Telomere Length, Oxidative Stress Markers, and Related miRNAs in Non-Invasive Samples of Mild COVID-19 Cases
Abstract
1. Introduction
2. Results
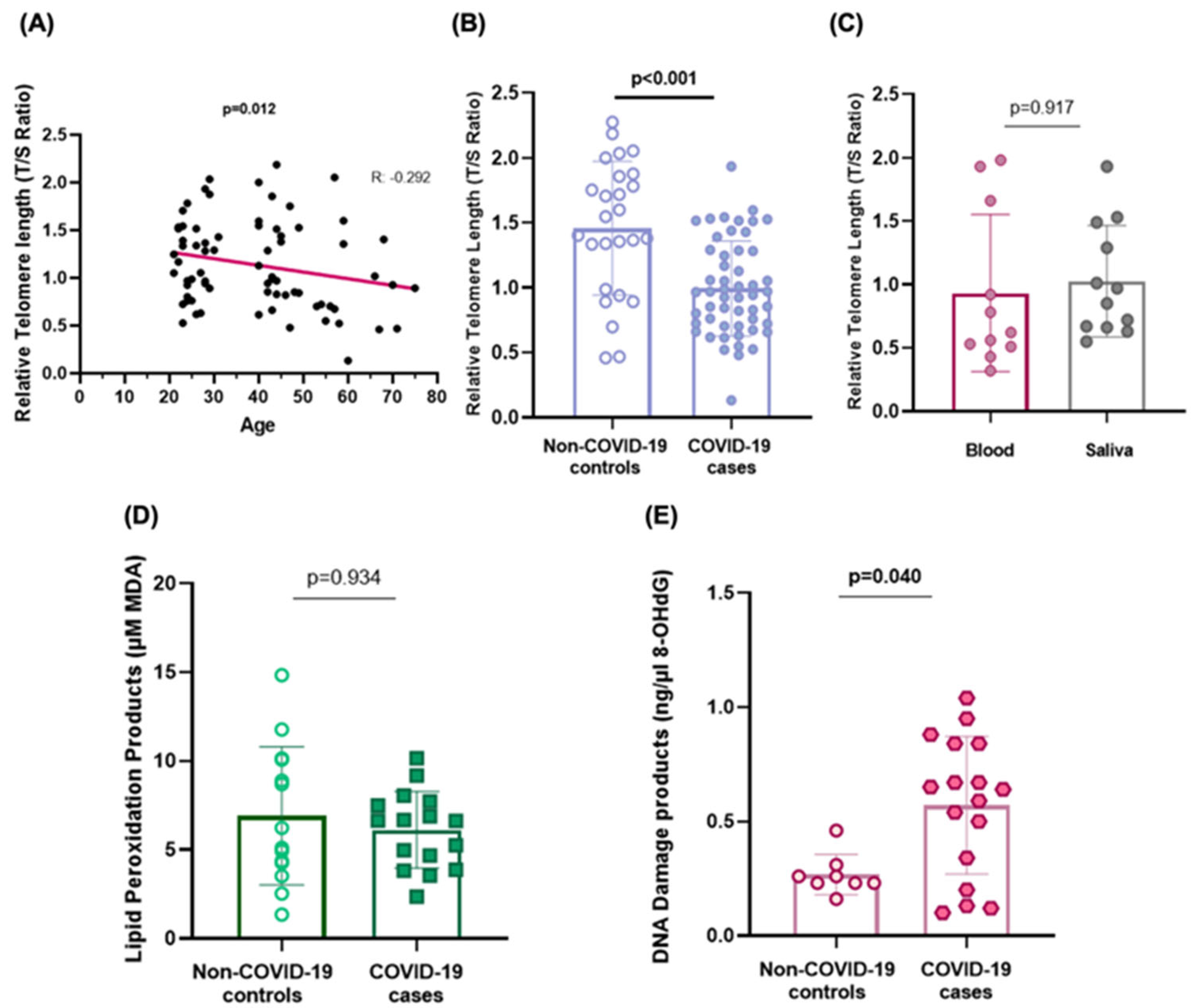
2.1. Telomere Shortening
2.2. Biomarkers of Oxidative Stress
2.3. Telomere and Oxidative Stress-Related microRNA Expression
2.4. Functional Annotation Analysis and Gene Target Prediction
3. Discussion
Limitations of the Study
4. Materials and Methods
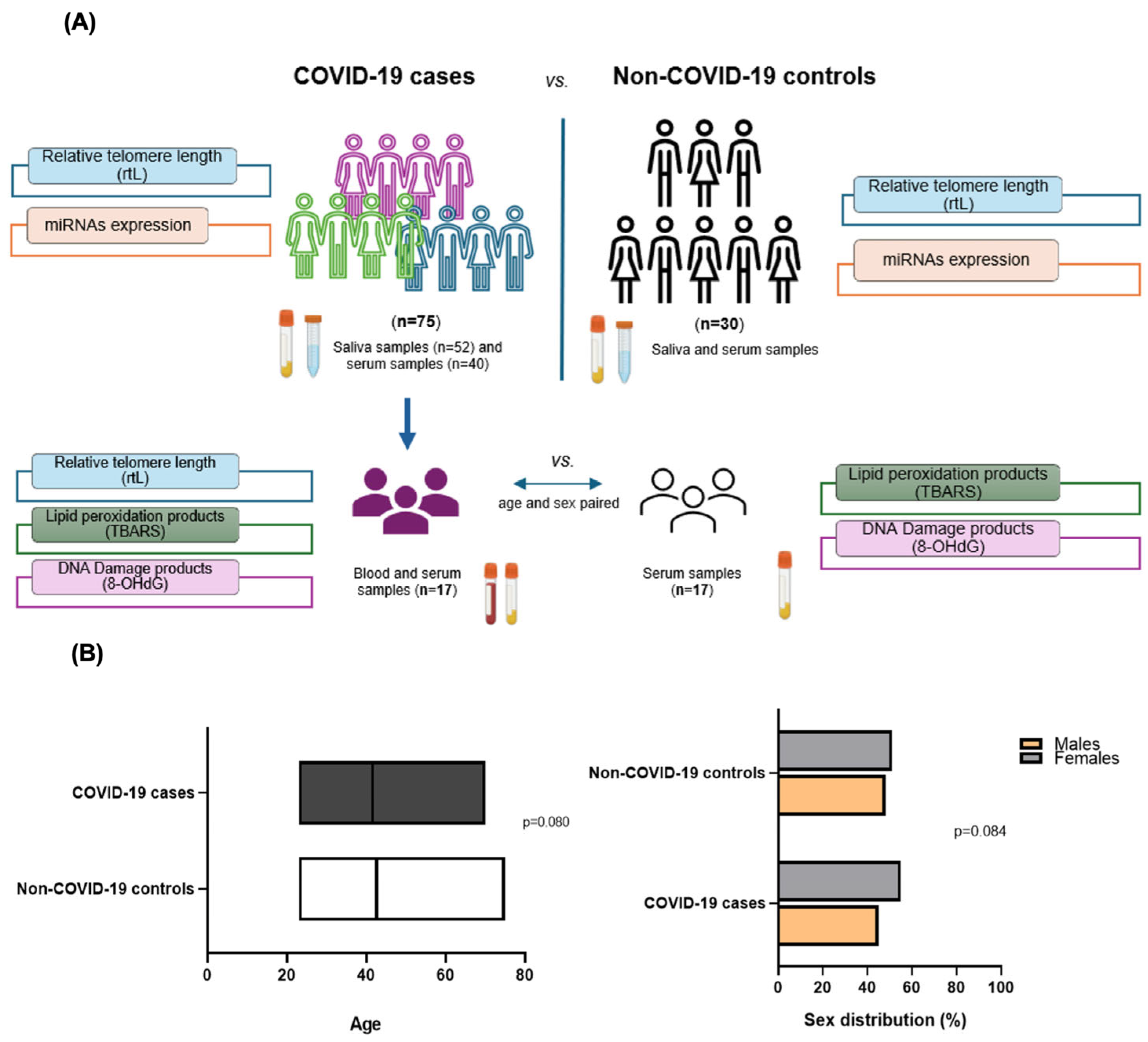
4.1. Selection of Individuals
4.2. Sampling
4.3. Telomere Length Measurement
4.4. Oxidative Stress Biomarkers Measurement
4.5. miRNAs Validation by qPCR
4.6. miRNAs In Silico Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TL | Telomere length |
| rTL | Relative telomere length |
| TBARS | Thiobarbituric acid |
| MDA | Malondialdehyde acid |
| 8-OHdG | 8′-hydroxy-2′-deoxyguanosine |
| miRNA | microRNA |
| WHO | World Health Organization |
| ROS | Reactive Oxygen species |
| SD | Standard deviation |
| CVD | Cardiovascular diseases |
References
- World Health Organization (WHO). (COVID-19 Epidemiological Update—15 July 2024, Edition 169). 2024. Available online: https://www.who.int/publications/m/item/covid-19-epidemiological-update-edition-169 (accessed on 30 October 2024).
