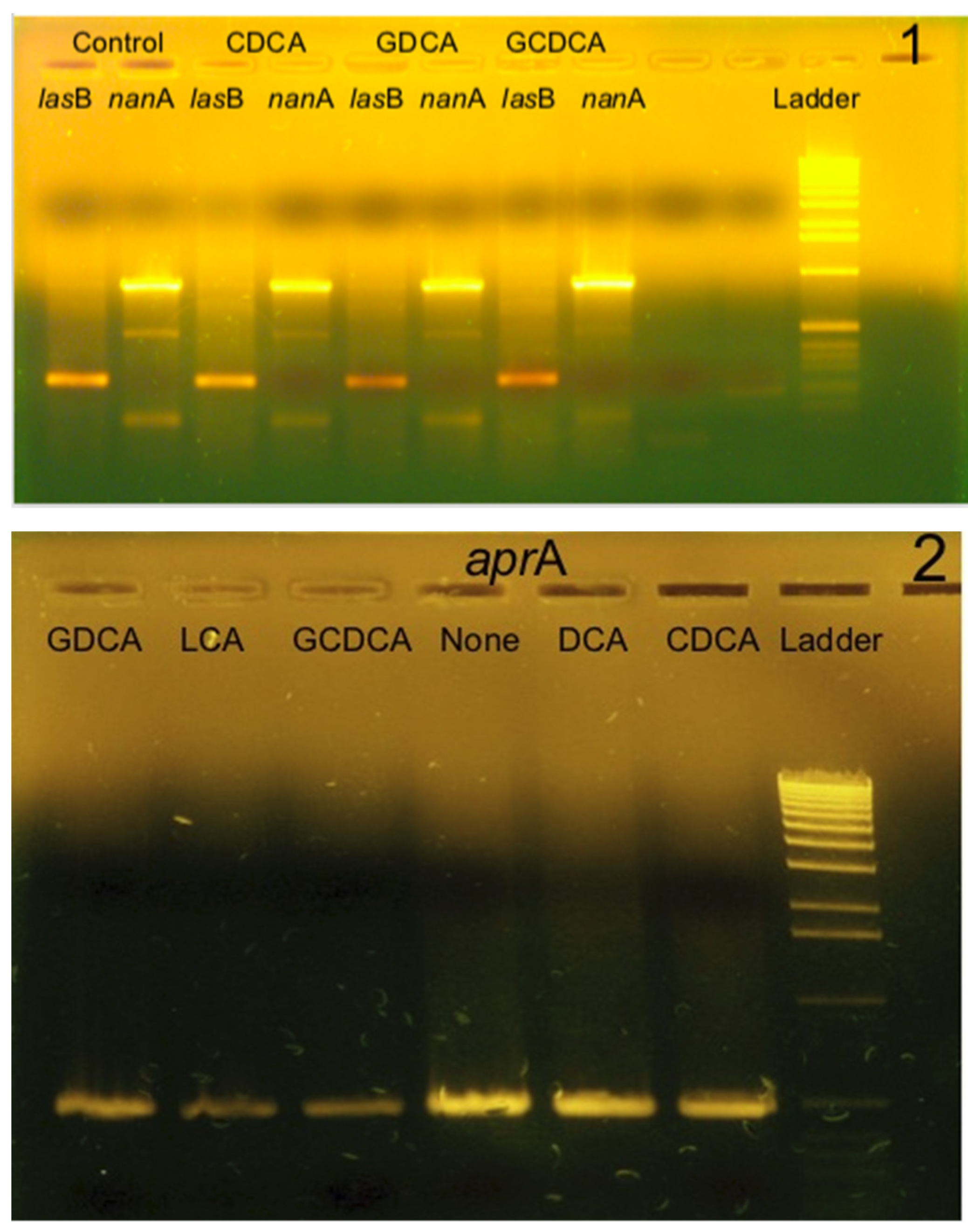
3.3.2. Study of the Transcription of Elastase, Alkaline Protease, and Pseudaminidase Genes by RT-PCR
3.4. Effect of Bile Acids on the Production of Pyocyanin by PAO1
3.5. Search for Potential Bile Acids Transporters in P. aeruginosa PAO1
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO (World Health Organisation). Global Status Report on Non Communicable Diseases 2010; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Falkowski, N.R.; Huffnagle, G.B.; Curtis, J.L. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio 2017, 8, e02287-16. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.C.; Milla, C.E.; Cornfield, D.N.; Quake, S.R. Quantitative Analysis of the Human Airway Microbial Ecology Reveals a Pervasive Signature for Cystic Fibrosis. Sci. Transl. Med. 2012, 4, 153ra130. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.J.; Allgaier, M.; Taylor, B.; Baek, M.S.; Huang, Y.J.; Daly, R.A.; Karaoz, U.; Andersen, G.L.; Brown, R.; Fujimura, K.E.; et al. Airway Microbiota and Pathogen Abundance in Age-Stratified Cystic Fibrosis Patients. PLoS ONE 2010, 5, e11044. [Google Scholar] [CrossRef] [PubMed]
- Boutin, S.; Graeber, S.Y.; Weitnauer, M.; Panitz, J.; Stahl, M.; Clausznitzer, D.; Kaderali, L.; Einarsson, G.; Tunney, M.M.; Elborn, J.S.; et al. Comparison of Microbiomes from Different Niches of Upper and Lower Airways in Children and Adolescents with Cystic Fibrosis. PLoS ONE 2015, 10, e0116029. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.D.N.; Viscogliosi, E.; Delhaes, L. The lung mycobiome: An emerging field of the human respiratory microbiome. Front. Microbiol. 2015, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.G. The Cystic Fibrosis Lung Microbiome. Ann. Am. Thorac. Soc. 2014, 11, 61. [Google Scholar] [CrossRef]
- Little, W.; Black, C.; Smith, A. Clinical Implications of Polymicrobial Synergism Effects on Antimicrobial Susceptibility. Pathogens 2021, 10, 144. [Google Scholar] [CrossRef]
- Zolin, A.; Orenti, A.; Naehrlich, L.; Jung, A.; van Rens, J.; Fox, A.; Krasnyk, M.; Cosgriff, R.; Hatziagorou, E.; Mei-Zahav, M.; et al. ECFS Patient Registry Annual Data Report 2018; European Cystic Fibrosis Society: Karup, Denmark, 2020. [Google Scholar]
- Guss, A.M.; Roeselers, G.; Newton, I.L.G.; Young, C.R.; Klepac-Ceraj, V.; Lory, S.; Cavanaugh, C.M. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2010, 5, 20–29. [Google Scholar] [CrossRef]
- Chmiel, J.F.; Davis, P.B. State of the Art. Why do the lungs of patients with cystic fibrosis become infected and why can’t they clear the infection? Respir. Res. 2003, 4, 8. [Google Scholar] [CrossRef]
