Five New Species of Marquandomyces (Clavicipitaceae, Ascomycota) from Asia
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Materials
2.2. Morphological Observations
2.3. DNA Extraction, PCR Amplification, and Sequencing
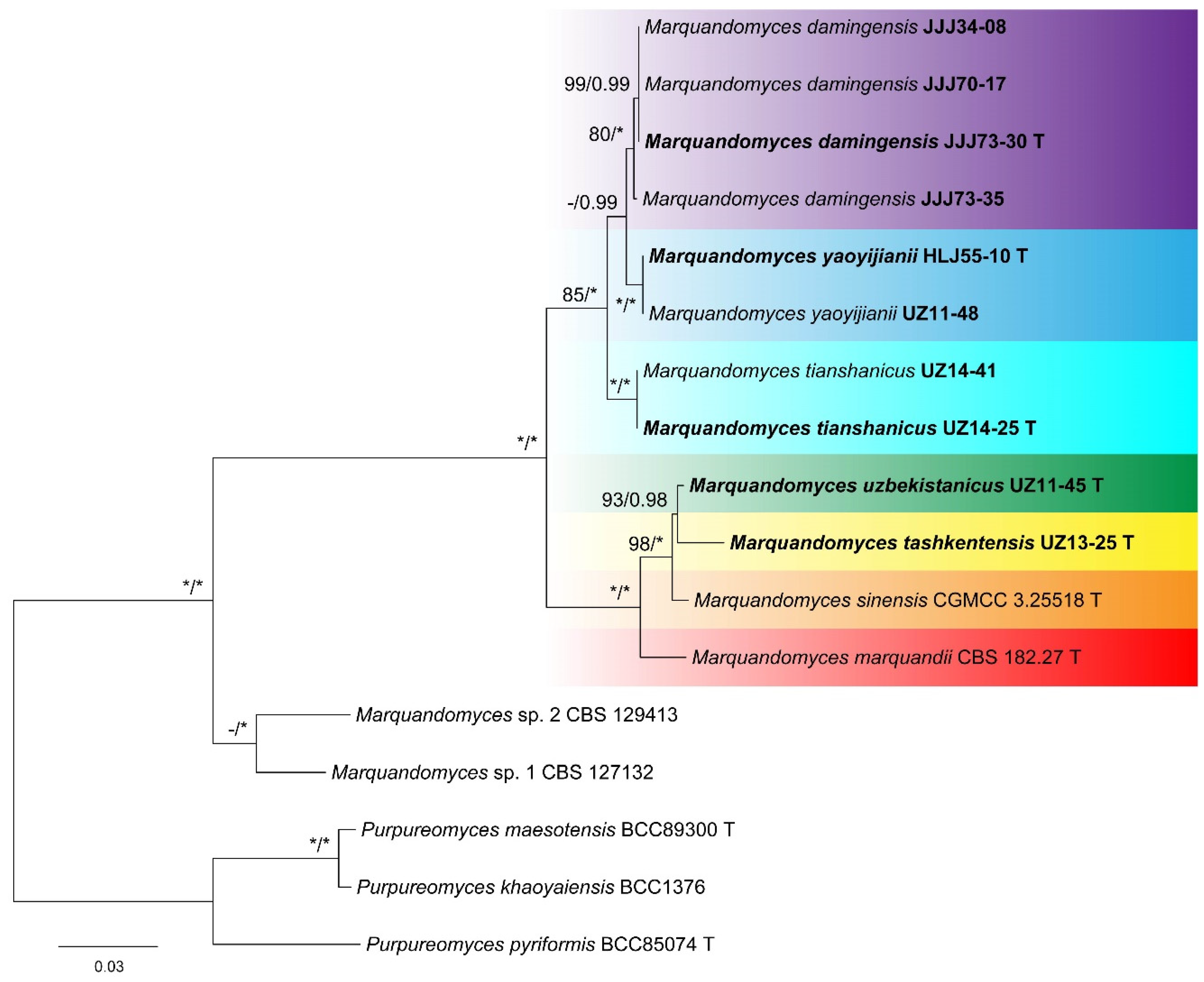
2.4. Phylogenetic Analyses
3. Results
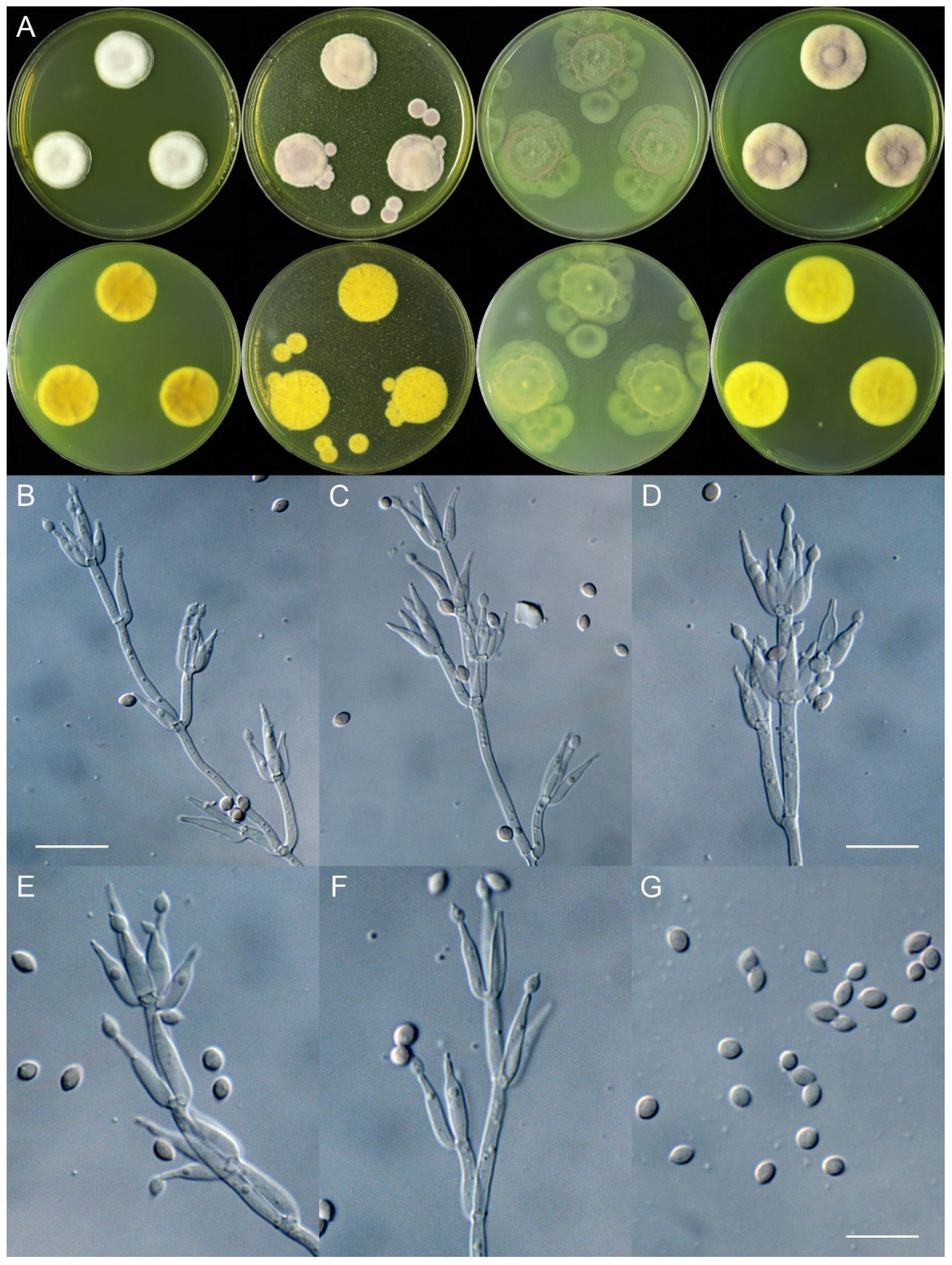
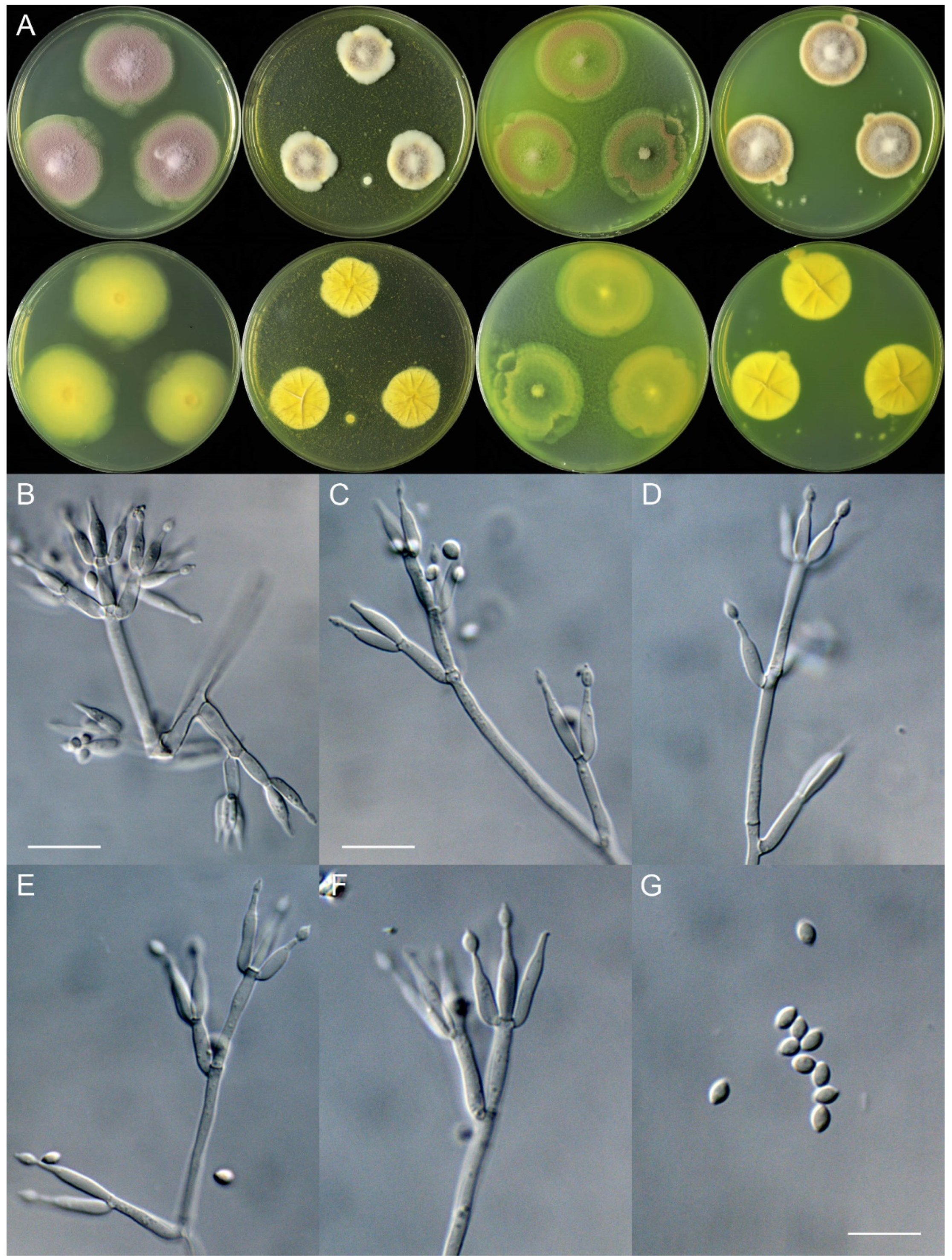
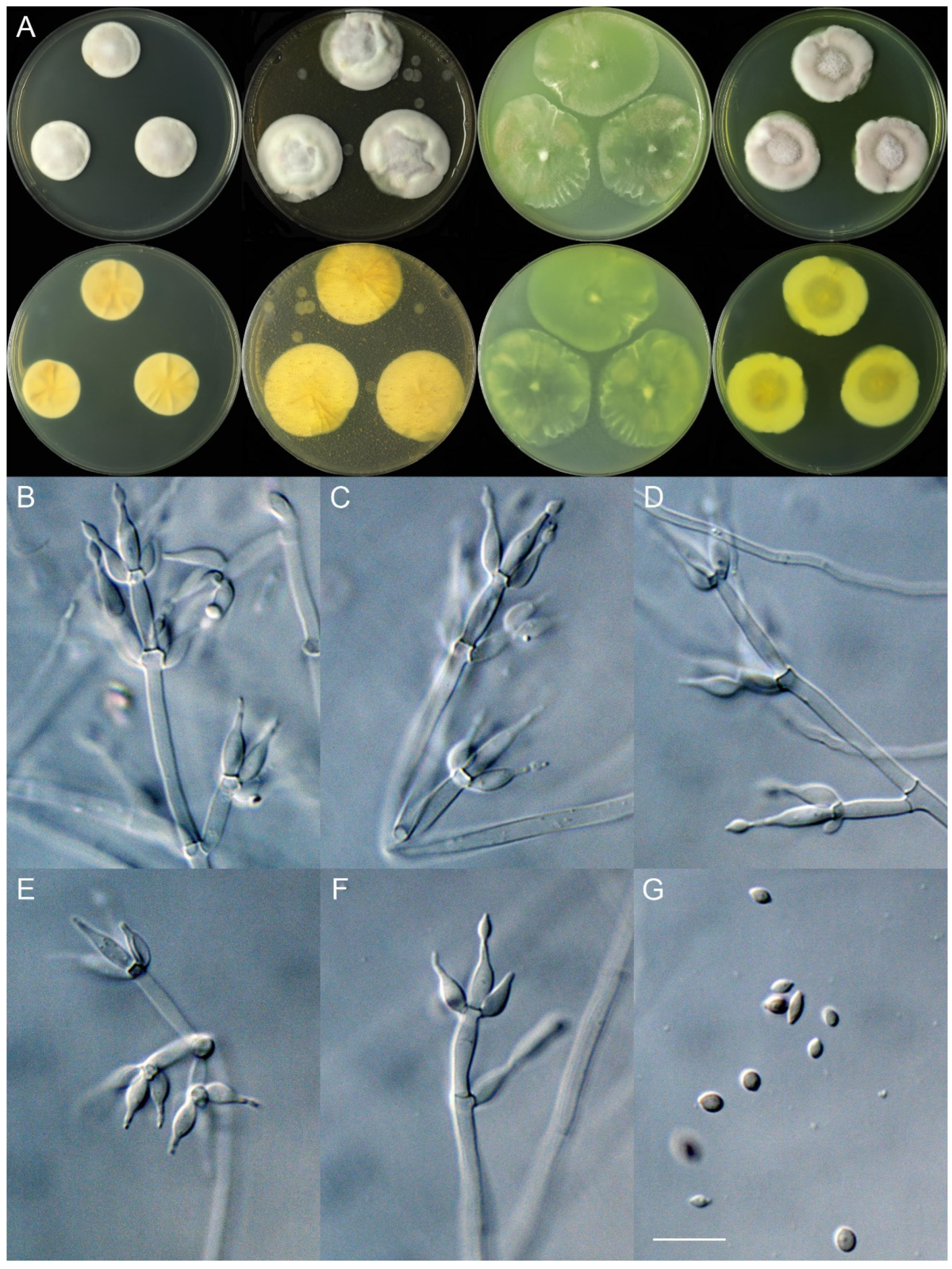
4. Taxonomy
| 1. Distributed worldwide | M. marquandii |
| 1. Distributed in Asia | 2 |
| 2. Chlamydospore-like structures present | M. sinensis |
| 2. Chlamydospore-like structures absent | 3 |
| 3. Conidia en masse on PDA 25 °C cinnamon-colored | M. uzbekistanicus |
| 3. Conidia en masse on PDA 25 °C purplish vinaceous | 4 |
| 4. Sporulation on OA 25 °C dense | 5 |
| 4. Sporulation on OA 25 °C sparse | 6 |
| 5. Growth rate on CYA 25 °C fast (more than 35 mm) | M. tianshanicus |
| 5. Growth rate on CYA 25 °C slow (no more than 30 mm) | M. damingensis |
| 6. Growth rate on MEA 25 °C fast (no less than 30 mm) | M. yaoyijianii |
| 6. Growth rate on MEA 25 °C slow (less than 25 mm) | M. tashkentensis |
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mongkolsamrit, S.; Khonsanit, A.; Thanakitpipattana, D.; Tasanathai, K.; Noisripoom, W.; Lamlertthon, S.; Himaman, W.; Houbraken, J.; Samson, R.A.; Luangsa-Ard, J. Revisiting Metarhizium and the description of new species from Thailand. Stud. Mycol. 2020, 95, 171–251. [Google Scholar] [CrossRef] [PubMed]
