Uropathogenic Escherichia coli Associated with Risk of Urosepsis—Genetic, Proteomic, and Metabolomic Studies
Abstract
1. Introduction
2. Results
2.1. Distribution of Virulence Factors in the Control Group vs. the Urosepsis Group Isolates
2.2. Binary Classification Results for Urosepsis Diagnostics
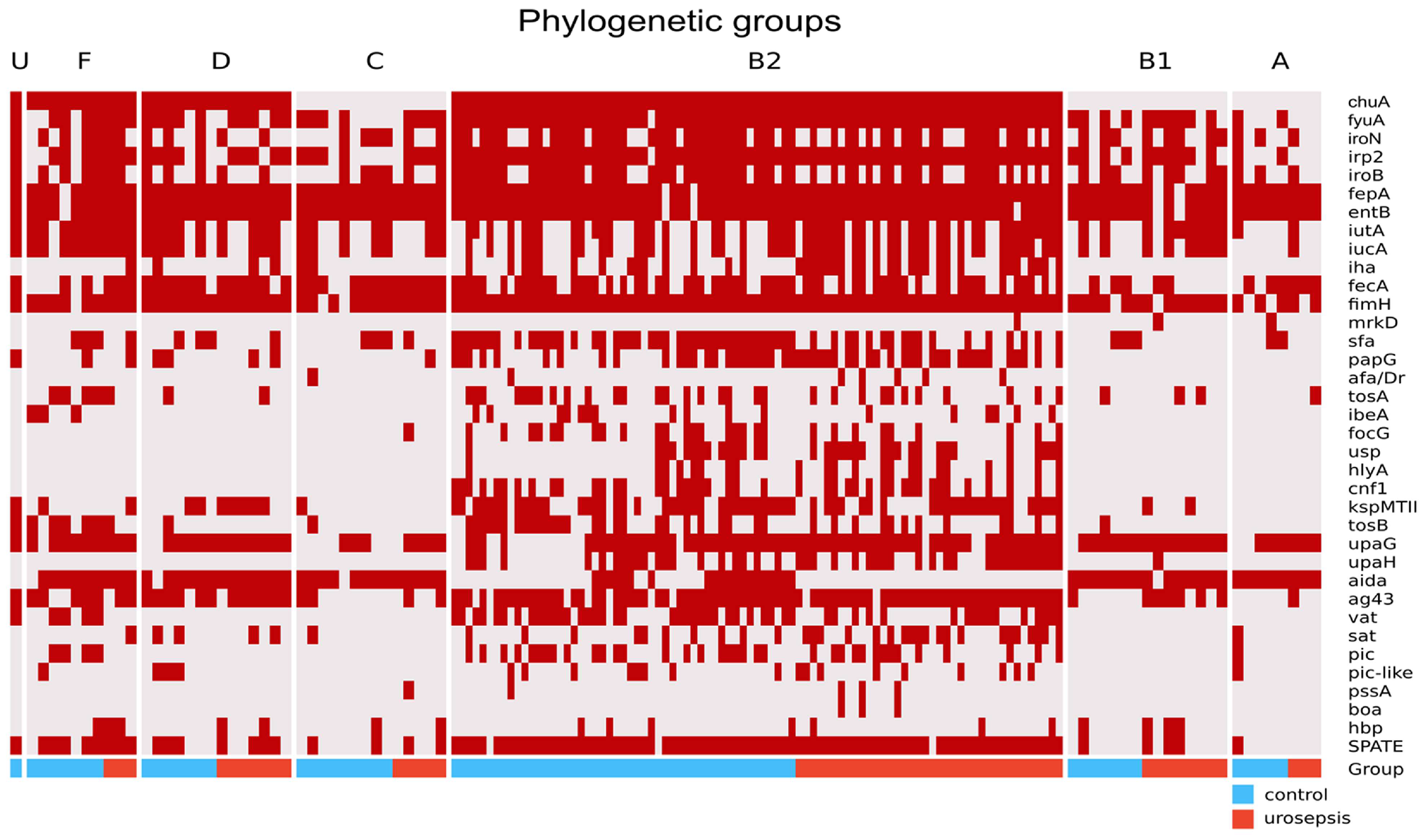
2.3. Phylogenetic Groups and VFs Concerning Urosepsis vs. Control Groups
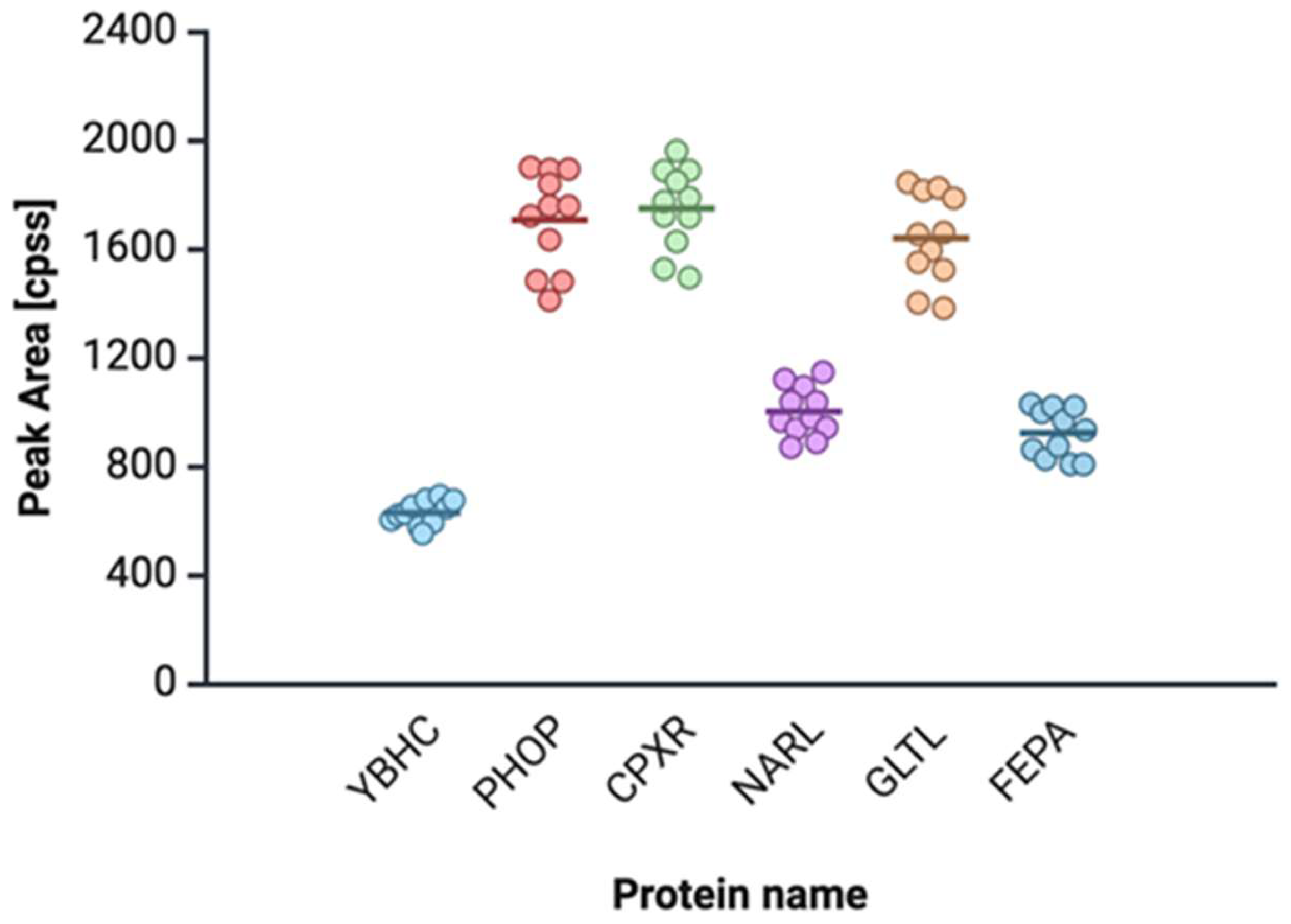
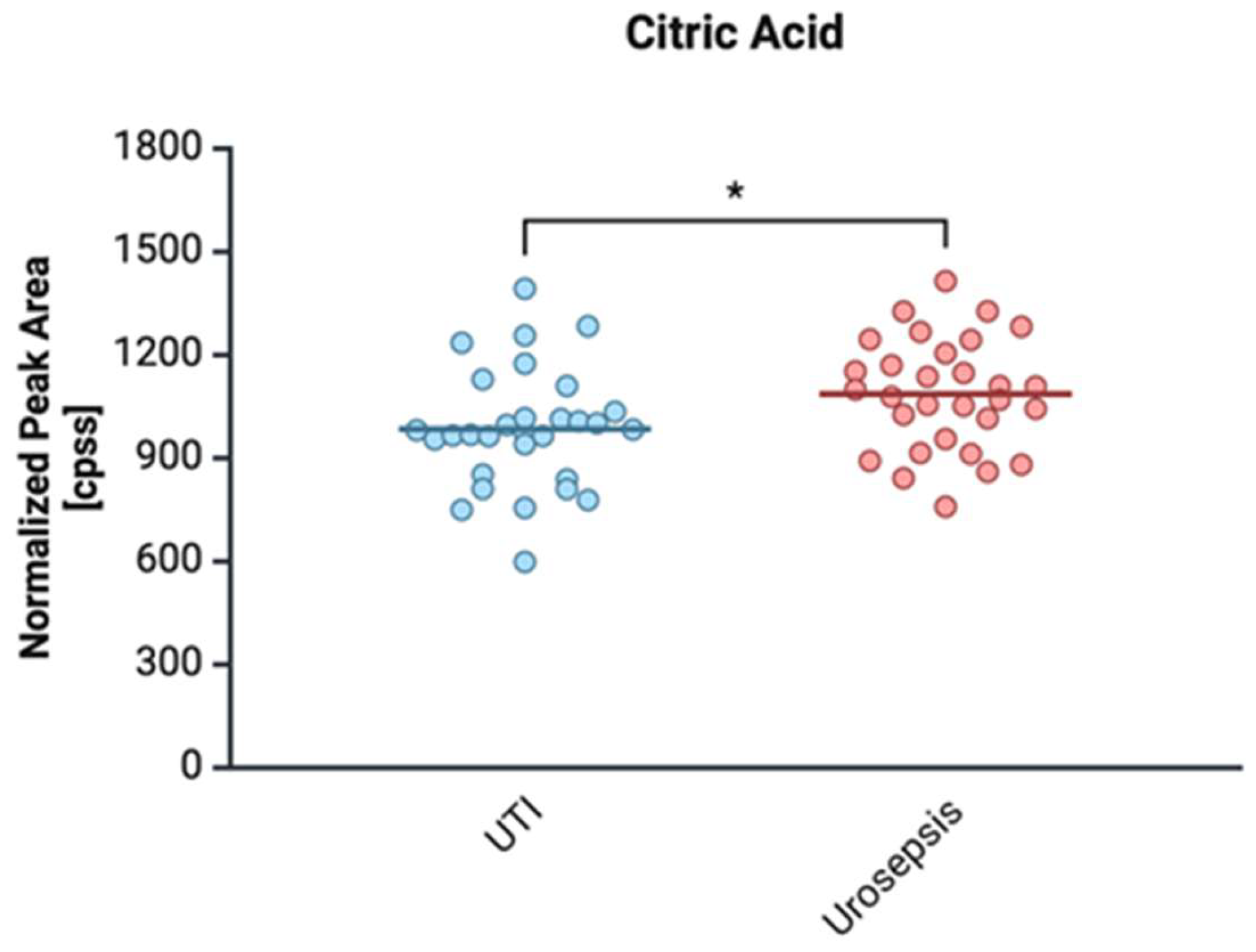
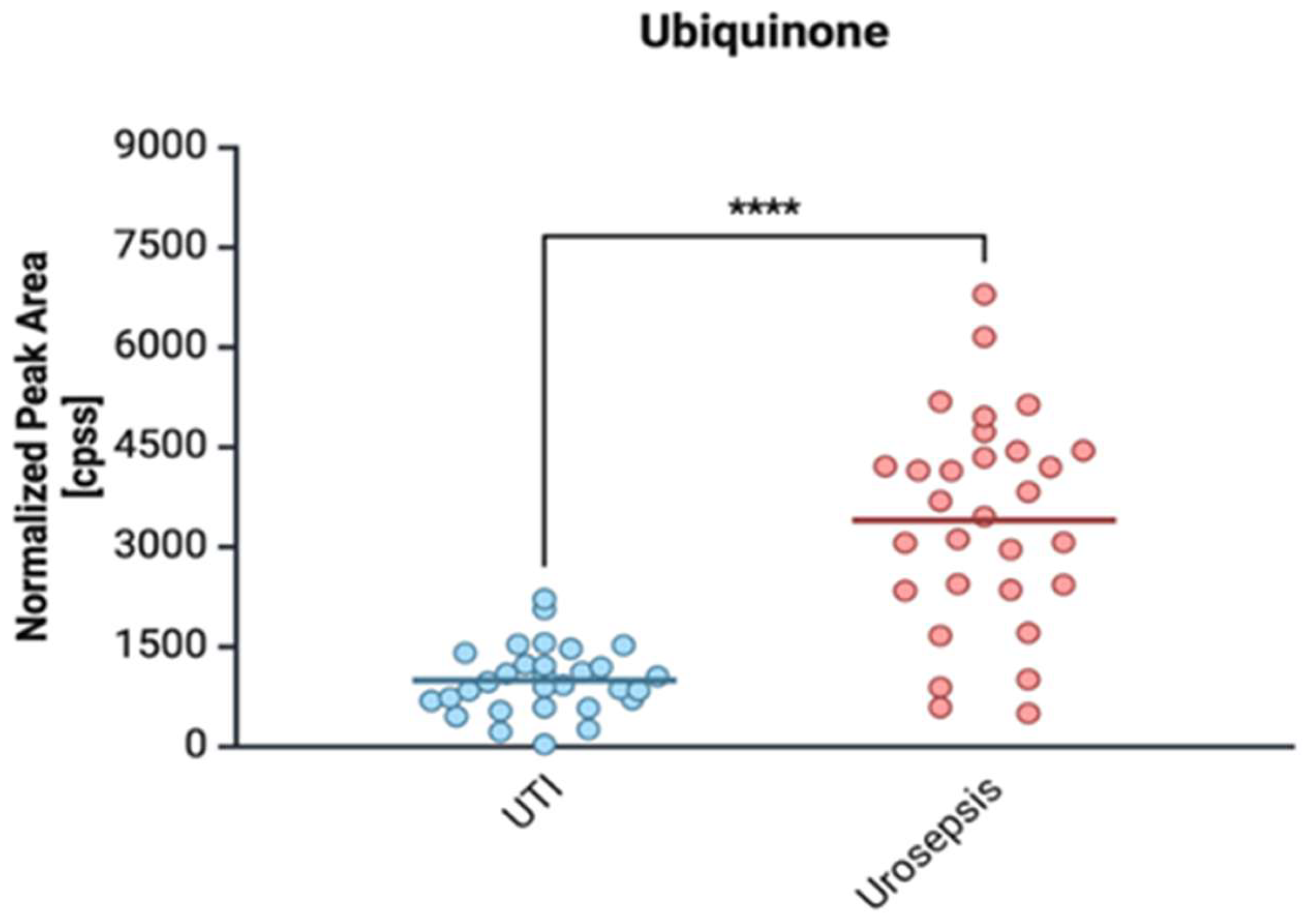
2.4. Proteomic and Metabolomic Profiling of Urosepsis Strains
2.4.1. Citrate
2.4.2. (S) Malate
2.4.3. Ubiquinone (Coenzyme Q10)
3. Discussion
3.1. Comparison of the Virulence of Strains Isolated from Urosepsis and the Control Group
3.2. Proteomic and Metabolomic Studies
4. Materials and Methods
4.1. Selection of Strains for Genetic and Proteomic Studies
4.2. Genetic Studies
4.2.1. Phylogenetic Group Determination of E. coli Isolates
4.2.2. Detection of Virulence Gene by PCR
4.2.3. PCR/RFLP for SPATE Genes
4.3. Proteomic Analysis
4.4. Metabolomic Analysis
4.5. Statistical and Pathways Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| UPEC | Uropathogenic Escherichia coli |
| Vfs | Virulence factors |
| ExPEC | Extraintestinal Pathogenic Escherichia coli |
| SPATE | Serine Protease Autotransporters of Enterobacteriaceae |
| AT | Autotransporter |
| PhoP | PhoP two-component system |
| CpxR | CpxR two-component system |
| GLTL | glutamate-aspartate transport system ATP-binding protein |
| YbhC | pectinesterase |
| NarL | Nitrate/nitrite response regulator protein NarL |
| FepA | Ferrienterobactin receptor FepA |
References
- He, Y.; Zhao, J.; Wang, L.; Han, C.; Yan, R.; Zhu, P. Epidemiological Trends and Predictions of Urinary Tract Infections in the Global Burden of Disease Study 2021. Sci. Rep. 2025, 15, 4702. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, H.; Zheng, Y.; Qu, S.; Wang, H.; Yi, F. Disease Burden and Long-Term Trends of Urinary Tract Infections: A Worldwide Report. Front. Public Health 2022, 10, 888205. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, 10-1128. [Google Scholar] [CrossRef]
- Kim, B.; Kim, J.-H.; Lee, Y. Virulence Factors Associated with Escherichia coli Bacteremia and Urinary Tract Infection. Ann. Lab. Med. 2022, 42, 203–212. [Google Scholar] [CrossRef]
- Desvaux, M.; Dalmasso, G.; Beyrouthy, R.; Barnich, N.; Delmas, J.; Bonnet, R. Pathogenicity Factors of Genomic Islands in Intestinal and Extraintestinal Escherichia coli. Front. Microbiol. 2020, 11, 2065. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the Various Phylogenetic Classification Schemes for Escherichia coli and the Correspondence among Schemes. Microbiology 2015, 161, 980–988. [Google Scholar] [CrossRef]
- Tourret, J.; Denamur, E. Population Phylogenomics of Extraintestinal Pathogenic Escherichia coli. Urin. Tract Infect. Mol. Pathog. Clin. Manag. 2017, 4, 207–233. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia coli Isolated from Different Sources: Recent Reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A. Characterization and Rapid Identification of Phylogroup G in Escherichia coli, a Lineage with High Virulence and Antibiotic Resistance Potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.J. Type 1 Pilus-Mediated Bacterial Invasion of Bladder Epithelial Cells. EMBO J. 2000, 19, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Klemm, P.; Hancock, V.; Schembri, M.A. Fimbrial Adhesins from Extraintestinal Escherichia coli. Environ. Microbiol. Rep. 2010, 2, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Lillington, J.; Geibel, S.; Waksman, G. Biogenesis and Adhesion of Type 1 and P Pili. Biochim. Et Biophys. Acta Gen. Subj. 2014, 1840, 2783–2793. [Google Scholar] [CrossRef]
