Clinical Stratification of High-Grade Ovarian Serous Carcinoma Using a Panel of Six Biomarkers
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Animal Studies
2.3. Immunohistochemical (IHC) and Histochemical Staining (HC)
2.4. Statistical Analysis
3. Results
3.1. Selection of Class-Specific Biomarkers, Development of SOPs for Detection, a Reference Human Tissue Library and Guidelines for Scoring
- (i)
- Score for Marker Frequency (SFreq)-percentage expression in total tumor cells of tissue section on a scale of 0–3 (0: absent, 1: 1–10%, 2: 11–50%, and 3: ≥51% marker-positive),
- TCF21: cardiac myocytes, ovarian stromal cells, and germinal cells of testis represented SFreq 0, 1, and 3 respectively; SFreq = 2 could not be identified in healthy tissues.
- E-cadherin: cardiac myocytes, liver hepatocytes, and prostate epithelial cells represented SFreq 0, 2, and 3 respectively; healthy tissues representing SFreq = 1 could not be identified.
- PARP1: mucosa of the small intestine, cardiac myocytes, germinal basal cells of testis represented SFreq as 0, 1, and 3 respectively; healthy tissues representing SFreq = 2 could not be identified.
- Slug: cardiac myocytes, smooth muscles of the appendix, lymphocytes of the small intestine represented SFreq 0, 1, and 2 respectively; healthy tissues representing SFreq = 3 could not be identified.
- HA: cartilage and sub-mucosa of the small intestine represented SFreq as 2 and 3 respectively; healthy tissues representing SFreq = 0 or 1 could not be identified.
- ANXA2: cardiac myocytes, the somatic muscle of the small intestine, epithelial cells of the gall bladder represented SFreq 0, 1, and 3 respectively; healthy tissues representing SFreq = 2 could not be identified.
- (ii)
- Score for marker intensity (SInt)-intensity of brown stain for IHC and blue for HC in positively stained tissue sections. A scale of 0–3 was established, 0: absent, 1: weak, 2: moderate, and 3: strong intensity of marker-positive cells,
- TCF21: cardiac myocytes, ovarian stromal cells, germinal basal cells of testis represented SInt 0, 1, and 2 respectively; SInt = 3 could not be identified in healthy tissues.
- E-cadherin: cardiac myocytes, epithelial cells of the small intestine, epithelial cells of prostate represented SInt 0, 2, and 3 respectively; healthy tissues representing SInt = 1 could not be identified.
- PARP1: mucosa of the small intestine, cardiac myocytes, and germinal basal cells of testis represented SInt 0, 1, and 2 respectively; healthy tissues representing SFreq = 3 could not be identified.
- Slug: cardiac myocytes, smooth muscle of the appendix, and lymphocytes of the small intestine represented SInt 0, 1, and 2 respectively; healthy tissues representing SInt = 3 could not be identified.
- HA: Intensity for hyaluronan was measured as blue color intensity developed by Alcian blue in comparison to hyaluronidase digested tissue section. Sub-mucosa of the small intestine and cartilage tissues represented SInt 1 and 2 respectively; healthy tissues representing SInt = 0 or 3 could not be identified.
- ANXA2: cardiac myocytes and epithelial cells of gall bladder represented SInt 0 and 2 respectively; healthy tissues representing SInt = 1 or 3 could not be identified.
- (iii)
- Score for Marker Localization (SLoc)-representing sub-cellular location of marker in the tissue section on a scale of 0–2, 0: Absent, 1: mislocalized (cellular localization does not correspond to known functionality, for example, cytoplasmic location for TCF21, PARP1, Slug, E-cadherin, ANXA2 or HA), 2: normal localization (for example, nuclear expression of TCF21, PARP1 or Slug, membrane for E-cadherin, membrane or cytoplasmic for ANXA2 and extracellular expression of HA.
- TCF21: cardiac myocytes, liver hepatocytes, germinal basal cells of testis represented SLoc 0, 1, and 2 respectively.
- E-cadherin: cardiac myocytes, prostate epithelial cells represented SLoc 0 and 2 respectively; healthy tissues representing SLoc = 1 could not be identified.
- PARP1: mucosa of the small intestine, germinal basal cells of testis represented SLoc 0 and 2 respectively; healthy tissues representing SLoc = 1 could not be identified.
- Slug: cardiac myocytes, the somatic muscle of the appendix, lymphocytes of the small intestine, represented SLoc 0, 1, and 2 respectively.
- HA: cartilage represented SLoc of score 2; healthy tissues representing SLoc = 1 could not be identified. A further consensus was reached in the pathology review to consider extracellular staining in tumor nests that is eliminated following hyaluronidase treatment as a proper localization, while distant stroma-associated HA was considered as mislocalization.
- ANXA2: cardiac myocytes, stromal cells of the gall bladder, epithelial cells of the gall bladder represented SLoc as 0, 1, and 2 respectively.