- Marbanmendoza, N.; Garciae, R.; Dicklow, M.B.; Zuckerman, B.M. Studies on Paecilomyces marquandii from nematode suppressive chinampa soils. J. Chem. Ecol. 1992, 18, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Abawi, G.S.; Zuckerman, B.M. Efficacy of Bacillus thuringiensis, Paecilomyces marquandii, and Streptomyces costaricanus with and without organic amendments against Meloidogyne hapla infecting lettuce. J. Nematol. 2000, 32, 70–77. [Google Scholar]
- Ahuja, A.; Ghosh, S.B.; D’Souza, S.F. Isolation of a starch utilizing, phosphate solubilizing fungus on buffered medium and its characterization. Bioresour. Technol. 2007, 98, 3408–3411. [Google Scholar] [CrossRef]
- Ahuja, A.; D’Souza, S.F. Bioprocess for solubilization of rock phosphate on starch based medium by Paecilomyces marquandii immobilized on polyurethane foam. Appl. Biochem. Biotechnol. 2009, 152, 1–5. [Google Scholar] [CrossRef]
- Posada, R.H.; Heredia-Abarca, G.; Sieverding, E.; de Prager, M.S. Solubilization of iron and calcium phosphates by soil fungi isolated from coffee plantations. Arch. Agron. Soil Sci. 2013, 59, 185–196. [Google Scholar] [CrossRef]
- Baron, N.C.; Souza Pollo, A.D.; Rigobelo, E.C. Purpureocillium lilacinum and Metarhizium marquandii as plant growth-promoting fungi. PeerJ 2020, 8, e9005. [Google Scholar] [CrossRef]
- Slaba, M.; Dlugonski, J. Selective recovery of Zn2+ from waste slag from a metal-processing plant by the microscopic fungus Verticillium marquandii. Biotechnol. Lett. 2000, 22, 1699–1704. [Google Scholar] [CrossRef]
- Slaba, M.; Dlugonski, J. Zinc and lead uptake by mycelium and regenerating protoplasts of Verticillium marquandii. World J. Microbiol. Biotechnol. 2004, 20, 323–328. [Google Scholar] [CrossRef]