- Melican, K.; Sandoval, R.M.; Kader, A.; Josefsson, L.; Tanner, G.A.; Molitoris, B.A.; Richter-Dahlfors, A. Uropathogenic Escherichia coli P and Type 1 Fimbriae Act in Synergy in a Living Host to Facilitate Renal Colonization Leading to Nephron Obstruction. PLoS Pathog. 2011, 7, e1001298. [Google Scholar] [CrossRef]
- Lane, M.C.; Alteri, C.J.; Smith, S.N.; Mobley, H.L.T. Expression of Flagella Is Coincident with Uropathogenic Escherichia coli Ascension to the Upper Urinary Tract. Proc. Natl. Acad. Sci. USA 2007, 104, 16669–16674. [Google Scholar] [CrossRef]
- Dobrindt, U.; Blum-Oehler, G.; Hartsch, T.; Gottschalk, G.; Ron, E.Z.; Funfstuck, R. S-Fimbria-Encoding Determinant Sfa I Is Located on Pathogenicity Island III 536 of Uropathogenic Escherichia coli Strain 536. Infect. Immun. 2001, 69, 4248–4256. [Google Scholar] [CrossRef]
- Govindarajan, D.K.; Kandaswamy, K. Virulence Factors of Uropathogens and Their Role in Host Pathogen Interactions. Cell Surf. 2022, 8, 100075. [Google Scholar] [CrossRef]
- Rahdar, M.; Rashki, A.; Miri, H.R.; Rashki Ghalehnoo, M. Detection of Pap, Sfa, Afa, Foc, and Fim Adhesin-Encoding Operons in Uropathogenic Escherichia coli Isolates Collected from Patients with Urinary Tract Infection. Jundishapur J. Microbiol. 2015, 8, e22647. [Google Scholar] [CrossRef]
- Behzadi, P. Uropathogenic Escherichia coli and Fimbrial Adhesins Virulome. In Urinary Tract Infection—The Result of the Strength of the Pathogen, or the Weakness of the Host; InTech Publishers: London, UK, 2018. [Google Scholar] [CrossRef]
- Zamani, H.; Salehzadeh, A. Biofilm Formation in Uropathogenic Escherichia coli: Association with Adhesion Factor Genes. Turk. J. Med. Sci. 2018, 48, 162–167. [Google Scholar] [CrossRef]
- Khonsari, M.S.; Behzadi, P.; Foroohi, F. The Prevalence of Type 3 Fimbriae in Uropathogenic Escherichia coli Isolated from Clinical Urine Samples. Meta Gene 2021, 28, 100881. [Google Scholar] [CrossRef]
- Behzadi, P.; Urbán, E.; Gajdács, M. Association between Biofilm-Production and Antibiotic Resistance in Uropathogenic Escherichia coli (UPEC): An in Vitro Study. Diseases 2020, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Hacker, J. Glycolipid Receptors of F1C Fimbrial Adhesin of Uropathogenic Escherichia coli. In Genes and Proteins Underlying Microbial Urinary Tract Virulence. Advances in Experimental Medicine and Biology; Emoődy, L., Pál, T., Hacker, J., Blum-Oehler, G., Eds.; Springer: Boston, MA, USA, 2002; Volume 485. [Google Scholar] [CrossRef]
- Alvarez-Fraga, L.; Phan, M.-D.; Goh, K.G.K.; Nhu, N.T.K.; Hancock, S.J.; Allsopp, L.P.; Peters, K.M.; Forde, B.M.; Roberts, L.W.; Sullivan, M.J.; et al. Differential Afa/Dr Fimbriae Expression in the Multidrug-Resistant Escherichia coli ST131 Clone. mBio 2022, 13, e03519-21. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Wang, Y.; Qiu, Z.; Zhu, S.; Guo, K.; Chen, W. Necroptosis, Pyroptosis, Ferroptosis in Sepsis and Treatment. Shock 2022, 57, 161–171. [Google Scholar] [CrossRef]
- Torres, A.G.; Redford, P.; Welch, R.A.; Payne, S.M. TonB-Dependent Systems of Uropathogenic Escherichia coli: Aerobactin and Heme Transport and TonB Are Required for Virulence in the Mouse. Infect. Immun. 2001, 69, 6179–6185. [Google Scholar] [CrossRef]