- Muhlebach, M.S.; Sha, W.; MacIntosh, B.; Kelley, T.J.; Muenzer, J. Metabonomics reveals altered metabolites related to inflammation and energy utilization at recovery of cystic fibrosis lung exacerbation. Metab. Open 2019, 3, 100010. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Flynn, S.; Woods, D.F.; Dunphy, N.; Chróinín, M.N.; Mullane, D.; Stick, S.; Adams, C.; O’Gara, F. Bile signalling promotes chronic respiratory infections and antibiotic tolerance. Sci. Rep. 2016, 6, 29768. [Google Scholar] [CrossRef]
- Esteller, A. Physiology of bile secretion. World J. Gastroenterol. 2008, 14, 5641–5649. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P. Role of the intestinal bile acid transporters in bile acid and drug disposition. Handb. Exp. Pharmacol. 2001, 201, 169–203. [Google Scholar]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorakova, K.; Garewal, H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res./Rev. Mutat. Res. 2005, 589, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Colombo, C.; Russo, M.C.; Zazzeron, L.; Romano, G. Liver disease in cystic fibrosis. J. Pediatr. Gastroenterol. Nutr. 2006, 43 (Suppl. S1), S49–S55. [Google Scholar] [CrossRef]
- Bernstein, C.; Bernstein, H.; Payne, C.M.; Beard, S.E.; Schneider, J. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 1999, 39, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.Y.; Bertenthal, D.; Nilles, M.L.; Bertrand, K.P.; Nikaido, H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 2003, 48, 1609–1619. [Google Scholar] [CrossRef]
- Reen, F.J.; Woods, D.F.; Mooij, M.J.; Adams, C.; O’Gara, F. Respiratory pathogens adopt a chronic lifestyle in response to bile. PLoS ONE 2012, 9, e45978. [Google Scholar] [CrossRef] [PubMed]
- Laursen, J.B.; Nielsen, J. Phenazine natural pruducts: Biosynthesis, Synthetic analogues and biological activity. Chem. Rev. 2004, 104, 1663–1686. [Google Scholar] [CrossRef] [PubMed]
- Denning, M.; Railsback, M.A.; Rasmussen, G.T.; Cox, C.D.; Britigan, B.E. Pseudomonas pyocyanine alters calcium signaling in human airway epithelial cells. Am. J. Physiol. 1998, 274, L893–L900. [Google Scholar]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Morihara, K.; Tsuzuki, H.; Oka, T.; Inoue, H.; Ebata, M. Pseudomonas aeruginosa elastase: Isolation, crystallization, and preliminary characterization. J. Biol. Chem. 1965, 240, 3295–3304. [Google Scholar] [CrossRef] [PubMed]