- Salimi, S.; Hamlyn, J.M. COVID-19 and crosstalk with the hallmarks of aging. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, e34–e41. [Google Scholar] [CrossRef] [PubMed]
- Renu, K.; Prasanna, P.L.; Gopalakrishnan, A.V. Coronaviruses pathogenesis, comorbidities and multi-organ damage—A review. Life Sci. 2020, 255, 117839. [Google Scholar] [CrossRef] [PubMed]
- Ragab, D.; Salah Eldin, H.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020, 11, 1446. [Google Scholar] [CrossRef]
- Gajate-Arenas, M.; García-Pérez, O.; Chao-Pellicer, J.; Domínguez-De-Barros, A.; Dorta-Guerra, R.; Lorenzo-Morales, J.; Córdoba-Lanús, E. Differential expression of antiviral and immune-related genes in individuals with COVID-19 asymptomatic or with mild symptoms. Front. Cell. Infect. Microbiol. 2023, 13, 1173213. [Google Scholar] [CrossRef]
- Kumar, A.; Prasoon, P.; Kumari, C.; Pareek, V.; Faiq, M.A.; Narayan, R.K.; Kulandhasamy, M.; Kant, K. SARS-CoV-2-specific virulence factors in COVID-19. J. Med. Virol. 2021, 93, 1343–1350. [Google Scholar] [CrossRef]
- Amati, F.; Vancheri, C.; Latini, A.; Colona, V.L.; Grelli, S.; D’Apice, M.R.; Balestrieri, E.; Passarelli, C.; Minutolo, A.; Loddo, S.; et al. Expression profiles of the SARS-CoV-2 host invasion genes in nasopharyngeal and oropharyngeal swabs of COVID-19 patients. Heliyon 2020, 6, e05143. [Google Scholar] [CrossRef]
- Gajate Arenas, M.; Fricke-Galindo, I.; García-Pérez, O.; Domínguez-de-Barros, A.; Pérez Rubio, G.; Dorta-Guerra, R.; Buendía-Roldán, I.; Chávez-Galán, L.; Lorenzo-Morales, J.; Falfán-Valencia, R.; et al. The Immune Response of OAS1, IRF9, and IFI6 Genes in the Pathogenesis of COVID-19. Int. J. Mol. Sci. 2024, 25, 4632. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Levy, M.Z.; Allsopp, R.C.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere end-replication problem and cell aging. J. Mol. Biol. 1992, 225, 951–960. [Google Scholar] [CrossRef]
- Rossiello, F.; Jurk, D.; Passos, J.F.; d’Adda di Fagagna, F. Telomere dysfunction in ageing and age-related diseases. Nat. Cell Biol. 2022, 24, 135–147. [Google Scholar] [CrossRef]
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.C.; Wang, C.B.; Bernardini, S. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388. [Google Scholar] [CrossRef]
- Córdoba-Lanús, E.; Cazorla-Rivero, S.; García-Bello, M.A.; Mayato, D.; Gonzalvo, F.; Ayra-Plasencia, J.; Celli, B.; Casanova, C. Telomere length dynamics over 10-years and related outcomes in patients with COPD. Respir. Res. 2021, 22, 56. [Google Scholar] [CrossRef]
- Iskandar, M.; Xiao Barbero, M.; Jaber, M.; Chen, R.; Gomez-Guevara, R.; Cruz, E.; Westerheide, S. A Review of Telomere Attrition in Cancer and Aging: Current Molecular Insights and Future Therapeutic Approaches. Cancers 2025, 17, 257. [Google Scholar] [CrossRef]
- Sanchez-Vazquez, R.; Guío-Carrión, A.; Zapatero-Gaviria, A.; Martínez, P.; Blasco, M.A. Shorter telomere lengths in patients with severe COVID-19 disease. Aging 2021, 13, 1–15. [Google Scholar] [CrossRef]
- Aviv, A.; Shay, J.W. Reflections on telomere dynamics and ageing-related diseases in humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160436. [Google Scholar] [CrossRef]
- Haridoss, M.; Ayyasamy, L.; Bagepally, B.S. Is COVID-19 severity associated with telomere length? A systematic review and meta-analysis. Virus Genes 2023, 59, 489–498. [Google Scholar] [CrossRef]
- Lorente, L.; Martín, M.M.; González-Rivero, A.F.; Pérez-Cejas, A.; Cáceres, J.J.; Perez, A.; Ramos-Gómez, L.; Solé-Violán, J.; Marcos y Ramos, J.A.; Ojeda, N.; et al. DNA and RNA oxidative damage and mortality of patients with COVID-19. Am. J. Med. Sci. 2021, 361, 585–590. [Google Scholar] [CrossRef]
- Di Minno, A.; Turnu, L.; Porro, B.; Squellerio, I.; Cavalca, V.; Tremoli, E.; Di Minno, M.N.D. 8-Hydroxy-2-deoxyguanosine levels and heart failure: A systematic review and meta-analysis of the literature. Nutr. Metab. Cardiovasc. Dis. 2017, 27, 201–208. [Google Scholar] [CrossRef]