- Li, C.; Pan, D.; Li, M.; Wang, Y.; Song, L.; Yu, D. Aerobactin-Mediated Iron Acquisition Enhances Biofilm Formation, Oxidative Stress Resistance, and Virulence of Yersinia Pseudotuberculosis. Front. Microbiol. 2021, 12, 699913. [Google Scholar] [CrossRef]
- Wang, H.; Xu, Q.; Chen, K.; Kwan, B.; Ye, L.; Yang, X.; Xie, M. A Siderophore-Encoding Plasmid Encodes High-Level Virulence in Escherichia coli. Microbiol. Spectr. 2022, 10, e0252821. [Google Scholar] [CrossRef]
- Arnold, E. Non-Classical Roles of Bacterial Siderophores in Pathogenesis. Front. Cell. Infect. Microbiol. 2024, 14, 1465719. [Google Scholar] [CrossRef]
- Nipič, D.; Podlesek, Z.; Budič, M.; črnigoj, M.; Žgur-Bertok, D. Escherichia coli Uropathogenic-Specific Protein, Usp, Is a Bacteriocin-like Genotoxin. J. Infect. Dis. 2013, 208, 1545–1552. [Google Scholar] [CrossRef]
- Dhakal, B.K.; Mulvey, M.A. The UPEC Pore-Forming Toxin α-Hemolysin Triggers Proteolysis of Host Proteins to Disrupt Cell Adhesion, Inflammatory, and Survival Pathways. Cell Host Microbe 2012, 11, 58–69. [Google Scholar] [CrossRef]
- Landraud, L.; Gauthier, M.; Fosse, T.; Boquet, P. Frequency of Escherichia coli Strains Producing the Cytotoxic Necrotizing Factor (CNF1) in Nosocomial Urinary Tract Infections. Lett. Appl. Microbiol. 2000, 30, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Boquet, P. The Cytotoxic Necrotizing Factor 1 (CNF1) from Escherichia coli. Toxicon 2001, 39, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, A.; Travaglione, S.; Fiorentini, C. Escherichia coli Cytotoxic Necrotizing Factor 1 (CNF1): Toxin Biology, in Vivo Applications and Therapeutic Potential. Toxins 2010, 2, 283–296. [Google Scholar] [CrossRef]
- Valle, J.; Mabbett, A.N.; Ulett, G.C.; Toledo-Arana, A.; Wecker, K.; Totsika, M. UpaG, a New Member of the Trimeric Autotransporter Family of Adhesins in Uropathogenic Escherichia coli. J. Bacteriol. 2008, 190, 4147–4161. [Google Scholar] [CrossRef]
- Vo, J.L.; Martínez Ortiz, G.C.; Subedi, P.; Keerthikumar, S.; Mathivanan, S.; Paxman, J.J. Autotransporter Adhesins in Escherichia coli Pathogenesis. Proteomics 2017, 17, 1600431. [Google Scholar] [CrossRef]
- Habouria, H.; Pokharel, P.; Maris, S.; Garénaux, A.; Bessaiah, H.; Houle, S. Three New Serine-Protease Autotransporters of Enterobacteriaceae (SPATEs) from Extra-Intestinal Pathogenic Escherichia coli and Combined Role of SPATEs for Cytotoxicity and Colonization of the Mouse Kidney. Virulence 2019, 10, 568–587. [Google Scholar] [CrossRef]
- Pokharel, P.; Habouria, H.; Bessaiah, H.; Dozois, C.M. Serine Protease Autotransporters of the Enterobacteriaceae (SPATEs): Out and about and Chopping It Up. Microorganisms 2019, 7, 594. [Google Scholar] [CrossRef]
- Buckles, E.L.; Wang, X.; Lane, M.C.; Lockatell, C.V.; Johnson, D.E.; Rasko, D.A.; Mobley, H.L.T.; Donnenberg, M.S. Role of the K2 Capsule in Escherichia coli Urinary Tract Infection and Serum Resistance. J. Infect. Dis. 2009, 199, 1689–1697. [Google Scholar] [CrossRef]
- Demšar, J.; Curk, T.A.; Erjavec, T.; Hočevar, M.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, A.; Starič, M.; Stajdohar, L.; et al. Zupan, Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Brownlee, J. Information Gain and Mutual Information for Machine Learning. Available online: https://machinelearningmastery.com/information-gain-and-mutual-information/ (accessed on 26 February 2020).
- Berrar, D.; Dubitzky, W. Information Gain. In Encyclopedia of Systems Biology; Springer: New York, NY, USA, 2013; pp. 1022–1023. [Google Scholar] [CrossRef]