- Slaba, M.; Dlugonski, J. Efficient Zn2+ and Pb2+ uptake by filamentous fungus Paecilomyces marquandii with engagement of metal hydrocarbonates precipitation. Int. Biodeter. Biodegr. 2011, 65, 954–960. [Google Scholar] [CrossRef]
- Slaba, M.; Szewczyk, R.; Piatek, M.A.; Dlugonski, J. Alachlor oxidation by the filamentous fungus Paecilomyces marquandii. J. Hazard. Mater. 2013, 261, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Slaba, M.; Rozalska, S.; Bernat, P.; Szewczyk, R.; Piatek, M.A.; Dlugonski, J. Efficient alachlor degradation by the filamentous fungus Paecilomyces marquandii with simultaneous oxidative stress reduction. Bioresour. Technol. 2015, 197, 404–409. [Google Scholar] [CrossRef]
- Szewczyk, R.; Sobon, A.; Slaba, M.; Dlugonski, J. Mechanism study of alachlor biodegradation by Paecilomyces marquandii with proteomic and metabolomic methods. J. Hazard. Mater. 2015, 291, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A. Paecilomyces and some allied Hyphomycetes. Stud. Mycol. 1974, 6, 1–119. [Google Scholar]
- Luangsa-ard, J.J.; Hywel-Jones, N.L.; Samson, R.A. The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia 2004, 96, 773–780. [Google Scholar] [CrossRef]
- Luangsa-ard, J.J.; Hywel-Jones, N.L.; Manoch, L.; Samson, R.A. On the relationships of Paecilomyces sect. Isarioidea species. Mycol. Res. 2005, 109, 581–589. [Google Scholar] [CrossRef]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Kepler, R.M.; Humber, R.A.; Bischoff, J.F.; Rehner, S.A. Clarification of generic and species boundaries for Metarhizium and related fungi through multigene phylogenetics. Mycologia 2014, 106, 811–829. [Google Scholar] [CrossRef]