- Satała, J.; Woźniak, A.; Fabiś, M.; Gorzelak-Pabis, P.; Pawlos, A.; Fabis, J.; Broncel, M.; Wozniak, E. Severe COVID-19 classified by simple covid risk index is associated with higher levels of advanced oxidation protein products and 8-hydroxy 2 deoxyguanosine. Epidemiol. Infect. 2023, 151, e140. [Google Scholar] [CrossRef]
- Coronel, P.M.V.; Pereira, I.C.; Basilio, D.C.L.S.; Espinoca, I.T.; de Souza, K.F.S.; Ota, R.S.N.; Borges de Almeida, E.; Paredes-Gamero, E.J.; Wilhelm Filho, D.; Trentin Perdomo, R.; et al. Biomarkers of oxidative stress and inflammation in subjects with COVID-19: Characterization and prognosis of the disease. Microb. Pathog. 2023, 184, 106339. [Google Scholar] [CrossRef]
- Marnett, L.J. Lipid peroxidation- DNA damage by malondialdehyde. Mutat. Res. 1999, 424, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Neves, F.F.; Pott-Junior, H.; Yamashita, K.M.C.; de Sousa Santos, S.; Cominetti, M.R.; de Melo Freire, C.C.; Ferreira da Cunha, A.; Jordao Junior, A.A. Do the oxidative stress biomarkers predict COVID-19 outcome? An in-hospital cohort study. Free Radic. Biol. Med. 2023, 207, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, R.; Li, B.; Zhang, J.; Liu, P.; Li, B.; Li, F.; Zhang, W.; Lyu, X.; Hu, M. Oxidative stress indexes as biomarkers of the severity in COVID-19 patients. Int. J. Med. Sci. 2024, 21, 3034. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Czajkowsky, D.M. SARS-CoV-2 infection and oxidative stress: Pathophysiological insight into thrombosis and therapeutic opportunities. Cytokine Growth Factor Rev. 2022, 63, 44–57. [Google Scholar] [CrossRef]
- Tahamtan, A.; Inchley, C.S.; Marzban, M.; Tavakoli-Yaraki, M.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. The role of microRNAs in respiratory viral infection: Friend or foe? Rev. Med. Virol. 2016, 26, 389–407. [Google Scholar] [CrossRef]
- Ogalur, I.; Pat, Y.; Ardicli, O.; Barletta, E.; Cevhertas, L.; Fernandez-Santamaria, R.; Huang, M.; Bel Imam, M.; Koch, J.; Ma, S.; et al. Advances and highlights in biomarkers of allergic diseases. Allergy 2021, 76, 3659–3686. [Google Scholar] [CrossRef]
- Córdoba-Lanús, E.; Domínguez de-Barros, A.; Oliva, A.; Mayato, D.; Gonzalvo, F.; Remírez-Sanz, A.; Zulueta, J.J.; Celli, B.; Casanova, C. Circulating miR-206 and miR-1246 as Markers in the Early Diagnosis of Lung Cancer in Patients with Chronic Obstructive Pulmonary Disease. Int. J. Mol. Sci. 2023, 24, 12437. [Google Scholar] [CrossRef]
- Li, J.; Wang, Q.; Wen, R.; Liang, J.; Zhong, X.; Yang, W.; Su, D.; Tang, J. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J. Cell. Mol. Med. 2015, 19, 2793–2805. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, J.; Zheng, G.; Wang, Q.; Li, X.; Feng, Y.; Shang, F.; He, S.; Jiang, Q.; Shi, B.; et al. Co-Expression of miR155 or LSD1 shRNA Increases the Anti-Tumor Functions of CD19 CAR-T Cells. Front. Immunol. 2022, 12, 811364. [Google Scholar] [CrossRef]
- Wu, J.; Han, X.; Yang, X.; Li, Y.; Liang, Y.; Sun, G.; Wang, R.; Wan, P.; Xie, S.; Feng, J.; et al. MiR-138-5p suppresses the progression of lung cancer by targeting SNIP1. Thorac. Cancer 2023, 14, 612–623. [Google Scholar] [CrossRef]
- Dinami, R.; Ercolani, C.; Petti, E.; Piazza, S.; Ciani, Y.; Sestito, R.; Sacconi, A.; Biagioni, F.; le Sage, C.; Agami, R.; et al. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res. 2014, 74, 4145–4156. [Google Scholar] [CrossRef] [PubMed]
- Asadpour-Behzadi, A.; Kariminik, A.; Kheirkhah, B. MicroRNA-155 is a main part of proinflammatory puzzle during severe coronavirus disease 2019 (COVID-19). Allergol. Immunopathol. 2023, 51, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Al-Nuaimi, B.N.; Al-Azzawi, R.H. Association between MicroRNA-155 Expression and Pro inflammatory Cytokines in Severe COVID-19 Patients. Int. J. Med. Toxicol. Legal Med. 2024, 27, 3. [Google Scholar]