- Owrangi, B.; Masters, N.; Kuballa, A.; O’Dea, C.; Vollmerhausen, T.L.; Katouli, M. Invasion and Translocation of Uropathogenic Escherichia coli Isolated from Urosepsis and Patients with Community-Acquired Urinary Tract Infection. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018, 37, 833–839. [Google Scholar] [CrossRef]
- Subashchandrabose, S.; Mobley, H.L.T. Virulence and Fitness Determinants of Uropathogenic Escherichia coli. Microbiol. Spectr. 2015, 235–261. [Google Scholar] [CrossRef] [PubMed]
- Floyd, K.A.; Moore, J.L.; Eberly, A.R.; Good, J.A.D.; Shaffer, C.L.; Zaver, H. Adhesive Fiber Stratification in Uropathogenic Escherichia coli Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili. PLOS Pathog. 2015, 11, e1004697. [Google Scholar] [CrossRef] [PubMed]
- Greene, S.E.; Hibbing, M.E.; Janetka, J.; Chen, S.L.; Hultgren, S.J. Human Urine Decreases Function and Expression of Type 1 Pili in Uropathogenic Escherichia coli. mBio 2015, 6, e00820-15. [Google Scholar] [CrossRef]
- Mulvey, M.A.; Klumpp, D.J.; Stapleton, A.E. Urinary Tract Infections: Molecular Pathogenesis and Clinical Management; ASM Press: Washington, DC, USA, 2017; ISBN 9781555817404. [Google Scholar]
- Schwartz, D.J.; Chen, S.L.; Hultgren, S.J.; Seed, P.C. Population Dynamics and Niche Distribution of Uropathogenic Escherichia coli during Acute and Chronic Urinary Tract Infection. Infect. Immun. 2011, 79, 4250–4259. [Google Scholar] [CrossRef]
- Nagamatsu, K.; Hannan, T.J.; Guest, R.L.; Kostakioti, M.; Hadjifrangiskou, M.; Binkley, J. Dysregulation of Escherichia coliα-Hemolysin Expression Alters the Course of Acute and Persistent Urinary Tract Infection. Proc. Natl. Acad. Sci. USA 2015, 112, E871–E880. [Google Scholar] [CrossRef]
- Totsika, M.; Wells, T.J.; Beloin, C.; Valle, J.; Allsopp, L.P.; King, N.P. Molecular Characterization of the EhaG and UpaG Trimeric Autotransporter Proteins from Pathogenic Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 2179–2189. [Google Scholar] [CrossRef]
- Kjærgaard, K.; Schembri, M.A.; Hasman, H.; Klemm, P. Antigen 43 from Escherichia coli Induces Inter- and Intraspecies Cell Aggregation and Changes in Colony Morphology of Pseudomonas Fluorescens. J. Bacteriol. 2000, 182, 4789–4796. [Google Scholar] [CrossRef]
- Sherlock, O.; Dobrindt, U.; Jensen, J.B.; Munk Vejborg, R.; Klemm, P. Glycosylation of the Self-Recognizing Escherichia coli Ag43 Autotransporter Protein. J. Bacteriol. 2006, 188, 1798–1807. [Google Scholar] [CrossRef]
- Vo, J.L.; Ortiz, G.C.M.; Totsika, M.; Lo, A.W.; Hancock, S.J.; Whitten, A.E. Variation of Antigen 43 Self-Association Modulates Bacterial Compacting within Aggregates and Biofilms. Biofilms Microbiomes 2022, 8, 20. [Google Scholar] [CrossRef]
- Allsopp, L.P.; Beloin, C.; Moriel, D.G.; Totsika, M.; Ghigo, J.-M.; Schembri, M.A. Functional Heterogeneity of the UpaH Autotransporter Protein from Uropathogenic Escherichia coli. J. Bacteriol. 2012, 194, 5769–5782. [Google Scholar] [CrossRef]
- Guyer, D.M.; Radulovic, S.; Jones, F.-E.; Mobley, H.L.T. Sat, the Secreted Autotransporter Toxin of Uropathogenic Escherichia coli, Is a Vacuolating Cytotoxin for Bladder and Kidney Epithelial Cells. Infect. Immun. 2002, 70, 4539–4546. [Google Scholar] [CrossRef] [PubMed]