3.2. Establishment of Scoring Guidelines for Stratification Using a Panel of Xenograft
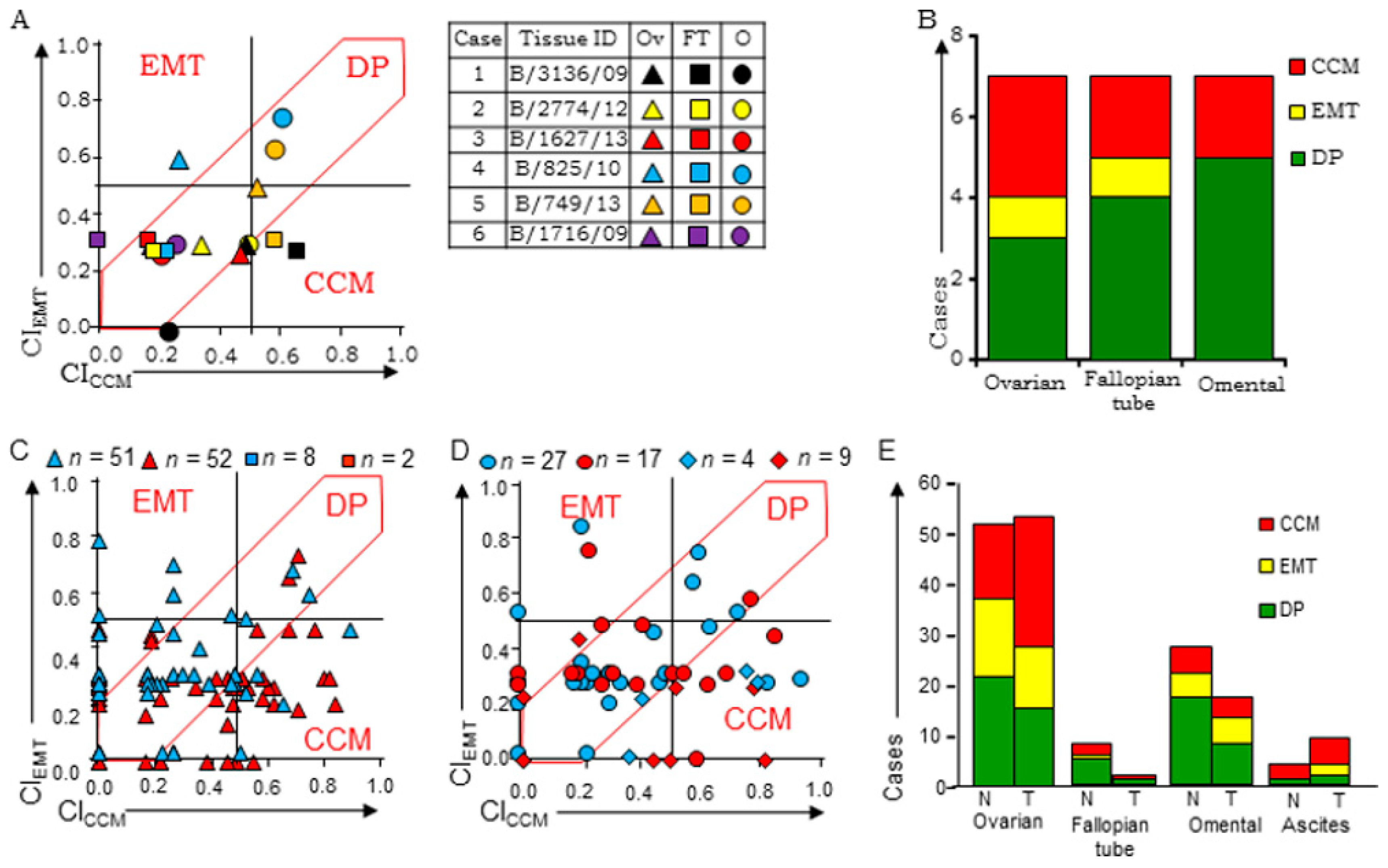
3.3. Evaluation of Stratification Guidelines in TMAs
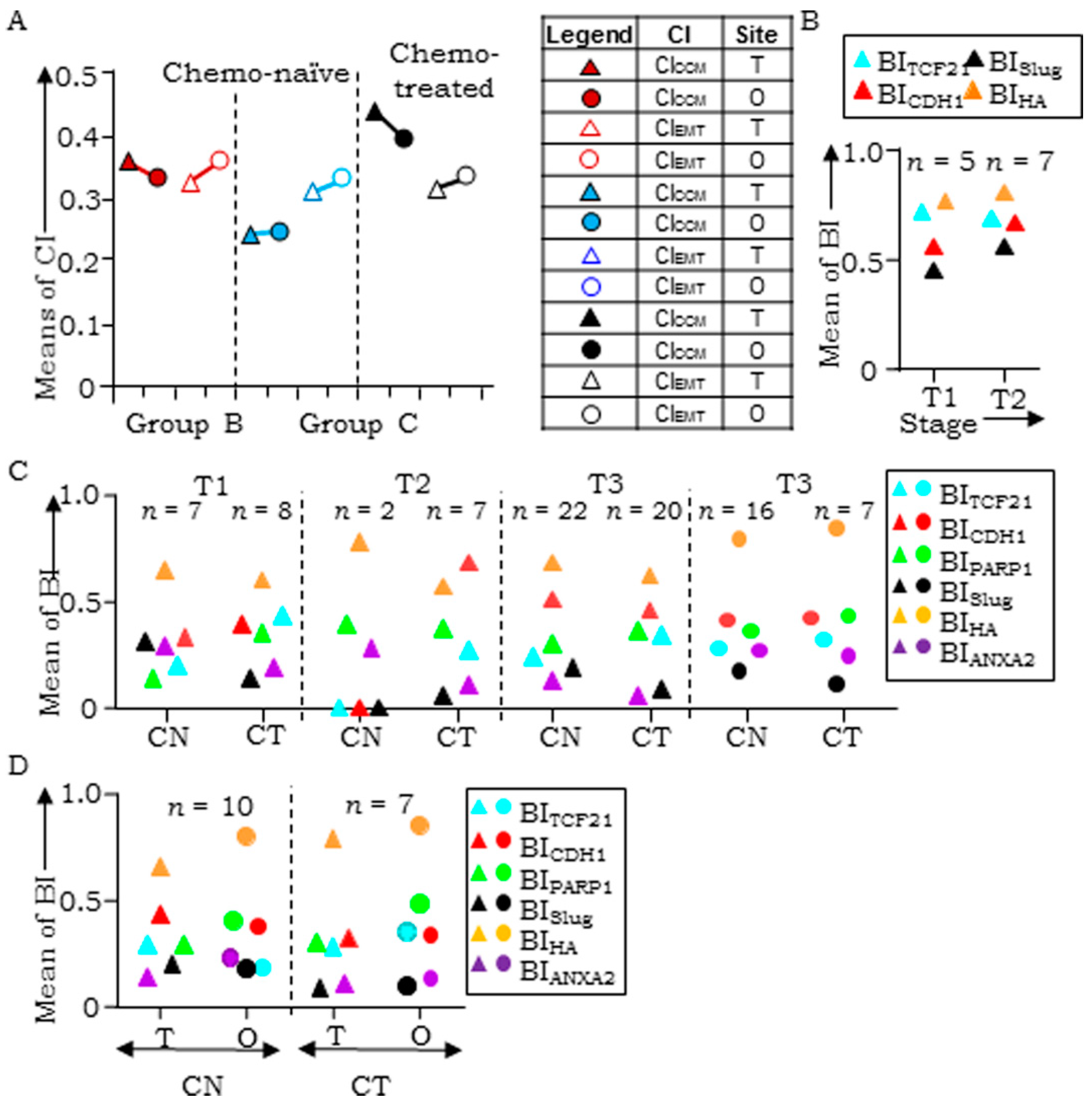
3.4. Evaluation of Clinical Samples Associates CCM-Markers with Metastases and Chemotherapy
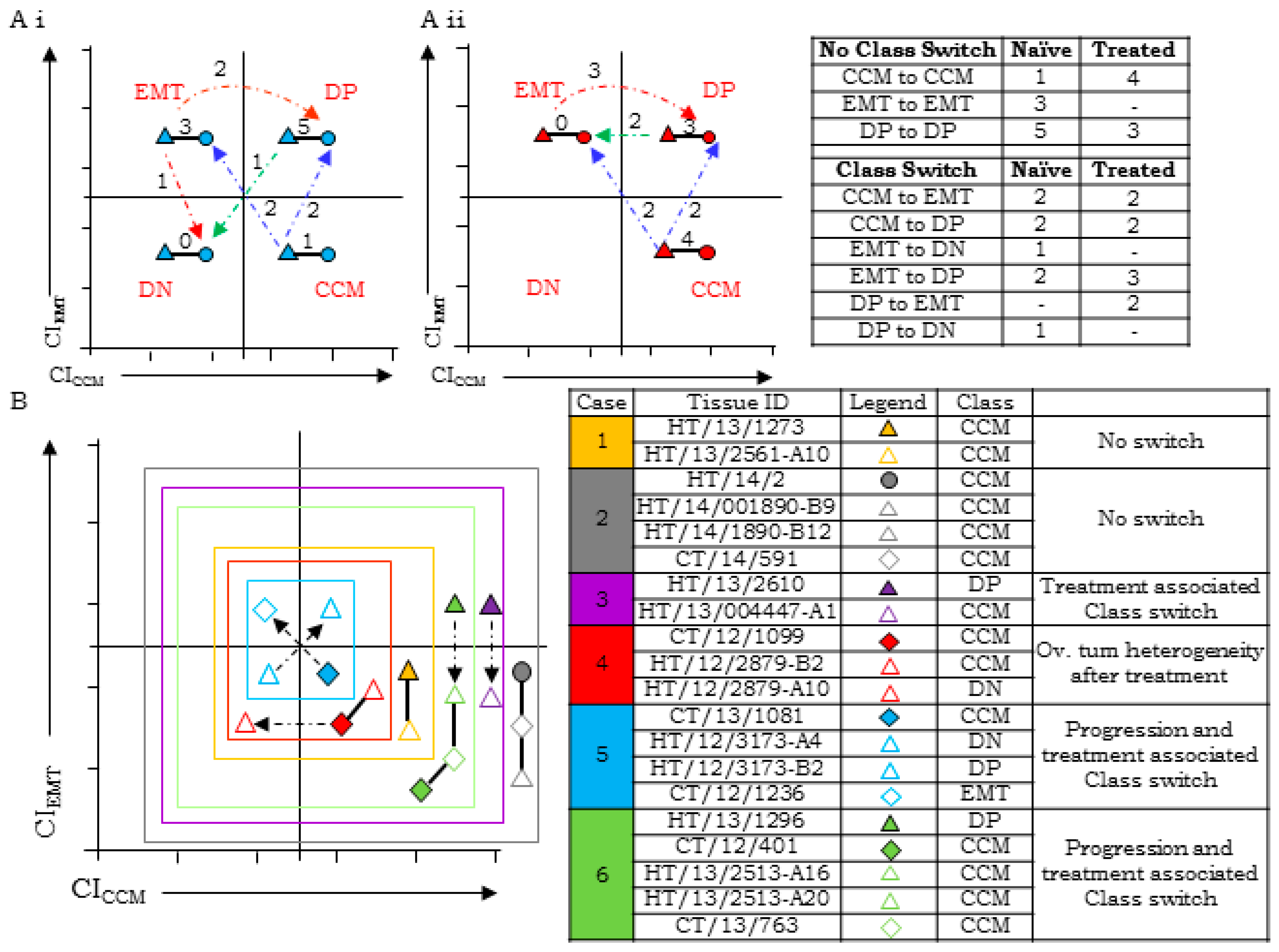
3.5. HGSC Tumors at Different Sites Exhibit Molecular Heterogeneity and Class-Switching
3.6. Disease Progression is Inclined Towards Enrichment of CCM-Markers
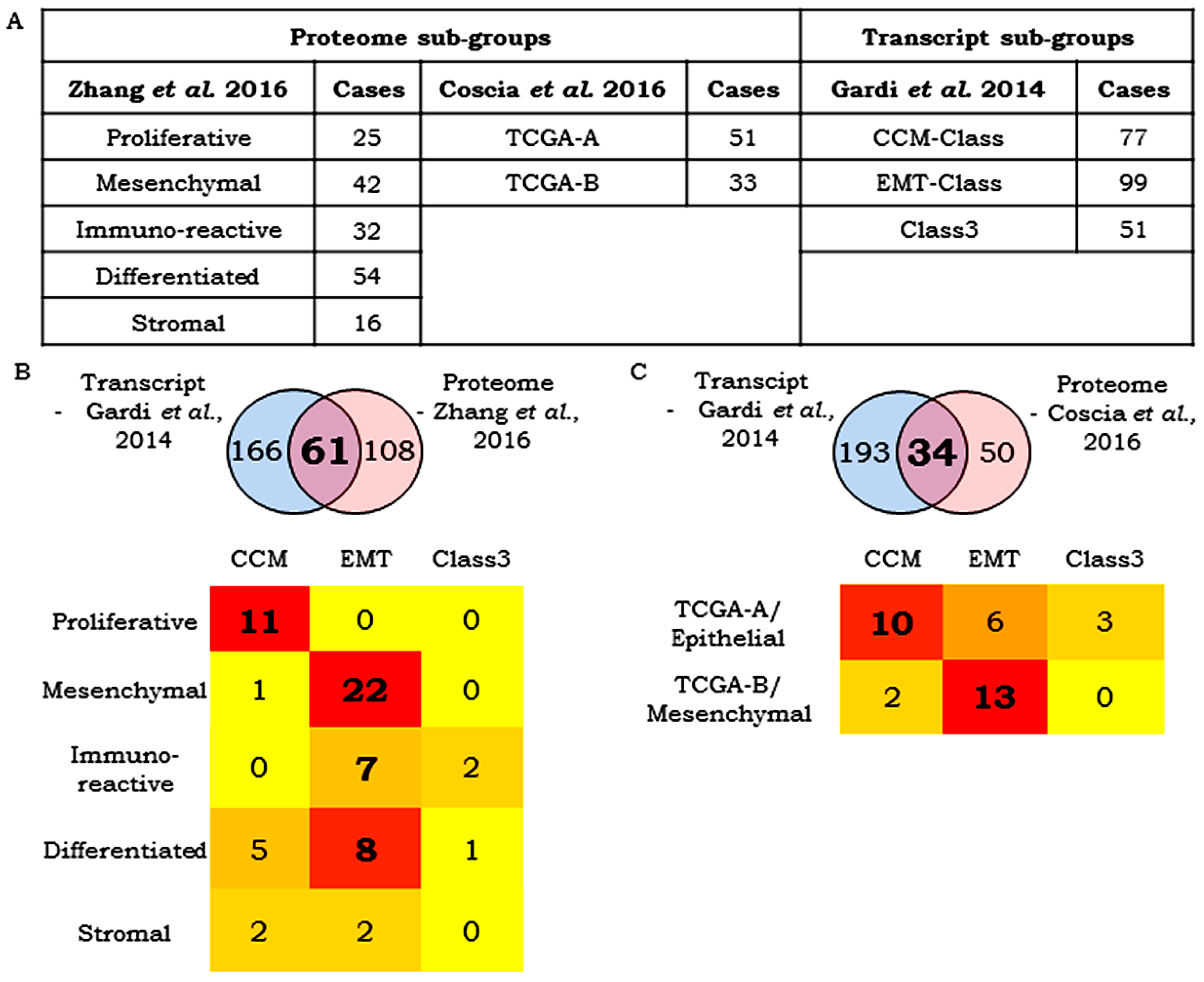
3.7. Correlation Between Transcript- and Protein-Based Stratification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weigelt, B.; Reis-Filho, J.S.; Swanton, C. Genomic analyses to select patients for adjuvant chemotherapy: Trials and tribulations. Ann. Oncol. 2012, 23, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Gorgun, G.; Calabrese, E.; Hideshima, T.; Ecsedy, J.; Perrone, G.; Mani, M.; Ikeda, H.; Bianchi, G.; Hu, Y.; Cirstea, D.; et al. A novel aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell cycle arrest in multiple experimental multiple myeloma model. Blood 2010, 115, 5202–5213. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, M.G.; Ecsedy, J.A.; Chakravarty, A.; Silverman, L.; Zhang, M.; Hoar, K.M.; Stroud, S.G.; Chen, W.; Shinde, V.; Huck, J.J.; et al. Characterization of alisertib (MLN8237), an investigational small-molecule inhibitor of Aurora A kinase using novel in vivo pharmacodynamic assays. Clin. Cancer Res. 2011, 17, 7614–7624. [Google Scholar] [CrossRef] [PubMed]
- Cidon, E.U.; Ellis, S.G.; Inam, Y.; Adeleke, S.; Zarif, S.; Geldart, T. Molecular targeted agents for gastric cancer: A step forward towards personalized therapy. Cancers 2013, 5, 64–91. [Google Scholar] [CrossRef] [PubMed]
- Muro, K.; Chung, H.C.; Shankaran, V.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Golan, T.; Le, D.T.; Burtness, B.; et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): A multicentre, open-label, phase 1b trial. Lancet Oncol. 2016, 17, 717–726. [Google Scholar] [CrossRef]