- Luangsa-ard, J.J.; Mongkolsamrit, S.; Thanakitpipattana, D.; Khonsanit, A.; Tasanathai, K.; Noisripoom, W.; Humber, R.A. Clavicipitaceous entomopathogens: New species in Metarhizium and a new genus Nigelia. Mycol. Prog. 2017, 16, 369–391. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Han, Y.F.; Liang, Z.Q.; Liu, A.Y.; Chu, H.L. Two new records of Paecilomyces in China. J. Fung. Res. 2003, 1, 49–51. [Google Scholar]
- Zhang, Z.Y.; Pan, H.; Tao, G.; Li, X.; Han, Y.F.; Feng, Y.; Tong, S.Q.; Ding, C.Y. Culturable mycobiota from Guizhou wildlife park in China. Mycosphere 2024, 15, 654–763. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Xia, Y.W.; Wang, L.; Li, T.H.; Zhuang, W.Y. A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa 2016, 267, 187–200. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Qin, W.T.; Zhuang, W.Y. Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycol. Prog. 2017, 16, 73–81. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Zeng, Z.Q.; Zhuang, W.Y. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci. Rep. 2017, 7, 8233. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Wang, X.C.; Zhuang, W.Y.; Cheng, X.H.; Zhao, P. New species of Talaromyces (Fungi) isolated from soil in southwestern China. Biology 2021, 10, 745. [Google Scholar] [CrossRef]
- Wang, X.C.; Zhuang, W.Y. New Species of Aspergillus (Aspergillaceae) from tropical islands of China. J. Fungi 2022, 8, 225. [Google Scholar] [CrossRef]
- Wang, X.C.; Zhuang, W.Y. New Species of Talaromyces (Trichocomaceae, Eurotiales) from Southwestern China. J. Fungi 2022, 8, 647. [Google Scholar] [CrossRef]