- Restieri, C.; Garriss, G.; Locas, M.-C.; Dozois, C.M. Autotransporter-Encoding Sequences Are Phylogenetically Distributed among Escherichia coli Clinical Isolates and Reference Strains. Appl. Environ. Microbiol. 2007, 73, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Moseley, S.L.; Roberts, P.L.; Stamm, W.E. Aerobactin and Other Virulence Factor Genes among Strains of Escherichia coli Causing Urosepsis: Association with Patient Characteristics. Infect. Immun. 1988, 56, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Burzyńska, M.; Wityk, P.; Laskowska, A.; Stanistawska, S.A.; Raczak, G.J.; Waszczuk, J.M.; Bronk, M.; Markuszewski, M.J. Iron uptake by Escherichia coli in urinary tract infections and urosepsis—Genetic and proteomic studies. Authorea 2023. [Google Scholar] [CrossRef]
- Li, H.; Wang, X.; Yang, Q.; Cheng, L.; Zeng, H.-L. Identification of Iron Metabolism-Related Genes as Diagnostic Signatures in Sepsis by Blood Transcriptomic Analysis. Cent. Eur. J. Biol. 2023, 18, 20220549. [Google Scholar] [CrossRef]
- Johnson, J.R.; Jelacic, S.; Schoening, L.M.; Clabots, C.; Shaikh, N.; Mobley, H.L.T. The IrgA Homologue Adhesin Iha Is an Escherichia coli Virulence Factor in Murine Urinary Tract Infection. Infect. Immun. 2005, 73, 965–971. [Google Scholar] [CrossRef]
- Léveillé, S.; Caza, M.; Johnson, J.R.; Clabots, C.; Sabri, M.; Dozois, C.M. Iha from an Escherichia coli Urinary Tract Infection Outbreak Clonal Group a Strain Is Expressed in Vivo in the Mouse Urinary Tract and Functions as a Catecholate Siderophore Receptor. Infect. Immun. 2006, 74, 3427–3436. [Google Scholar] [CrossRef]
- Magistro, G.; Hoffmann, C.; Schubert, S. The Salmochelin Receptor IroN Itself, but Not Salmochelin-Mediated Iron Uptake Promotes Biofilm Formation in Extraintestinal Pathogenic Escherichia coli (ExPEC). Int. J. Med. Microbiol. 2015, 305, 435–445. [Google Scholar] [CrossRef]
- Frick-Cheng, A.E.; Sintsova, A.; Smith, S.N.; Pirani, A.; Snitkin, E.S. Harry Ferric Citrate Uptake Is a Virulence Factor in Uropathogenic Escherichia coli. mBio 2022, 13, e0103522. [Google Scholar] [CrossRef]
- Skorokhodova, A.Y.; Stasenko, A.A.; Krasilnikova, N.V.; Gulevich, A.Y.; Debabov, V.G. Engineering Escherichia coli for Efficient Aerobic Conversion of Glucose to Malic Acid through the Modified Oxidative TCA Cycle. Fermentation 2022, 8, 738. [Google Scholar] [CrossRef]
- Kim, J.W.; Jang, H.-D.; Bae, J.H.; Lee, J.G. Effects of Coenzyme Q10 on Bladder Dysfunction Induced by Chronic Bladder Ischemia in a Rat Model. J. Urol. 2013, 189, 2371–2376. [Google Scholar] [CrossRef] [PubMed]
- Floyd, K.A.; Mitchell, C.A.; Eberly, A.R.; Colling, S.J.; Zhang, E.W.; DePas, W.; Chapman, M.R.; Conover, M.; Rogers, B.R.; Hultgren, S.J.; et al. The UbiI (VisC) Aerobic Ubiquinone Synthase Is Required for Expression of Type 1 Pili, Biofilm Formation, and Pathogenesis in Uropathogenic Escherichia coli. J. Bacteriol. 2016, 198, 2662–2672. [Google Scholar] [CrossRef] [PubMed]
- Mayorga, H.; Rolz, C. Pectinesterase Activity as a Function of PH, Enzyme, and Cation Concentrations. J. Agric. Food Chem. 1971, 19, 179–181. [Google Scholar] [CrossRef]
- Di, R.; Vakkalanka, M.S.; Onumpai, C.; Chau, H.K.; White, A.; Rastall, R.A. Pectic Oligosaccharide Structure-Function Relationships: Prebiotics, Inhibitors of Escherichia coli O157:H7 Adhesion and Reduction of Shiga Toxin Cytotoxicity in HT29 Cells. Food Chem. 2017, 227, 245–254. [Google Scholar] [CrossRef]
- Shubakov, A.; Mikhailova, E. The Study of the Growth of Escherichia coli on Pectins. Int. J. Biomed. 2019, 9, 366–369. [Google Scholar] [CrossRef]
- Han, J.; Gao, X.; Luo, X.; Zhu, L.; Zhang, Y.; Dong, P. The Role of PhoP/PhoQ System in Regulating Stress Adaptation Response in Escherichia coli O157:H7. Food Microbiol. 2023, 112, 104244. [Google Scholar] [CrossRef]
- Forst, S.; Inouye, M. Environmentally Regulated Gene Expression for Membrane Proteins in Escherichia coli. Annu. Rev. Cell Biol. 1988, 4, 21–42. [Google Scholar] [CrossRef]