- Magaki, S.; Hojat, S.A.; Wei, B.; So, A.; Yong, W.H. An Introduction to the Performance of Immunohistochemistry. In Biobanking. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; pp. 289–298. [Google Scholar]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Díaz del Arco, C.; Estrada Muñoz, L.; Molina Roldán, E.; Cerón Nieto, M.Á.; Ortega Medina, L.; García Gómez de las Heras, S.; Fernández Aceñero, M.J. Immunohistochemical classification of gastric cancer based on new molecular biomarkers: A potential predictor of survival. Virchows Arch. 2018, 473, 687–695. [Google Scholar] [CrossRef]
- Kim, J.; Kong, J.K.; Yang, W.; Cho, H.; Chay, D.B.; Lee, B.H.; Cho, S.J.; Hong, S.; Kim, J.-H. DNA Mismatch Repair Protein Immunohistochemistry and MLH1 Promotor Methylation Testing for Practical Molecular Classification and the Prediction of Prognosis in Endometrial Cancer. Cancers 2018, 10, 279. [Google Scholar] [CrossRef]
- Karnezis, A.N.; Leung, S.; Magrill, J.; McConechy, M.K.; Yang, W.; Chow, C.; Kobel, M.; Lee, C.-H.; Huntsman, D.G.; Talhouk, A.; et al. Evaluation of endometrial carcinoma prognostic immunohistochemistry markers in the context of molecular classification. J. Pathol. Clin. Res. 2017, 3, 279–293. [Google Scholar] [CrossRef]
- Ten Hoorn, S.; Trinh, A.; de Jong, J.; Koens, L.; Vermeulen, L. Classification of Colorectal Cancer in Molecular Subtypes by Immunohistochemistry. In Methods in molecular biology (Clifton, N.J.); Beaulieu, J.-F., Ed.; Humana Press: New York, NY, USA, 2018. [Google Scholar]
- Kim, S.; Moon, B.-I.; Lim, W.; Park, S.; Cho, M.S.; Sung, S.H. Feasibility of Classification of Triple Negative Breast Cancer by Immunohistochemical Surrogate Markers. Clin. Breast Cancer 2018, 18, e1123–e1132. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Wu, Y.; Ma, K.; Fan, Y.; Dong, D.; Geng, N.; Li, E. Molecular classification of esophagogastric junction carcinoma correlated with prognosis. Onco. Targets. Ther. 2017, 10, 4765–4772. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Beaulande, M.; Hamila, M.; Erb, G.; Hofman, V.; Hofman, P. Automated chromogenic multiplexed immunohistochemistry assay for diagnosis and predictive biomarker testing in non-small cell lung cancer. Lung Cancer 2018, 124, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA. Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C.; Hennessy, B.; Mills, G.B. The biology of ovarian cancer: New opportunities for translation. Nat. Rev. Cancer 2009, 9, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Carden, C.P.; Kaye, S.B. Beyond chemotherapy: Targeted therapies in ovarian cancer. Nat. Rev. Cancer 2009, 9, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel Molecular Subtypes of Serous and Endometrioid Ovarian Cancer Linked to Clinical Outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- TCGA Network; Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; DiSaia, P.; Gabra, H.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Yang, D.; Sun, Y.; Hu, L.; Zheng, H.; Ji, P.; Pecot, C.V.; Zhao, Y.; Reynolds, S.; Cheng, H.; Rupaimoole, R.; et al. Integrated Analyses Identify a Master MicroRNA Regulatory Network for the Mesenchymal Subtype in Serous Ovarian Cancer. Cancer Cell 2013, 23, 186–199. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.M.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef]
- Kanchi, K.L.; Johnson, K.J.; Lu, C.; McLellan, M.D.; Leiserson, M.D.M.; Wendl, M.C.; Zhang, Q.; Koboldt, D.C.; Xie, M.; Kandoth, C.; et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat. Commun. 2014, 5, 3156. [Google Scholar] [CrossRef] [PubMed]
- Gardi, N.L.; Deshpande, T.U.; Kamble, S.C.; Budhe, S.R.; Bapat, S.A. Discrete molecular classes of ovarian cancer suggestive of unique mechanisms of transformation and metastases. Clin. Cancer Res. 2014, 20, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, T.; Zhang, Z.; Chan, D.W.; Rodland, K.D.; Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Coscia, F.; Watters, K.M.; Curtis, M.; Eckert, M.A.; Chiang, C.Y.; Tyanova, S.; Montag, A.; Lastra, R.R.; Lengyel, E.; Mann, M. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat. Commun. 2016, 7, 12645. [Google Scholar] [CrossRef] [PubMed]
- Winget, M.D.; Baron, J.A.; Spitz, M.R.; Brenner, D.E.; Warzel, D.; Kincaid, H.; Thornquist, M.; Feng, Z. Development of common data elements: The experience of and recommendations from the early detection research network. Int. J. Med. Inform. 2003, 70, 41–48. [Google Scholar] [CrossRef]
- Shariat, S.; Lotan, Y.; Vickers, A.; Karakiewicz, P.; Schmitz-Dräger, B.; Goebell, P.; Malats, N. Statistical consideration for clinical biomarker research in bladder cancer. Urol. Oncol. 2010, 28, 389–400. [Google Scholar] [CrossRef]