- Wang, X.C.; Zhang, Z.K.; Zhuang, W.Y. Species Diversity of Penicillium in Southwest China with discovery of forty-three new species. J. Fungi 2023, 9, 1150. [Google Scholar] [CrossRef]
- Song, H.; Ding, Y.J.; Zhuang, W.Y.; Ding, G.Z.; Wang, X.C. Three new species of Penicillium from East and Northeast China. J. Fungi 2024, 10, 342. [Google Scholar] [CrossRef]
- Wang, X.C.; Zhuang, W.Y.; Zhao, R.L. Species diversity of Helvella lacunosa clade (Pezizales, Ascomycota) in China and description of sixteen new species. J. Fungi 2023, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Bio. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Held, B.W.; Salomon, C.E.; Blanchette, R.A. Diverse subterranean fungi of an underground iron ore mine. PLoS ONE 2020, 15, e0234208. [Google Scholar] [CrossRef]
- Pattanaik, A.; Sukla, L.B.; Devi, N.; Pradhan, N.; Pradhan, D. Tungsten dissolution from Hutti Goldmine overburden by Aspergillus niger. Geomicrobiol. J. 2022, 39, 496–501. [Google Scholar] [CrossRef]


| Species | Strain | Locality | Substrate | ITS | LSU | TEF | Reference |
|---|---|---|---|---|---|---|---|
| Marquandomyces damingensis X.C. Wang, L.Y. Peng & W.Y. Zhuang, sp. nov. | JJJ34-08 | China: Hebei | soil | PQ484185 | PQ484199 | PQ469016 | This study |
| JJJ70-17 | China: Hebei | soil | PQ484186 | PQ484200 | PQ469017 | This study | |
| JJJ73-30 = CGMCC 3.28567 T | China: Hebei | soil | PQ484187 | PQ484201 | PQ469018 | This study | |
| JJJ73-35 | China: Hebei | soil | PQ484188 | PQ484202 | PQ469019 | This study | |