- Heck, L.W.; Morihara, K.; McRae, W.B.; Miller, E.J. Specific cleavage of human type III and IV collagens by Pseudomonas aeruginosa elastase. Infect. Immun. 1986, 51, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Heck, L.W.; Morihara, K.; Abrahamson, D.R. Degradation of soluble laminin and depletion of tissue-associated basement membrane laminin by Pseudomonas aeruginosa elastase and alkaline protease. Infect. Immun. 1986, 54, 149–153. [Google Scholar] [CrossRef]
- Azghani, A.O.; Gray, L.D.; Johnson, A.R. A bacterial protease perturbs the paracellular barrier function of transporting epithelial monolayers in culture. Infect. Immun. 1993, 61, 2681–2686. [Google Scholar] [CrossRef] [PubMed]
- Doring, G.; Goldstein, W.; Roll, A.; Schiotz, P.O.; Hoiby, N.; Botzenhart, K. Role of Pseudomonas aeruginosa exoenzymes in lung infections of patients with cystic fibrosis. Infect. Immun. 1985, 49, 557–562. [Google Scholar] [CrossRef]
- Heck, L.W.; Alarcon, P.G.; Kulhavy, R.M.; Morihara, K.; Russell, M.W.; Mestecky, J.F. Degradation of IgA proteins by Pseudomonas aeruginosa elastase. J. Immunol. 1990, 144, 2253–2257. [Google Scholar] [CrossRef]
- Braun, P.; Ockhuijsen, C.; Eppens, E.; Koster, M.; Bitter, W.; Tommassen, J. Maturation of Pseudomonas aeruginosa elastase. Formation of the disulfide bonds. J. Biol. Chem. 2001, 276, 26030–26035. [Google Scholar] [CrossRef]
- Rumbaugh, K.P.; Griswold, J.A.; Hamood, A.N. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2000, 2, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, M.M.; Khodi, S.; Mirhosseini, A. Quorum sensing in bacteria and a glance on Pseudomonas aeruginosa clinical microbiology. Clin. Microbial. 2014, 3, 156. [Google Scholar] [CrossRef]
- Goldberg, J.B.; Ohman, D.E. Activation of an elastase precursor by the lasA gene product of Pseudomonas aeruginosa. J. Bacteriol. 1987, 169, 4532–4539. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, J.B.; Ohman, D.E. Cloning and transcriptional regulation of the elastase lasA gene in mucoid and nonmucoid Pseudomonas aeruginosa. J. Bacteriol. 1987, 169, 1349–1351. [Google Scholar] [CrossRef] [PubMed]
- Schad, P.A.; Iglewski, B.H. Nucleotide sequence and expression in Escherichia coli of the Pseudomonas aeruginosa lasA gene. J. Bacteriol. 1988, 170, 2784–2789. [Google Scholar] [CrossRef] [PubMed]
- Kessler, E.; Safrin, M.; Gustin, J.K.; Ohman, D.E. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J. Biol. Chem. 1998, 273, 30225–30231. [Google Scholar] [CrossRef]
- Kessler, E.; Safrin, M.; Olson, J.C.; Ohman, D.E. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 1993, 268, 7503–7508. [Google Scholar] [CrossRef]