- Kothe, S.; Muller, J.; Bohmer, S.-A.; Tschongov, T.; Fricke, M.; Koch, S.; Thiede, C.; Requardt, R.P.; Rubio, I.; Bohmer, F.D. Features of Ras activation by a mislocalized oncogenic tyrosine kinase: FLT3 ITD signals through K-Ras at the plasma membrane of acute myeloid leukemia cells. J. Cell Sci. 2013, 126, 4746–4755. [Google Scholar] [CrossRef]
- Barham, W.; Chen, L.; Tikhomirov, O.; Onishko, H.; Gleaves, L.; Stricker, T.P.; Blackwell, T.S.; Yull, F.E. Aberrant activation of NF-κB signaling in mammary epithelium leads to abnormal growth and ductal carcinoma in situ. BMC Cancer 2015, 1–17. [Google Scholar] [CrossRef]
- Guttilla, I.K.; Phoenix, K.N.; Hong, X.; Tirnauer, J.S.; Claffey, K.P.; White, B.A. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res. Treat. 2012, 132, 75–85. [Google Scholar] [CrossRef]
- Sidney, L.E.; McIntosh, O.D.; Hopkinson, A. Phenotypic change and induction of cytokeratin expression during in vitro culture of corneal stromal cells. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7225–7235. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.Y.; Antony, J.; Tan, T.Z.; Tan, D.S. Targeting the AXL signaling pathway in ovarian cancer. Mol. Cell Oncol. 2016, 4, e1263716. [Google Scholar] [CrossRef] [PubMed]
- Smigiel, J.M.; Parameswaran, N.; Jackson, M.W. Targeting Pancreatic Cancer Cell Plasticity: The Latest in Therapeutics. Cancers 2018, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Banyard, J.; Chung, I.; Wilson, A.M.; Vetter, G.; Le Béchec, A.; Bielenberg, D.R.; Zetter, B.R. Regulation of epithelial plasticity by miR-424 and miR-200 in a new prostate cancer metastasis model. Sci. Rep. 2013, 3, 3151. [Google Scholar] [CrossRef] [PubMed]
- Quagliata, L.; Schlageter, M.; Quintavalle, C.; Tornillo, L.; Terracciano, L. Identification of New Players in Hepatocarcinogenesis: Limits and Opportunities of Using Tissue Microarray (TMA). Microarrays 2014, 3, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Ku, M.; Kang, M.; Suh, J.; Yang, J. Effects for Sequential Treatment of siAkt and Paclitaxel on Gastric Cancer Cell Lines. Int. J. Med. Sci. 2016, 13, 708–716. [Google Scholar] [CrossRef]