- Liu, X.; Ferenci, T. An Analysis of Multifactorial Influences on the Transcriptional Control of OmpF and OmpC Porin Expression under Nutrient Limitation. Microbiology 2001, 147, 2981–2989. [Google Scholar] [CrossRef]
- Pratt, L.A.; Hsing, W.; Gibson, K.E.; Silhavy, T.J. From Acids to OsmZ: Multiple Factors Influence Synthesis of the OmpF and OmpC Porins in Escherichia coli. Mol. Microbiol. 1996, 20, 911–917. [Google Scholar] [CrossRef]
- Cohen, S.P.; McMurry, L.M.; Hooper, D.C.; Wolfson, J.S.; Levy, S.B. Cross-Resistance to Fluoroquinolones in Multiple-Antibiotic-Resistant (Mar) Escherichia coli Selected by Tetracycline or Chloramphenicol: Decreased Drug Accumulation Associated with Membrane Changes in Addition to OmpF Reduction. Antimicrob. Agents Chemother. 1989, 33, 1318–1325. [Google Scholar] [CrossRef]
- Dorel, C.; Vidal, O.; Prigent-Combaret, C.; Vallet, I.; Lejeune, P. Involvement of the Cpx Signal Transduction Pathway of E. Coli in Biofilm Formation. FEMS Microbiol. Lett. 1999, 178, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Hung, D.L. Cpx Signaling Pathway Monitors Biogenesis and Affects Assembly and Expression of P Pili. EMBO J. 2001, 20, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Mora-Rillo, M.; Fernández-Romero, N.; Navarro-San Francisco, C.; Díez-Sebastián, J.; Romero-Gómez, M.P.; Arnalich Fernández, F.; Arribas López, J.R.; Mingorance, J. Impact of Virulence Genes on Sepsis Severity and Survival in Escherichia coli Bacteremia. Virulence 2015, 6, 93–100. [Google Scholar] [CrossRef]
- Kompaniiets, D.; He, L.; Wang, D.; Zhou, W.; Yang, Y.; Hu, Y. Structural Basis for Transcription Activation by the Nitrate-Responsive Regulator NarL. Nucleic Acids Res. 2024, 52, 1471–1482. [Google Scholar] [CrossRef]
- Niu, H.; Li, T.; Du, Y.; Lv, Z.; Cao, Q.; Zhang, Y. Glutamate Transporters GltS, GltP and GltI Are Involved in Escherichia coli Tolerance in Vitro and Pathogenicity in Mouse Urinary Tract Infections. Microorganisms 2023, 11, 1173. [Google Scholar] [CrossRef]
- Robinson, A.E.; Heffernan, J.R.; Henderson, J.P. The Iron Hand of Uropathogenic Escherichia coli: The Role of Transition Metal Control in Virulence. Future Microbiol. 2018, 13, 745–756. [Google Scholar] [CrossRef]
- Guérin, F.; Galimand, M.; Tuambilangana, F.; Courvalin, P.; Cattoir, V. Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes is driven by inactivation of its local repressor FepR. PLoS ONE 2014, 9, e106340. [Google Scholar] [CrossRef]
- Biville, F.; Cwerman, H.; Létoffé, S.; Rossi, M.S.; Drouet, V.; Ghigo, J.M.; Wandersman, C. Haemophore-mediated signalling in Serratia marcescens: A new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in haem acquisition. Mol. Microbiol. 2004, 53, 1267–1277. [Google Scholar] [CrossRef]
- Cassat, J.E.; Skaar, E.P. Iron in infection and immunity. Cell Host Microbe 2013, 13, 509–519. [Google Scholar] [CrossRef]
- Baghal, S.M.; Gargari, S.L.; Rasooli, I. Production and immunogenicity of recombinant ferric enterobactin protein (FepA). Int. J. Infect. Dis. 2010, 14 (Suppl. S3), e166–e170. [Google Scholar] [CrossRef] [PubMed]
- Passari, A.K.; Ruiz-Villafán, B.; Cruz-Bautista, R.; Díaz-Domínguez, V.; Rodríguez-Sanoja, R.; Sanchez, S. Opportunities and challenges of microbial siderophores in the medical field. Appl. Microbiol. Biotechnol. 2023, 107, 6751–6759. [Google Scholar] [CrossRef] [PubMed]