- Grossi, I.; Radeghieri, A.; Paolini, L.; Porrini, V.; Pilotto, A.; Padovani, A.; Marengoni, A.; Barbon, A.; Belluci, A.; Pizzi, M.; et al. MicroRNA-34a-5p expression in the plasma and in its extracellular vesicle fractions in subjects with Parkinson’s disease: An exploratory study. Int. J. Mol. Med. 2020, 47, 533–546. [Google Scholar] [CrossRef]
- McDonald, J.T.; Enguita, F.J.; Taylor, D.; Griffin, R.J.; Priebe, W.; Emmett, M.R.; Sajadi, M.M.; Harris, A.D.; Clement, J.; Dybas, J.M.; et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep. 2021, 37, 109839. [Google Scholar] [CrossRef]
- Barreda-Manso, M.A.; Nieto-Díaz, M.; Soto, A.; Muñoz-Galdeano, T.; Reigada, D.; Maza, R.M. In Silico and In Vitro Analyses Validate Human MicroRNAs Targeting the SARS-CoV-2 3′-UTR. Int. J. Mol. Sci. 2021, 22, 6094. [Google Scholar] [CrossRef]
- Qiao, X.R.; Wang, L.; Liu, M.; Tian, Y.; Chen, T. MiR-210-3p attenuates lipid accumulation and inflammation in atherosclerosis by repressing IGF2. Biosci. Biotechnol. Biochem. 2020, 84, 321–329. [Google Scholar] [CrossRef]
- Baig, M.S.; Deepanshu, P.P.; Alam, P.; Krishnan, A. In silico analysis reveals hypoxia-induced miR-210-3p specifically targets SARS-CoV-2 RNA. J. Biomol. Struct. Dyn. 2023, 41, 2305–12327. [Google Scholar] [CrossRef]
- Holmannova, D.; Borsky, P.; Parova, H.; Stverakova, T.; Vosmik, M.; Hruska, L.; Fiala, Z.; Borska, L. Non-Genomic Hallmarks of Aging-The Review. Int. J. Mol. Sci. 2023, 24, 15468. [Google Scholar] [CrossRef]
- Whang, Z.; Deng, Z.; Tutton, S.; Lieberman, P.M. The Telomeric Response to Viral Infection. Viruses 2017, 9, 218. [Google Scholar] [CrossRef]
- Helby, J.; Nordestgaard, B.G.; Benfield, T.; Bojesen, S.E. Shorter leukocyte telomere length is associated with higher risk of infections: A prospective study of 75,309 individuals from the general population. Hematologica 2017, 102, 1457. [Google Scholar] [CrossRef]
- Dos Santos, G.A.; Pimenta, R.; Viana, N.I.; Guimarães, V.R.; Romão, P.; Candido, P.; de Camargo, J.A.; Hatanaka, D.M.; Queiroz, P.G.; Teruya, A.; et al. Shorter leukocyte telomere length is associated with severity of COVID-19 infection. Biochem. Biophys. Rep. 2021, 27, 101056. [Google Scholar] [CrossRef]
- Goldman, E.A.; Eick, G.N.; Compton, D.; Kowal, P.; Snodgrass, J.J.; Eisenberg, D.T.A.; Sterner, K.N. Evaluating minimally invasive sample collection methods for telomere length measurement. Am. J. Hum. Biol. 2018, 30, e23062. [Google Scholar] [CrossRef]
- Yildiz, H.; Alp, H.H.; Ekin, S.; Arisoy, A.; Gunbatar, H.; Asker, S.; Cilingir, B.M.; Sunnetcioglu, A.; Celikel, M.; Esen, N.; et al. Analysis of endogenous oxidative damage markers and association with pulmonary involvement severity in patients with SARS-CoV-2 pneumonia. Infect. Dis. Now. 2021, 51, 429–434. [Google Scholar] [CrossRef]
- Vazquez-Agra, N.; Marques-Afonso, A.T.; Cruces-Sande, A.; Novo-Veleiro, I.; Pose-Reino, A.; Mendez-Alvarez, E.; Soto-Otero, R.; Hermida-Ameijeiras, A. Assessment of oxidative stress markers in elderly patients with SARS-CoV-2 infection and potential prognostic implications in the medium and long term. PLoS ONE 2022, 17, e0268871. [Google Scholar] [CrossRef]
- Lage, S.L.; Amaral, E.P.; Hilligan, K.L.; Laidlaw, E.; Rupert, A.; Namasivayan, S.; Rocco, J.; Galindo, F.; Kellogg, A.; Kumar, P.; et al. Persistent Oxidative Stress and Inflammasome Activation in CD14 high CD16− Monocytes from COVID-19 Patients. Front. Immunol. 2022, 12, 799558. [Google Scholar] [CrossRef]
- Cekerevac, I.; Turnic, T.N.; Draginic, N.; Andjic, M.; Zivkovic, V.; Simovic, S.; Susa, R.; Novkovic, L.; Mijailovic, Z.; Andjelkovic, M.; et al. Predicting Severity and Intrahospital Mortality in COVID-19: The Place and Role of Oxidative Stress. Oxid. Med. Cell. Longev. 2021, 2021, 6615787. [Google Scholar] [CrossRef]
- Jankauskas, S.S.; Kansakar, U.; Sardu, C.; Varzideh, F.; Avvisato, R.; Wang, X.; Matarese, A.; Marfella, R.; Ziosi, M.; Gambardella, J.; et al. COVID-19 Causes Ferroptosis and Oxidative Stress in Human Endothelial Cells. Antioxidants 2023, 12, 326. [Google Scholar] [CrossRef]