- Su, H.; Lin, F.; Deng, X.; Shen, L.; Fang, Y.; Fei, Z.; Zhao, L.; Zhang, X.; Pan, H.; Xie, D.; et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J. Transl. Med. 2016, 14, 225. [Google Scholar] [CrossRef]
- Litviakov, N.V.; Cherdyntseva, N.V.; Tsyganov, M.M.; Slonimskaya, E.M.; Ibragimova, M.K.; Kazantseva, P.V.; Kzhyshkowska, J.; Choinzonov, E.L. Deletions of multidrug resistance gene loci in breast cancer leads to the down-regulation of its expression and predict tumor response to neoadjuvant chemotherapy. Oncotarget 2016, 5, 7829–7841. [Google Scholar] [CrossRef]
- Negrei, C.; Hudita, A.; Ginghina, O.; Galateanu, B.; Voicu, S.N.; Stan, M.; Costache, M.; Fenga, C.; Drakoulis, N.; Tsatsakis, A.M. Colon Cancer Cells Gene Expression Signature As Response to 5-Fluorouracil, Oxaliplatin, and Folinic Acid Treatment. Front. Pharmacol. 2016, 7, 172. [Google Scholar] [CrossRef]
- Bjarnadottir, O.; Kimbung, S.; Johansson, I.; Veerla, S.; Jönsson, M.; Bendahl, P.-O.; Grabau, D.; Hedenfalk, I.; Borgquist, S. Global Transcriptional Changes Following Statin Treatment in Breast Cancer. Clin. Cancer Res. 2015, 21, 3402–3411. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.W.; Hotte, S.J.; Kollmannsberger, C.K.; Renouf, D.J.; Cescon, D.W.; Hedley, D.; Chow, S.; Moscow, J.; Chen, Z.; Perry, M.; et al. A phase I trial of ANG1/2-Tie2 inhibitor trebaninib (AMG386) and temsirolimus in advanced solid tumors (PJC008/NCI#9041). Invest. New Drugs 2016, 34, 104–111. [Google Scholar] [PubMed]
- Kim, G.; Ison, G.; McKee, A.E.; Zhang, H.; Tang, S.; Gwise, T.; Sridhara, R.; Lee, E.; Tzou, A.; Philip, R.; et al. FDA Approval Summary: Olaparib Monotherapy in Patients with Deleterious Germline BRCA-Mutated Advanced Ovarian Cancer Treated with Three or More Lines of Chemotherapy. Clin. Cancer Res. 2015, 21, 4257–4261. [Google Scholar] [CrossRef] [PubMed]
- AlHilli, M.M.; Becker, M.A.; Weroha, S.J.; Flatten, K.S.; Hurley, R.M.; Harrell, M.I.; Oberg, A.L.; Maurer, M.J.; Hawthorne, K.M.; Hou, X.; et al. In vivo anti-tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol. Oncol. 2016, 143, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Caiping, S.; Qing, Z.; Xiaojing, W. Sonic hedgehog-Gli1 signals promote epithelial-mesenchymal transition in ovarian cancer by mediating PI3K/AKT pathway. Med. Oncol. 