- Nouwens, A.S.; Beatson, S.A.; Whitchurch, C.B.; Walsh, B.J.; Schweizer, H.P.; Mattick, J.S.; Cordwell, S.J. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 2003, 149, 1311–1322. [Google Scholar] [CrossRef]
- Van Delden, C.; Iglewski, B.H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 1998, 4, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Gambello, M.J.; Kaye, S.; Iglewski, B.H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 1993, 61, 1180–1184. [Google Scholar] [CrossRef]
- Antunes, L.C.; Ferreira, R.B.; Buckner, M.M.; Finlay, B.B. Quorum sensing in bacterial virulence. Microbiology 2010, 156, 2271–2282. [Google Scholar] [CrossRef]
- Caldwell, C.C.; Chen, Y.; Goetzmann, H.S.; Hao, Y.; Borchers, M.T.; Hassett, D.J.; Young, L.R.; Mavrodi, D.; Thomashow, L.; Lau, G.W. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am. J. Pathol. 2009, 175, 2473–2488. [Google Scholar] [CrossRef]
- Heurlier, K.; Dénervaud, V.; Pessi, G.; Reimmann, C.; Haas, D. Negative Control of Quorum Sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J. Bact. 2003, 185, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Reimmann, C.; Beyeler, M.; Latifi, A.; Winteler, H.; Foglino, M.; Lazdunski, A.; Haas, D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 1997, 24, 309–319. [Google Scholar] [CrossRef]
- Coggan, K.A.; Wolfgang, M.C. Global Regulatory Pathways and Cross-talk Control Pseudomonas aeruginosa Environmental Lifestyle and Virulence Phenotype. Curr. Issues Mol. Biol. 2012, 14, 47–70. [Google Scholar] [CrossRef] [PubMed]
- Sabra, W.; Kim, E.-J.; Zeng, A.-P. Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology 2002, 148, 3195–3202. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chiang, J.Y.L. Bile Acid signaling in liver metabolism and diseases. J. Lipids 2012, 2012, 754067. [Google Scholar] [CrossRef]
- Padayatti, P.S.; Lee, S.C.; Stanfield, R.L.; Wen, P.-C.; Tajkhorshid, E.; Wilson, I.A.; Zhang, Q. Structural insights into the Lipid A transport pathway in MsbA. Structure 2019, 27, 1114–1123.e3. [Google Scholar] [CrossRef] [PubMed]
- Hulen, C.; Racine, P.-J.; Chevalier, S.; Feuilloley, M.; Lomri, N.-E. Identification of the PA1113 Gene Product as an ABC Transporter Involved in the Uptake of Carbenicillin in Pseudomonas aeruginosa PAO1. Antibiotics 2020, 9, 596. [Google Scholar] [CrossRef] [PubMed]
- Noda, K.-I.; Watanabe, K.; Maruhashi, K. Isolation of the Pseudomonas aeruginosa gene affecting uptake of Dibenzothiophene in n-tetradecane. J. Biosci. Bioeng. 2003, 95, 504–511. [Google Scholar] [CrossRef]