- Alwash, M.; Alilish, A.S.; Aboulkasem, S.; Aldeeb, M.M.; Aborwis, M.M. Prediction and Analysis of Targeting Libyan Severe Acute Respiratory Syndrome Corona Virus 2 isolates by Micro-RNA. Sci. J. Fac. Sci. 2023, 3, 44–50. [Google Scholar]
- Chen, Y.; Williams, V.; Filippova, M.; Filippov, V.; Duerksen-Hughes, P. Viral carcinogenesis: Factors inducing DNA damage and virus integration. Cancers 2014, 6, 2155–2186. [Google Scholar] [CrossRef]
- Gajate-Arenas, M.; Sirvent-Blanco, C.; García-Pérez, O.; Domínguez-De-Barros, A.; Piñero, J.E.; Lorenzo-Morales, J.; Córdoba-Lanús, E. miR-27a-5p, miR-21-5p, miR-1246 and miR-4508: A candidate microRNA signature in the protection and regulation of viral infection in mild COVID-19. Mol. Med. 2025, 31, 102. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Li, S.; Hu, Q.; Liao, D. Genetic correlations, shared risk genes and immunity landscapes between COVID-19 and venous thromboembolism: Evidence from GWAS and bulk transcriptome data. Inflamm. Res. 2024, 73, 619–640. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Feng, B.; Lu, L.; Han, S.; Chu, X.; Chen, L.; Wang, R. MiRNAs and E2F3: A complex network of reciprocal regulations in human cancers. Oncotarget 2017, 8, 60624–60639. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Chen, D.; Ding, W.; Wu, P.; Hou, H.; Bai, Y.; Zhou, Y.; Li, K.; Xiang, S.; Liu, P.; et al. The trans-omics landscape of COVID-19. Nat. Commun. 2021, 12, 4543. [Google Scholar] [CrossRef]
- Bost, P.; Giladi, A.; Liu, Y.; Bendjelal, Y.; Xu, G.; David, E.; Blecher-Gonen, R.; Cohen, M.; Medaglia, C.; Li, H.; et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 2020, 181, 1475–1488. [Google Scholar] [CrossRef]
- Ibáñez-Cabellos, J.S.; Pallardó, F.V.; García-Giménez, J.L.; Seco-Cervera, M. Oxidative Stress and Epigenetics: miRNA Involvement in Rare Autoimmune Diseases. Antioxidants 2023, 12, 800. [Google Scholar] [CrossRef]
- Lin, J.; Smith, D.; Esteves, K.; Drury, S. Telomere length measurement by qPCR- Summary of critical factors and recommendations for assay design. Psychoneuroendocrinology 2019, 99, 271–278. [Google Scholar] [CrossRef]
- Liu, N.; Landreh, M.; Cao, K.; Abe, M.; Hendriks, G.J.; Kennerdell, J.R.; Zhu, Y.; Wang, L.S.; Bonini, N.M. The micro-RNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 2012, 482, 519–523. [Google Scholar] [CrossRef]
- Boon, R.A.; Lekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Tréguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef]
- Lee, J.; Kemper, J.K. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging 2010, 2, 527–534. [Google Scholar] [CrossRef]
- Saulle, I.; Garziano, M.; Cappelletti, G.; Limanaqi, F.; Strizzi, S.; Vanetti, C.; Lo Caputo, S.; Poliseno, M.; Santantonio, T.A.; Clerici, M.; et al. Salivary miRNA Profiles in COVID-19 Patients with Different Disease Severities. Int. J. Mol. Sci. 2023, 24, 10992. [Google Scholar] [CrossRef] [PubMed]
- Abbasifard, M.; Ebrahimi, H.O.; Sharifi, G.T.K.; Bahrehmand, F.; Bagheri-Hosseinabadi, Z. Investigation of the circulatory microRNAs and their involvement in regulation of inflammation in patients with COVID-19. Hum. Immunol. 2025, 86, 111208. [Google Scholar] [CrossRef] [PubMed]
- Donyavi, T.; Bokharaei-Salim, F.; Baghi, H.B.; Khanaliha, K.; Janat-Makan, M.A.; Karimi, B.; Hahand, J.S.; Mirzaei, H.; Khatami, A.; Garshasbi, S.; et al. Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155-5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021, 97, 107641. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Meroni, M.; Casati, S.; Goldoni, R.; Vieira-Thomaz, D.; Seda-Kehr, N.; Galimberti, D.; Del-Fabbro, M.; Tartaglia, G.M. Salivary biomarkers: Novel noninvasive tools to diagnose chronic inflammation. Int. J. Oral Sci. 2023, 15, 27. [Google Scholar] [CrossRef]
- COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines; National Institutes of Health: Bethesda, MD, USA, 2 March 2024.