2015, 32, 368. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.H.; García-García, C.; Serra, V.; He, L.; Torres-Lockhart, K.; Prat, A.; Anton, P.; Cozar, P.; Guzmán, M.; Grueso, J.; et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012, 2, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Juvekar, A.; Burga, L.N.; Hu, H.; Lunsford, E.P.; Ibrahim, Y.H.; Balmañà, J.; Rajendran, A.; Papa, A.; Spencer, K.; Lyssiotis, C.A.; et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012, 2, 1048–1063. [Google Scholar] [CrossRef] [PubMed]
- González-Billalabeitia, E.; Seitzer, N.; Song, S.J.; Song, M.S.; Patnaik, A.; Liu, X.-S.; Epping, M.T.; Papa, A.; Hobbs, R.M.; Chen, M.; et al. Vulnerabilities of PTEN-TP53-deficient prostate cancers to compound PARP-PI3K inhibition. Cancer Discov. 2014, 4, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, C.; Zhang, Y.; Wang, M.; Jiang, N.; Xiang, L.; Li, T.; Roberts, T.M.; Zhao, J.J.; Cheng, H.; et al. Combined inhibition of PI3K and PARP is effective in the treatment of ovarian cancer cells with wild-type PIK3CA genes. Gynecol. Oncol. 2016, 142, 548–556. [Google Scholar] [CrossRef]
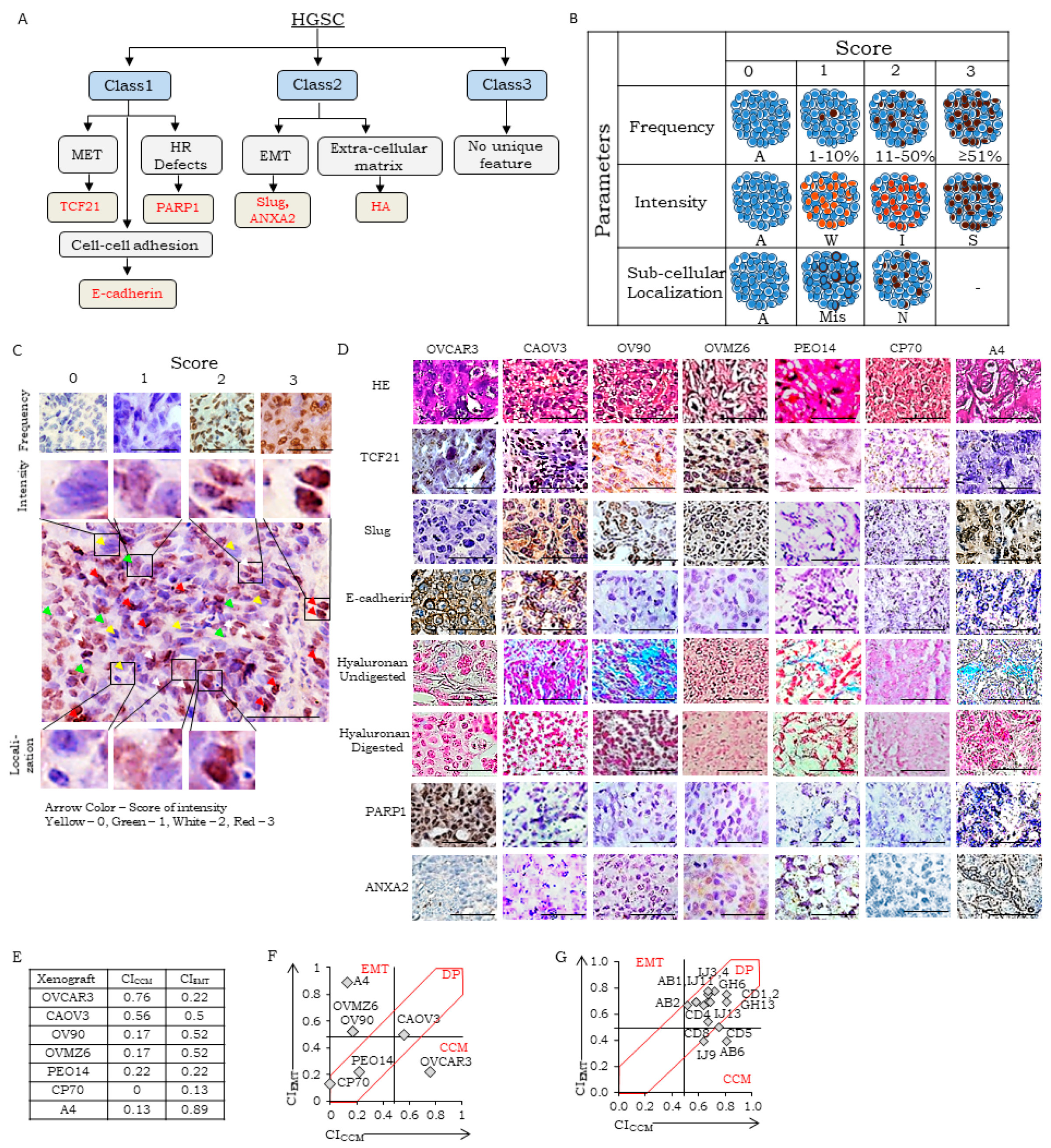
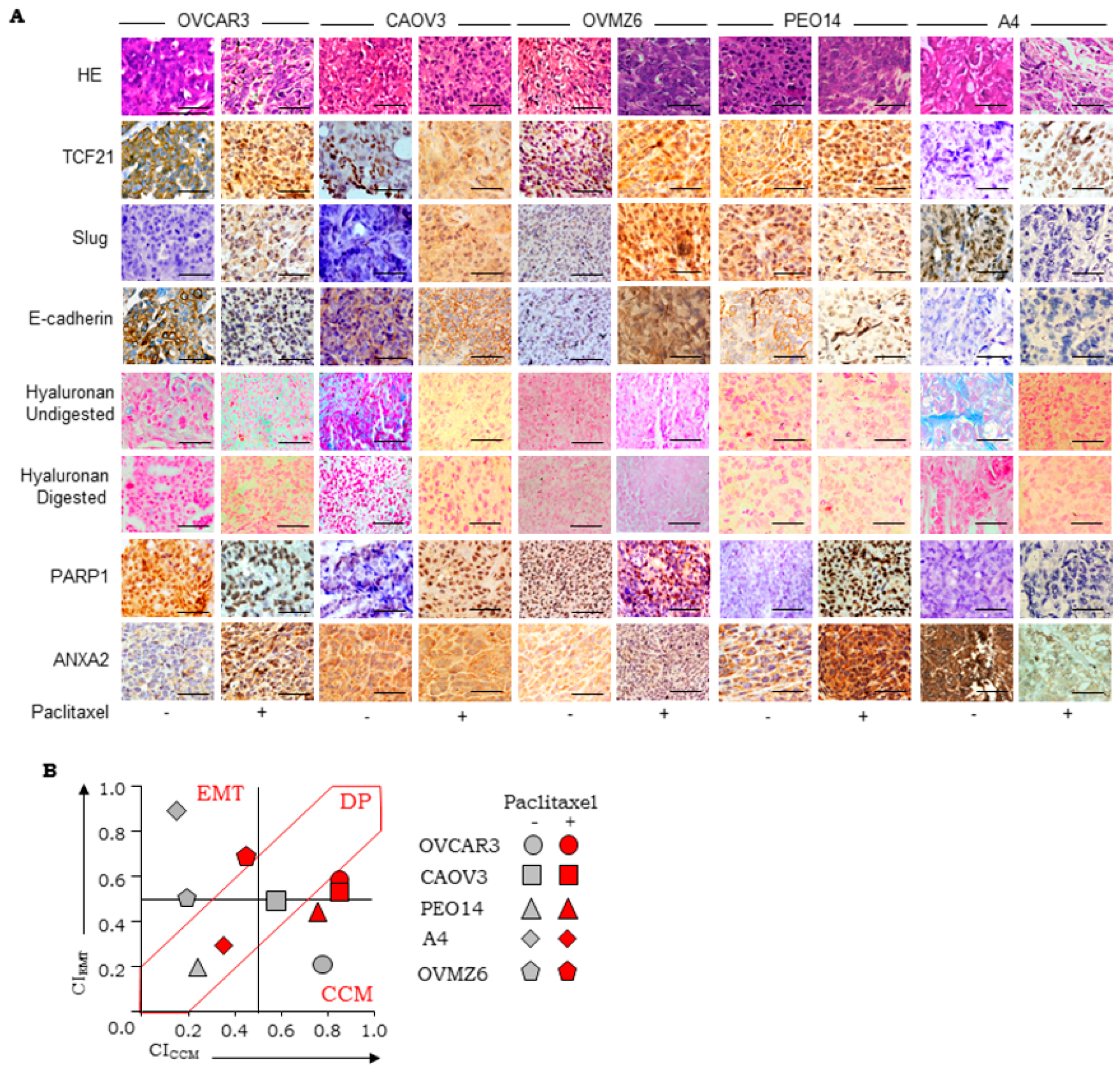
| CCM Markers | |||||||||||||
| Cell Line Derived Xenograft | TCF21 | E-cadherin | PARP1 | CICCM | |||||||||
| SFreq | SInt | SLoc | BITCF21 | SFreq | SInt | SLoc | BICDH1 | SFreq | SInt | SLoc | BIPARP1 | ||
| CAOV3 | 2 | 2 | 2 | 0.78 | 3 | 2 | 2 | 0.89 | 0 | 0 | 0 | 0 | 0.56 |
| OVMZ6 | 2 | 1 | 1 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.17 |
| CP70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OV90 | 2 | 1 | 1 | 0.5 | 1 | 2 | 1 | 0.5 | 0 | 0 | 0 | 0 | 0.17 |
| A4 | 1 | 1 | 1 | 0.39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 |
| OVCAR3 | 2 | 3 | 1 | 0.72 | 2 | 2 | 2 | 0.78 | 2 | 2 | 2 | 0.78 | 0.76 |
| PEO14 | 1 | 2 | 2 | 0.67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.22 |
| EMT Markers | |||||||||||||
| Cell Line Derived Xenograft | Slug | HA | ANXA2 | CIEMT | |||||||||
| SFreq | SInt | SLoc | BISlug | SFreq | SInt | SLoc | BIHA | SFreq | SInt | SLoc | BIAnxA2 | ||