- Möllmann, U.; Heinisch, L.; Bauernfeind, A.; Köhler, T.; Ankel-Fuchs, D. Siderophores as drug delivery agents: Application of the “Trojan Horse” strategy. Biometals 2009, 22, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Samet, A.; Leibner, J.; Sledzińska, A.; Kur, J. Evaluation of a PCR Melting Profile Technique for Bacterial Strain Differentiation. J. Clin. Microbiol. 2006, 44, 2327–2332. [Google Scholar] [CrossRef]
- Adamus-Bialek, W.; Wojtasik, A.; Majchrzak, M.; Sosnowski, M.; Parniewski, P. (CGG)4-Based PCR as a Novel Tool for Discrimination of Uropathogenic Escherichia coli Strains: Comparison with Enterobacterial Repetitive Intergenic Consensus-PCR. J. Clin. Microbiol. 2009, 47, 3937–3944. [Google Scholar] [CrossRef]
- Krawczyk, B.; Michalik, M.; Fordon, M.; Wysocka, M.; Samet, A.; Nowicki, B. Escherichia coli strains with virulent factors typical for uropathogens were isolated from sinuses from patients with chronic rhinosinusitis—Case report. Pathogens 2020, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Sahly, H.; Navon-Venezia, S.; Roesler, L.; Hay, A.; Carmeli, Y.; Podschun, R.; Hennequin, C.; Forestier, C.; Ofek, I. Extended-spectrum beta-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2008, 52, 3029–3034. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Le Bouguenec, C.; Archambaud, M.; Labigne, A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 1189–1193. [Google Scholar] [CrossRef]
- Blanco, M.; Blanco, J.E.; Alonso, M.P.; Mora, A.; Balsalobre, C.; Muñoa, F.; Juárez, A.; Blanco, J. Detection of pap, sfa and afa adhesin-encoding operons in uropathogenic Escherichia coli strains: Relationship with expression of adhesins and production of toxins. Res. Microbiol. 1997, 148, 745–755. [Google Scholar] [CrossRef]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef]
- Ochoa, S.A.; Cruz-Córdova, A.; Luna-Pineda, V.M.; Reyes-Grajeda, J.P.; Cázares-Domínguez, V.; Escalona, G.; Sepúlveda-González, M.E.; López-Montiel, F.; Arellano-Galindo, J.; López-Martínez, B.; et al. Multidrug- and Extensively Drug-Resistant Uropathogenic Escherichia coli Clinical Strains: Phylogenetic Groups Widely Associated with Integrons Maintain High Genetic Diversity. Front. Microbiol. 2016, 7, 2042. [Google Scholar] [CrossRef]
- Yamamoto, S.; Terai, A.; Yuri, K.; Kurazono, H.; Takeda, Y.; Yoshida, O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol. Med. Microbiol. 1995, 12, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Śledzińska, A.; Szemiako, K.; Samet, A.; Nowicki, B.; Kur, J. Characterisation of Escherichia coli isolates from the blood of haematological adult patients with bacteraemia: Translocation from gut to blood requires the cooperation of multiple virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1135. [Google Scholar] [CrossRef] [PubMed]
- Searle, L.J.; Méric, G.; Porcelli, I.; Sheppard, S.K.; Lucchini, S. Variation in siderophore biosynthetic gene distribution and production across environmental and faecal populations of Escherichia coli. PLoS ONE 2015, 10, e0117906. [Google Scholar] [CrossRef] [PubMed]
- Pilarczyk-Zurek, M.; Strus, M.; Adamski, P.; Heczko, P.B. The dual role of Escherichia coli in the course of ulcerative colitis. BMC Gastroenterol. 2016, 16, 128. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Pressler, U.; Staudenmaier, H.; Zimmermann, L.; Braun, V. Genetics of the iron dicitrate transport system of Escherichia coli. J. Bacteriol. 1988, 170, 2716. [Google Scholar] [CrossRef]
- Bauer, R.J.; Zhang, L.; Foxman, B.; Siitonen, A.; Jantunen, M.E.; Saxen, H.; Marrs, C.F. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection-usp, iha, and iroN(E. coli). J. Infect. Dis. J. Infect. Dis. 2002, 185, 1521. [Google Scholar] [CrossRef]