| Marquandomyces marquandii (Massee) Samson, Houbraken & Luangsa-ard 2020 | CBS 182.27 T | USA: Iowa | soil | MH854923 | EF468845 | EF468793 | [17,20] |
| Marquandomyces sinensis Zhi Y. Zhang & Y.F. Han 2024 | CGMCC 3.2551 T | China: Guizhou | soil | OR680543 | OR680607 | OR858937 | [22] |
| Marquandomyces tashkentensis X.C. Wang, L.Y. Peng, Gafforov & W.Y. Zhuang, sp. nov. | UZ13-25 = CGMCC 3.28568 T | Uzbekistan: Tashkent | soil | PQ484189 | PQ484203 | PQ469020 | This study |
| Marquandomyces tianshanicus X.C. Wang, L.Y. Peng, Gafforov & W.Y. Zhuang, sp. nov. | UZ14-25 = CGMCC 3.28569 T | Uzbekistan: Tashkent | soil | PQ484190 | PQ484204 | PQ469021 | This study |
| UZ14-41 | Uzbekistan: Tashkent | soil | PQ484191 | PQ484205 | PQ469022 | This study | |
| Marquandomyces uzbekistanicus X.C. Wang, L.Y. Peng, Gafforov & W.Y. Zhuang, sp. nov. | UZ11-45 = CGMCC 3.28570 T | Uzbekistan: Tashkent | soil | PQ484192 | PQ484206 | PQ469023 | This study |
| Marquandomyces yaoyijianii X.C. Wang, L.Y. Peng & W.Y. Zhuang, sp. nov. | HLJ55-10 = CGMCC 3.28571 T | China: Heilongjiang | soil | PQ484193 | PQ484207 | PQ469024 | This study |
| UZ11-48 | Uzbekistan: Tashkent | soil | PQ484194 | PQ484208 | PQ469025 | This study | |
| Marquandomyces sp. 1 | CBS 127132 | USA: Nebraska | soil | MT078882 | MT078857 | MT078849 | [1] |
| Marquandomyces sp. 2 | CBS 129413 | USA: Wisconsin | soil | MT561567 | MT078859 | MT078851 | [1] |
| Purpureomyces khaoyaiensis (Hywel-Jones) Luangsa-ard, Samson & Thanakitp. 2020 | BCC1376 | Thailand | Lepidoptera larva | KX983460 | KX983462 | KX983457 | [19] |