- Inagaki, T.; Moschetta, A.; Lee, Y.-K.; Peng, L.; Zhao, G.; Downes, M.; Yu, R.T.; Shelton, J.M.; Richardson, J.A.; Repa, J.J.; et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 3920–3925. [Google Scholar] [CrossRef]
- Rizzo, G.; Renga, B.; Mencarelli, A.; Pellicciari, R.; Fiorucci, S. Role of FXR in regulating bile acid homeostasis and relevance for human diseases. Curr. Drug Targets Immune Endocr. Metab. Disord. 2005, 5, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Attinkara, R.; Mwinyi, J.; Truninger, K.; Regula, J.; Gaj, P.; Rogler, G.; Kullack-Ublick, G.A.; Eloranta, J.J.; The Swiss IBD Cohort Study Group. Association of genetic variation in the NR1H4 gene, encoding the nuclear bile acid receptor FXR, with inflammatory bowel disease. BMC Res. Notes 2012, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.F.; Flynn, S.; Caparrós-Martín, J.A.; Stick, S.M.; Reen, F.J.; O’Gara, F. Systems biology and bile acid signalling in microbiome-host interactions in the cystic fibrosis lung. Antibiotics 2021, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Bayat, M.; Nahand, J.S.; Farsad-Akhatr, N.; Memar, M.Y. Bile effects on the Pseudomonas aeruginosa pathogenesis in cystic fibrosis patients with gastroesophageal reflux. Heliyon 2023, 9, e22111. [Google Scholar] [CrossRef] [PubMed]
- Behroozian, S.; Sampedro, I.; Dhodary, B.; Her, S.; Yu, Q.; Stanton, B.A.; Hill, J.E. Pseudomonas aeruginosa PAO1 is attracted to Bovine Bile in a novel, Cystic Fibrosis-derived bronchial epithelial cell model. Microorganisms 2022, 10, 716. [Google Scholar] [CrossRef]


| Bacteria Treatment | Adhesion Index |
|---|---|
| None | 2.95 ± 0.13 |
| CDCA | 2.98 ± 0.23 |
| GCDCA | 2.15 ± 0.2 |
| DCA | 2.11 ± 0.32 |
| GDCA | 2.16 ± 0.11 |
| LCA | 2.52 ± 0.13 |
| Treatment | Specific Protease Activity (mg Azocasein Hydrolyzed/min/mg of Secreted Proteins) | Inhibition (%) |
|---|---|---|
| None | 0.542 ± 0.082 | |
| CDCA | 0.423 ± 0.043 | 21.98 ± 2.25 (p = 0.017) |
| GCDCA | 0.432 ± 0.048 | 20.26 ± 2.24 (p = 0.038) |
| DCA | 0.527 ± 0.082 | 2.64 ± 0.41 (p = 0.014) |
| GDCA | 0.469 ± 0.068 | 13.33 ± 1.93 (p = 0.174) |
| LCA | 0.362 ± 0.045 | 33.25 ± 4.10 (p = 0.003) |
| Treatment | Specific Protease Activity (mg Azocasein Hydrolyzed/min/mg of Secreted Proteins) | Inhibition (%) |
|---|---|---|
| None | 1.329 ± 0.127 | |
| CDCA | 0.809 ± 0.236 | 39.14 ± 11.42 (p = 0.032) |
| GCDCA | 0.328 ± 0.031 | 75.35 ± 7.23 (p = 0.007) |
| DCA | 0.745 ± 0.176 | 43.99 ± 10.38 (p = 0.009) |