| CAOV3 | 3 | 1 | 1 | 0.61 | 3 | 2 | 2 | 0.89 | 0 | 0 | 0 | 0 | 0.5 |
| OVMZ6 | 2 | 1 | 2 | 0.67 | 0 | 0 | 0 | 0 | 3 | 2 | 2 | 0.89 | 0.52 |
| CP70 | 1 | 1 | 1 | 0.39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 |
| OV90 | 1 | 1 | 2 | 0.56 | 3 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0.52 |
| A4 | 3 | 3 | 2 | 1 | 3 | 2 | 2 | 0.89 | 3 | 1 | 2 | 0.78 | 0.89 |
| OVCAR3 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 0.67 | 0 | 0 | 0 | 0 | 0.22 |
| PEO14 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 0.67 | 0 | 0 | 0 | 0 | 0.22 |
| Group | Analyses | Samples (n) |
|---|---|---|
| A | Between-group analyses of tumors in chemo-naïve tumors (T vs. FT vs. O) | 6 |
| B | Within the group of single tumors derived from either ovarian or FT sites, omental deposits or cell blocks from tumor ascites in chemo-naïve (CN) cases and chemo-treated (CT) cases | CN–51 (T), 8 (FT), 27 (O), 4 (A); CT–52 (T), 2 (FT), 17 (O), 2 (A) |
| C | Within groups of primary tumor & omental tumors pairs from either chemo-naïve (CN) or chemo-treated (CT) cases | CN–17; CT–16 |
| D | Between-group analyses of tumor samples of the same case before and after chemotherapy | 6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamble, S.C.; Sen, A.; Dhake, R.D.; Joshi, A.N.; Midha, D.; Bapat, S.A. Clinical Stratification of High-Grade Ovarian Serous Carcinoma Using a Panel of Six Biomarkers. J. Clin. Med. 2019, 8, 330. https://doi.org/10.3390/jcm8030330
Kamble SC, Sen A, Dhake RD, Joshi AN, Midha D, Bapat SA. Clinical Stratification of High-Grade Ovarian Serous Carcinoma Using a Panel of Six Biomarkers. Journal of Clinical Medicine. 2019; 8(3):330. https://doi.org/10.3390/jcm8030330
Chicago/Turabian StyleKamble, Swapnil C., Arijit Sen, Rahul D. Dhake, Aparna N. Joshi, Divya Midha, and Sharmila A. Bapat. 2019. "Clinical Stratification of High-Grade Ovarian Serous Carcinoma Using a Panel of Six Biomarkers" Journal of Clinical Medicine 8, no. 3: 330. https://doi.org/10.3390/jcm8030330
APA StyleKamble, S. C., Sen, A., Dhake, R. D., Joshi, A. N., Midha, D., & Bapat, S. A. (2019). Clinical Stratification of High-Grade Ovarian Serous Carcinoma Using a Panel of Six Biomarkers. Journal of Clinical Medicine, 8(3), 330. https://doi.org/10.3390/jcm8030330