| Purpureomyces maesotensis Luangsa-ard, Noisrip. Thanakitp. & Samson 2020 | BCC89300 T | Thailand | Lepidoptera larva | MN781917 | MN781876 | MN781733 | [1] |
| Purpureomyces pyriformis Luangsa-ard, Noisrip., Himaman, Mongkols. & Thanakitp. & Samson 2020 | BCC85074 T | Thailand | Lepidoptera larva | MN781929 | MN781873 | MN781730 | [1] |
| Dataset | No. of Seq. | Length of Alignment (bp) | No. of Variable Sites | No. of Parsimony-Informative Sites | Model for BI |
|---|---|---|---|---|---|
| ITS | 17 | 609 | 185 | 141 | |
| LSU | 17 | 845 | 52 | 43 | |
| TEF | 17 | 394 | 42 | 27 | |
| ITS + LSU + TEF | 17 | 1848 | 279 | 211 | GTR + I + G |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, L.-Y.; Wang, Y.-F.; Song, H.; Urinboev, I.; Zhuang, W.-Y.; Gafforov, Y.; Wang, X.-C. Five New Species of Marquandomyces (Clavicipitaceae, Ascomycota) from Asia. J. Fungi 2025, 11, 180. https://doi.org/10.3390/jof11030180
Peng L-Y, Wang Y-F, Song H, Urinboev I, Zhuang W-Y, Gafforov Y, Wang X-C. Five New Species of Marquandomyces (Clavicipitaceae, Ascomycota) from Asia. Journal of Fungi. 2025; 11(3):180. https://doi.org/10.3390/jof11030180
Chicago/Turabian StylePeng, Lu-Yao, Yi-Fan Wang, He Song, Islomjon Urinboev, Wen-Ying Zhuang, Yusufjon Gafforov, and Xin-Cun Wang. 2025. "Five New Species of Marquandomyces (Clavicipitaceae, Ascomycota) from Asia" Journal of Fungi 11, no. 3: 180. https://doi.org/10.3390/jof11030180
APA StylePeng, L.-Y., Wang, Y.-F., Song, H., Urinboev, I., Zhuang, W.-Y., Gafforov, Y., & Wang, X.-C. (2025). Five New Species of Marquandomyces (Clavicipitaceae, Ascomycota) from Asia. Journal of Fungi, 11(3), 180. https://doi.org/10.3390/jof11030180