- Cawthon, R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009, 37, e21. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]

| KEGG Pathways | Genes | p-Value |
|---|---|---|
| miR-34a-5p | ||
| Fatty acid biosynthesis | FASN, ACSL4, ACSL1, ACACA | 9.94 × 10−8 |
| Cell Cycle | CDC6, CDKN2C, E2F1, CCNB1, SFN, CDK4, E2F2, CCNA2, HDAC1, MCM6, YWHAG, ORC1, MCM4, FZR1, MCM5, STAG2, CDKN2B, STATG1, CDK1, CDKN2A, CDK6, TGFB1, MCM7, TP53, ATM, CCND1, SMAD4, E2F5, E2F3, CDC23 | 8.76 × 10−5 |
| Pathways in cancer | BRAF, STAT3, PDGFRA, E2F1, ERBB2, NFKB1, MET, ADCY1, FIGF, GNA12, ROCK1, CK4, CXCL8, RAC2, E2F2, ADCY7, FGFR3, CRKL, STK4, MAP2K2, BIDM, AGTR1, GNAS, LAMA5, CRK, RUNX1, PIK3CB | 2.00 × 10−4 |
| Fatty acid metabolism | ASN, SCD5, ACOX1, ACOX3, ACAA2, PTPLA, CPT2, ACADVL, SCD, ACSL4, ACSL1, HSD17B12, MECR, ACACA | 2.13 × 10−4 |
| Viral carcinogenesis | STAT3, NFKB1, GTF2E2, CDK4, RASA2, CCNA2, GTFH1, PIK3CB, HDAC7, HDAC1, DLG1, VAC14, LTBR, PIK3R2, YWHAG, PXN, HIT1H4C, CDKN2B, CDK1, CDKN2A, CDK6, CREB3, CHD4, HIST1H2BD, HIST1H2BA, TP53 | 1.00 × 10−2 |
| MAPK signalling pathway | BRAF, BUSP4, PDGFRA, CACNA1A, HSPA1A, NFKB1, GNA12, MAP4K2, RAC2, FGFR3, CRKL, STK4, MAP2K2, RASA2, MAP3K3, CRK, MAP2K7, FGFR4, PTPRR, TAOK2, PPP3R1, RAF1, MAPK8IP1, MAP2K3, EGFR, PPP3CC, TAB2, TGFB1, MAP3K11, MAPK13, TP53, PPP3CA, AKT2, DUSP10, MAP3K14, JUN, RAPGEF2, MAPK8, MYC, PPM1A, RASGRP4, MAPKAPK3, PRKACA, CACNB1, CACNA1E, MAPT, FGF9, PRKX, DUSP8, CACNA2D4, FGF18, NF1, DUSP7, DUSP3, PRKCB, RPS6KA3, TNFRSF1A, MAP21, STMN1, MEF2C, FGF23, MKNK2, FGFR2, HSPA1B, NFATC3, FGFR1, DUSP16, MAPK1, NFATC1, FGF7, TGFBR2, ELK1, RRAS, DAXX, PDGFRB, ARRB1, PRKACB, TGFB3, CACNB3, NR4A1 | 3.94 × 10−2 |
| miR-138-5p | ||
| Hepatitis B | PCNA, CXCL8, CCNA2, SMAD3, BCL2, BIRC5, KRAS, STAT5B, DDX3X, TGF1B, AKT2, CASP3, CCND1, MYC, MMP9, CREB3L2, CCNE1, RELA, CREBBp | 1.05 × 10−3 |
| Pathways in cancer | DVL3, CXCL8, GNAS, ROCK2, SMAD3, BCL2, BIRC5, F2RL3, KRAS, STAT5B, TGFB1, EPAS1, LPAR4, AKT2, PLCG1, CASP3, CCND1, CTNNA1, SKP2, HIF1A, MYC, MMP9, DAPK1, HSP90AB1, GNG2, PTGS2, RARA, CCNE1, GNAI2, RELA, VEGFA, CREBBP, DVL2, PPARD, MDM2, BMP4, PDGFRB, COL4A1 | 6.41 × 10−3 |
| Notch signalling pathway | DVL3, APH1A, NOTCH2, HES1, JAG1, DTX1, SNW1, CREBBP, DVL2, MAML1 | 3.08 × 10−2 |
| p53 signalling pathway | CCNG1, GADD45A, CASP3, TSC2, CCND1, SHISA5, CCNE1, SERPINE1, GTSE1, MDM2, CCND3 | 3.08 × 10−2 |
| Viral carcinogenesis | CCNA2, HIST1H2BK, VAC14, PXN, KRAS, STAT5B, DDX3X, CHD4, HIST1H2BI, CASP3, CCND1, SKP2, SP100, CREB3L2, CCNE1, SNW1, RELA, CREBBP, MDM2, GTF2EI, CCND3, HIST1H2BJ | 4.42 × 10−2 |
| miR-155-5p | ||
| Hepatitis B | FOS, STAT3, NFKB1, CDK4, E2F2, CDK2, SMAD3, BCL2, DDB2, KRAS, CREB1, APAF1, MAVS, MYD88, CCND1, SMAD4, E2F3, PIK3R1, YWHAZ, TBK1, AKT3, PIK3CA, CDKN1A, STAT1, TNF, RELA, IL6, MAPK10 | 2.36 × 10−6 |
| TGF-beta signalling pathway | SMAD2, THBS1, PPP2CA, SMAD3, SMAD4, SMAD5, ACVR2A, GDF6, SP1, ACVR1C, PPP2CB, TNF, SMAD1, RPS6KB1 | 4.58 × 10−5 |
| FoxO signalling pathway | RS2, STAT3, SMAD2, SETD7, MAPK14, CDK2, PCK2, SMAD3, CAT, EGFR, KRAS, CCND1, SMAD4, S1PR1, GABARAPL1, PIK3R1, SOS1, AKT3, PIK3CA, FOXO3, USP7, CDKN1A, PLK1, SGK3, FOXO1, IL6, IL10, MDM2, MAPK10, C8orf44-SGK3 | 1.36 × 10−4 |