- Kotlowski, R.; Bernstein, C.N.; Sepehri, S.; Krause, D.O. High prevalence of Escherichia coli belonging to the B2 + D phylogenetic group in inflammatory bowel disease. Gut 2007, 56, 669–675. [Google Scholar] [CrossRef]
- Hozzari, A.; Behzadi, P.; Kerishchi Khiabani, P.; Sholeh, M.; Sabokroo, N. Clinical cases, drug resistance, and virulence genes profiling in Uropathogenic Escherichia coli. J. Appl. Genet. 2020, 61, 265–273. [Google Scholar] [CrossRef]
- Huang, S.-H.; Wass, C.; Fu, Q.; Prasadarao, N.V.; Stins, M.; Kim, K.S. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: Molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 1995, 63, 4470–4475. [Google Scholar] [CrossRef]


| Genes | Control Group n = 85 (%) | Urosepsis Group n = 64 (%) | p * |
|---|---|---|---|
| Fimbrial adhesins | |||
| fimG/H | 79/93% | 62/97% | 0.4669 |
| mrkD | 1/2% | 2/3% | 0.5771 |
| sfaD/E | 54/63% | 20/31% | 0.0001 |
| papG | 28/33% | 31/48% | 0.0641 |
| focG | 19/22% | 10/16% | 0.3676 |
| Afimbrial adhesins | |||
| Afa/Dr | 2/2% | 5/8% | 0.1005 |
| tosA | 26/31% | 16/25% | 0.4312 |
| Invasin | |||
| ibe | 14/16% | 6/9% | 0.2346 |
| Toxins | |||
| usp | 14/16% | 20/31% | 0.0478 |
| hlyA | 8/9% | 14/22% | 0.0385 |
| cnf1 | 24/28% | 19/30% | 0.8763 |
| Autotransporters | |||
| tosB | 35/41% | 13/20% | 0.002 |
| upaG | 57/67% | 60/94% | 0.00001 |
| upaH | 20/24% | 25/39% | 0.033 |
| ag43 | 57/67% | 53/83% | 0.014 |
| aidA | 52/61% | 25/39% | 0.003 |
| Gene of SPATE group | 60/71% | 49/70% | 1.00 |
| vat | 38/45% | 16/25% | 0.0161 |
| sat | 14/16% | 23/36% | 0.0077 |
| pic | 27/32% | 14/22% | 0.1514 |
| pic-like | 13/15% | 9/14% | 1.0 |
| pssA | 1/1% | 4/6% | 0.1184 |
| boa | 0/0% | 3/5% | 0.0594 |
| hbp | 7/8% | 11/17% | 0.1281 |
| Fe uptake | |||
| Enterobactin: | |||
| entB | 82/96% | 61/95% | 1.00 |
| fepA | 82/96% | 62/97% | 1.00 |
| Salmochelin: | |||
| iroB | 56/66% | 34/53% | 0.13 |
| iroN | 54/64% | 35/55% | 0.31 |
| Aerobactin: | |||
| iucA | 41/48% | 43/67% | 0.030 |
| iutA | 44/52% | 45/70% | 0.028 |
| Yersiniabactin: | |||
| irp-2 | 68/80% | 56/88% | 0.27 |
| fyuA | 68/80% | 56/88% | 0.27 |
| Other: | |||
| chuA | 64/75% | 48/75% | 1.00 |
| fecA | 57/67% | 43/67% | 1.00 |
| iha | 17/20% | 28/44% | 0.002 |
| Capsule | |||
| kspMTII | 41/48% | 32/50% | 1.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krawczyk, B.; Wityk, P.; Burzyńska, M.; Majchrzak, T.; Markuszewski, M.J. Uropathogenic Escherichia coli Associated with Risk of Urosepsis—Genetic, Proteomic, and Metabolomic Studies. Int. J. Mol. Sci. 2025, 26, 5681. https://doi.org/10.3390/ijms26125681
Krawczyk B, Wityk P, Burzyńska M, Majchrzak T, Markuszewski MJ. Uropathogenic Escherichia coli Associated with Risk of Urosepsis—Genetic, Proteomic, and Metabolomic Studies. International Journal of Molecular Sciences. 2025; 26(12):5681. https://doi.org/10.3390/ijms26125681
Chicago/Turabian StyleKrawczyk, Beata, Paweł Wityk, Magdalena Burzyńska, Tomasz Majchrzak, and Michał Jan Markuszewski. 2025. "Uropathogenic Escherichia coli Associated with Risk of Urosepsis—Genetic, Proteomic, and Metabolomic Studies" International Journal of Molecular Sciences 26, no. 12: 5681. https://doi.org/10.3390/ijms26125681
APA StyleKrawczyk, B., Wityk, P., Burzyńska, M., Majchrzak, T., & Markuszewski, M. J. (2025). Uropathogenic Escherichia coli Associated with Risk of Urosepsis—Genetic, Proteomic, and Metabolomic Studies. International Journal of Molecular Sciences, 26(12), 5681. https://doi.org/10.3390/ijms26125681