| GDCA | 0.489 ± 0.109 | 63.22 ± 14.10 (p = 0.0004) |
| LCA | 0.349 ± 0.113 | 73.74 ± 23.97 (p = 0.019) |
| Percentage of Inhibition of Gene Expression | |||
|---|---|---|---|
| Treatment | lasB | aprA | nanA |
| None | 0.00 | 0.00 | 0.00 |
| DCA | 3.2 ± 1.4 | 5.3 ± 7.1 | 4.7 ± 5 |
| GDCA | 38.7 ± 8.4 | 15.8 ± 3.2 | 4.2 ± 2.9 |
| CDCA | 28 ± 7 | 13.5 ± 2.5 | 2.7 ± 3.6 |
| GCDCA | 15 ± 5 | 20 ± 9 | 0.00 |
| LCA | 37 ± 17 | 45.7 ± 10.5 | 0.00 |
| Treatment | Pyocyanine Production (µg/109 Bacteria) | Inhibition (%) |
|---|---|---|
| None | 3.39 ± 0.26 | |
| CDCA | 1.93 ± 0.57 | 43 ± 0.3 (p = 0.25) |
| GCDCA | 1.34 ± 0.19 | 61 ± 0.9 (p = 0.05) |
| DCA | 1.57 ± 0.19 | 54 ± 0.6 (p = 0.17) |
| GDCA | 1.75 ± 0.28 | 49 ± 0.8 (p = 0.12) |
| LCA | 0.91 ± 0.18 | 73 ± 0.15 (p = 0.01) |
| Treatment | Pyocyanine Production (µg/109 Bacteria) | Inhibition (%) |
|---|---|---|
| None | 1.54 ± 0.18 | |
| CDCA | 1.00 ± 0.09 | 35 ± 0.3 (p = 0.004) |
| GCDCA | 0.85 ± 0.04 | 45 ± 0.2 (p = 0.05) |
| DCA | 1.03 ± 0.06 | 33 ± 0.2 (p = 0.022) |
| GDCA | 1.39 ± 0.12 | 10 ± 0.1 (p = 0.013) |
| LCA | 0.35 ± 0.02 | 77 ± 0.5 (p = 0.04) |
| Strain | Treatment | Pyocyanine Production (µg/109 Bacteria) | Inhibition (%) |
|---|---|---|---|
| PAO1 | none | 4.18 ± 0.04 | 0 |
| PAO1 | CDCA | 2.59 ± 0.06 | 38 ± 0.6 (p = 0.06) |
| PAO1 | LCA | 1.74 ± 0.04 | 58 ± 0.5 (p = 0.005) |
| PW3935 (gene PA1650) | none | 4.38 ± 0.35 | 0 |
| PW3935 (gene PA1650) | CDCA | 3.59 ± 0.09 | 20 ± 0.6 (p = 0.05) |
| PW3935 (gene PA1650) | LCA | 2.34 ± 0.06 | 47 ± 0.8 (p = 0.08) |
| PW6477 (gene PA3264) | none | 3.70 ± 0.05 | 0 |
| PW6477 (gene PA3264) | CDCA | 3.72 ± 0.02 | 0 (p = 0.05) |
| PW6477 (gene PA3264) | LCA | 2.46 ± 0.08 | 33 ± 0.8 (p = 0.08) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lomri, N.; Hulen, C. Effects of Several Bile Acids on the Production of Virulence Factors by Pseudomonas aeruginosa. Life 2024, 14, 1676. https://doi.org/10.3390/life14121676
Lomri N, Hulen C. Effects of Several Bile Acids on the Production of Virulence Factors by Pseudomonas aeruginosa. Life. 2024; 14(12):1676. https://doi.org/10.3390/life14121676
Chicago/Turabian StyleLomri, Noureddine, and Christian Hulen. 2024. "Effects of Several Bile Acids on the Production of Virulence Factors by Pseudomonas aeruginosa" Life 14, no. 12: 1676. https://doi.org/10.3390/life14121676
APA StyleLomri, N., & Hulen, C. (2024). Effects of Several Bile Acids on the Production of Virulence Factors by Pseudomonas aeruginosa. Life, 14(12), 1676. https://doi.org/10.3390/life14121676