| Apoptosis | NFKB1, IL1B, BCL2, PRKAR1B, DFFA, APAF1, MYD88, MAP3K14, TNFRSF10A, PIK3R1, CFLAR, PRKAR2A, TNFRSF10B, PRKAR1A, AKT3, PIK3CA, BIRC3, AIFM1, TNF, RELA, XIAP | 1.51 × 10−3 |
| TNF signalling pathway | FOS, NFKB1, IL1B, MAPK14, VCAM1, TAB2, RPS6KA5, ICAM1, CREB1, CEBPB, MAP3K14, JUNB, PIK3R1, CFLAR, AKT3, PIK3CA, BIRC3, TNF, RELA, IL6, TRAF3, MAPK10 | 4.10 × 10−3 |
| miR-182-5p | ||
| Fatty acid biosynthesis | FASN, ACSL4, ACACA | 3.27 × 10−11 |
| Viral carcinogenesis | STAT3, NFKB1, CDK4, NRAS, RASA2, YWHAE, HDAC3, HIST1H2BK, PIK3CB, CREB5, SYK, YWHAG, CCND2, BAX, PKM, CDKN1B, CDK1, CDK6, STAT5B, DDX3X, HIST1H2BD, HDAC9, TP53, EGR3, CREB1, EIF2AK2, SND1, CCND1, SKP2, TBPL2, DDB1, HIST1H4H, PIK3R1, RB1, YWHAZ, RAC1, CDC42, EP300, CREB3L2, CCNE1, PIK3CA, CREB3L1, BAK1, CDKN1A, TBP, RELA, GTF2A1, SRF, CREBBP, GRB2, MAPKAPK2, ATF4, PRKACB, HIST1H4I | 4.00 × 10−9 |
| Non-small cell lung cancer | BRAF, E2F1, CDK4, NRAS, PIK3CB, CDK6, TP53, PLCG1, CCND1, AKT1, PIK3R1, RB1, SOS1, PRKCB, AKT3, PIK3CA, FOXO3, PDPK1, GRB2 | 7.78 × 10−4 |
| FoxO signalling pathway | BRAF, STAT3, NRAS, PRKAA2, SIRT1, PIK3CB, SETD7, CCND2, CAT, CDKN1B, IGF1R, GADD45A, CCND1, SMAD4, SKP2, MAPK8, AKT1, IRS4, PLK2, PIK3R1, SOS1, IRS1, PRKAA1, CSNK1E, EP300, AKT3, PIK3CA, HOMER1, FOXO3, CDKN1A, PDPK1, PLK1, SGK3, FOXO1, PLK4, CREBBP, GRB2, BCL2L11 | 4.17 × 10−3 |
| miR-210-3p | ||
| Other types of O-glycan biosynthesis | ST3GAL3 | 8.99 × 10−4 |
| MicroRNAs in cancer | KIF23, E2F3, RASSF1 | 6.28 × 10−3 |
| Pancreatic secretion | CHRM3, RAB27B, ATP2B3 | 3.79 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domínguez-de-Barros, A.; Sirvent-Blanco, C.; García-Pérez, O.; Gajate-Arenas, M.; García-Ramos, A.; Migliazzo, C.; Piñero, J.E.; Lorenzo-Morales, J.; Córdoba-Lanús, E. Telomere Length, Oxidative Stress Markers, and Related miRNAs in Non-Invasive Samples of Mild COVID-19 Cases. Int. J. Mol. Sci. 2025, 26, 4934. https://doi.org/10.3390/ijms26104934
Domínguez-de-Barros A, Sirvent-Blanco C, García-Pérez O, Gajate-Arenas M, García-Ramos A, Migliazzo C, Piñero JE, Lorenzo-Morales J, Córdoba-Lanús E. Telomere Length, Oxidative Stress Markers, and Related miRNAs in Non-Invasive Samples of Mild COVID-19 Cases. International Journal of Molecular Sciences. 2025; 26(10):4934. https://doi.org/10.3390/ijms26104934
Chicago/Turabian StyleDomínguez-de-Barros, Angélica, Candela Sirvent-Blanco, Omar García-Pérez, Malena Gajate-Arenas, Alma García-Ramos, Claudia Migliazzo, José E. Piñero, Jacob Lorenzo-Morales, and Elizabeth Córdoba-Lanús. 2025. "Telomere Length, Oxidative Stress Markers, and Related miRNAs in Non-Invasive Samples of Mild COVID-19 Cases" International Journal of Molecular Sciences 26, no. 10: 4934. https://doi.org/10.3390/ijms26104934
APA StyleDomínguez-de-Barros, A., Sirvent-Blanco, C., García-Pérez, O., Gajate-Arenas, M., García-Ramos, A., Migliazzo, C., Piñero, J. E., Lorenzo-Morales, J., & Córdoba-Lanús, E. (2025). Telomere Length, Oxidative Stress Markers, and Related miRNAs in Non-Invasive Samples of Mild COVID-19 Cases. International Journal of Molecular Sciences, 26(10), 4934. https://doi.org/10.3390/ijms26104934








