Genetic and Evolutionary Analysis of Ake Chicken: New Insights into China’s Sole Indigenous Naked-Neck Chicken Breed
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Relationships
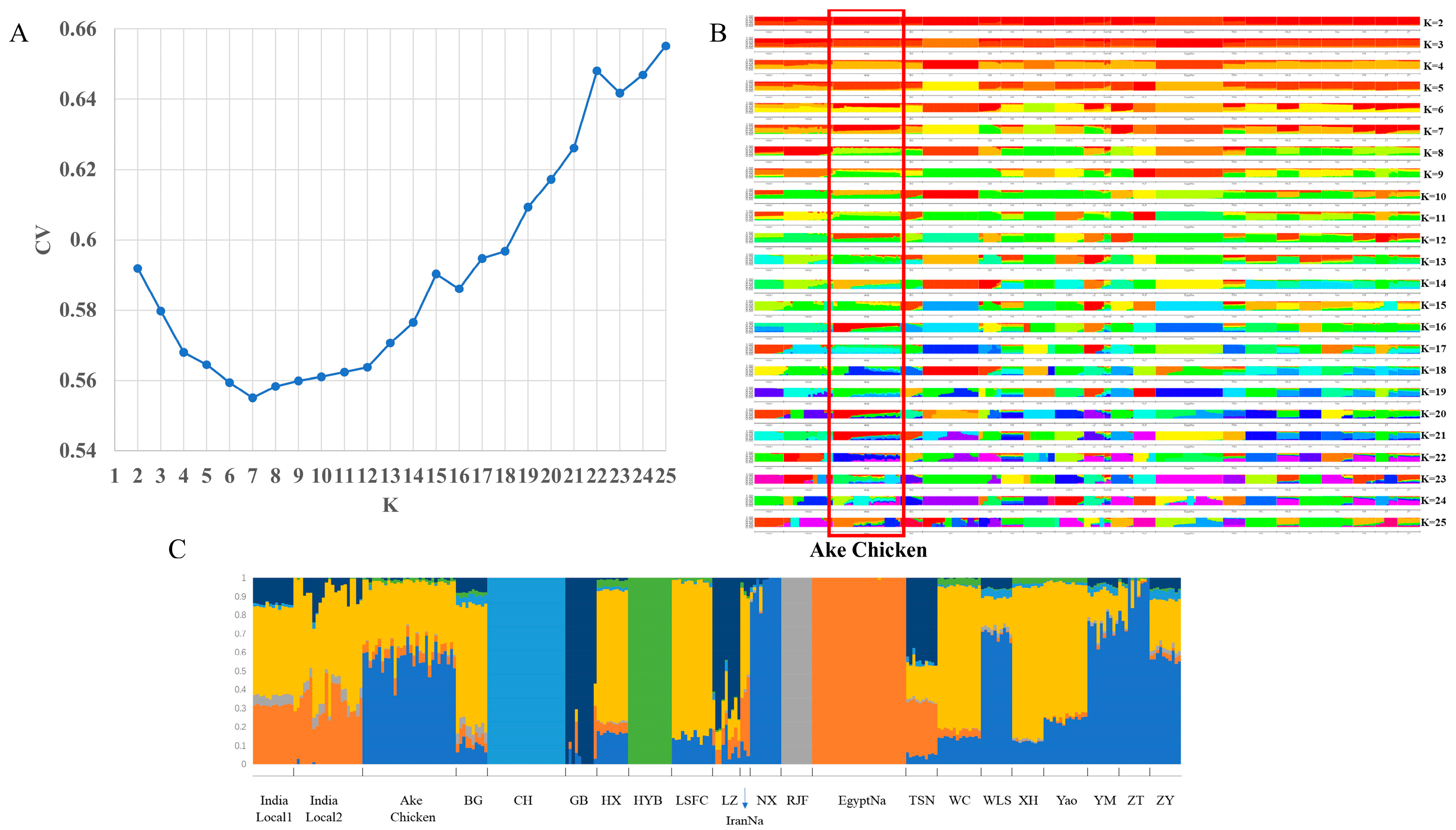
2.2. Population Structure
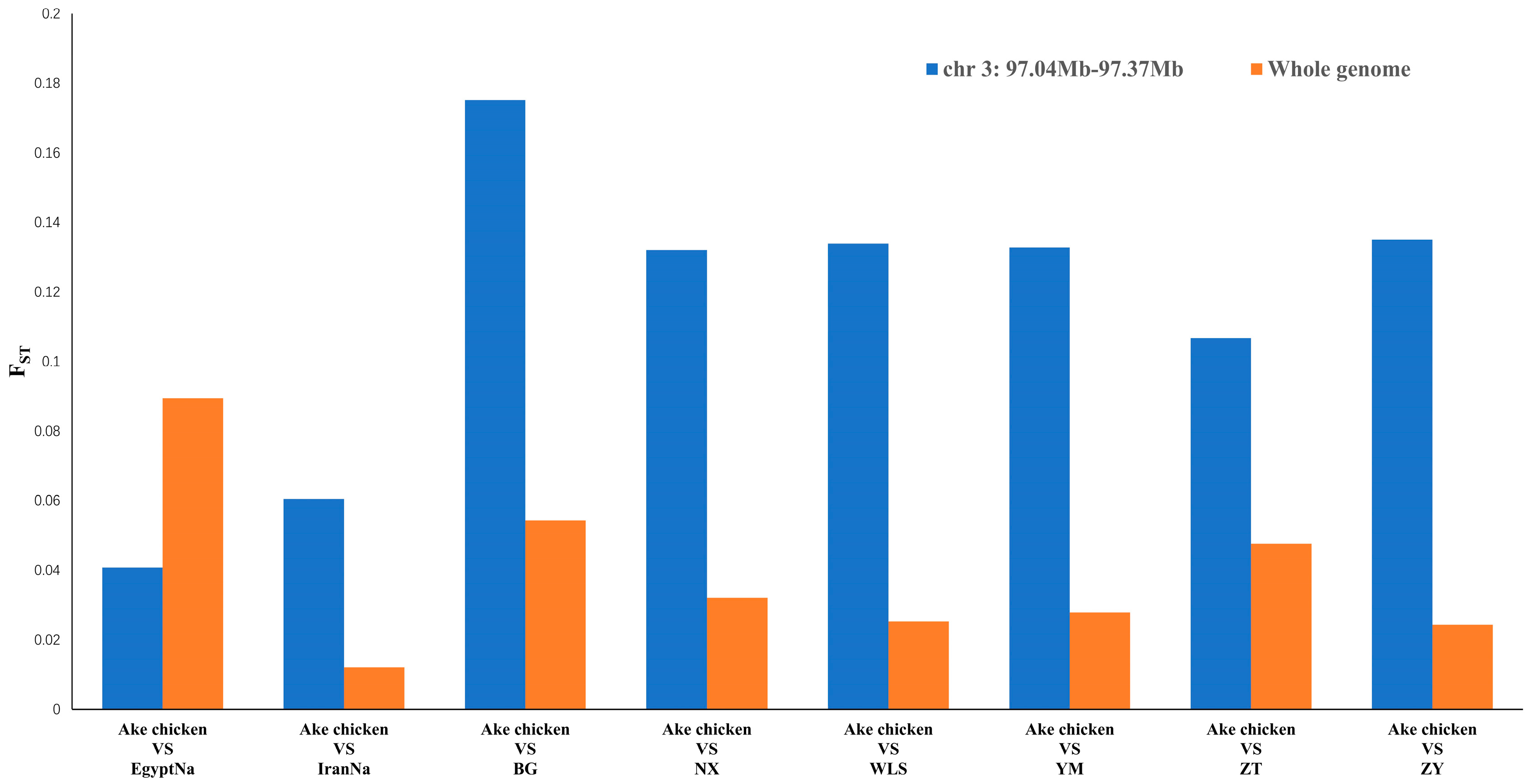
2.3. Selective Sweep in the Naked-Neck Trait Locus
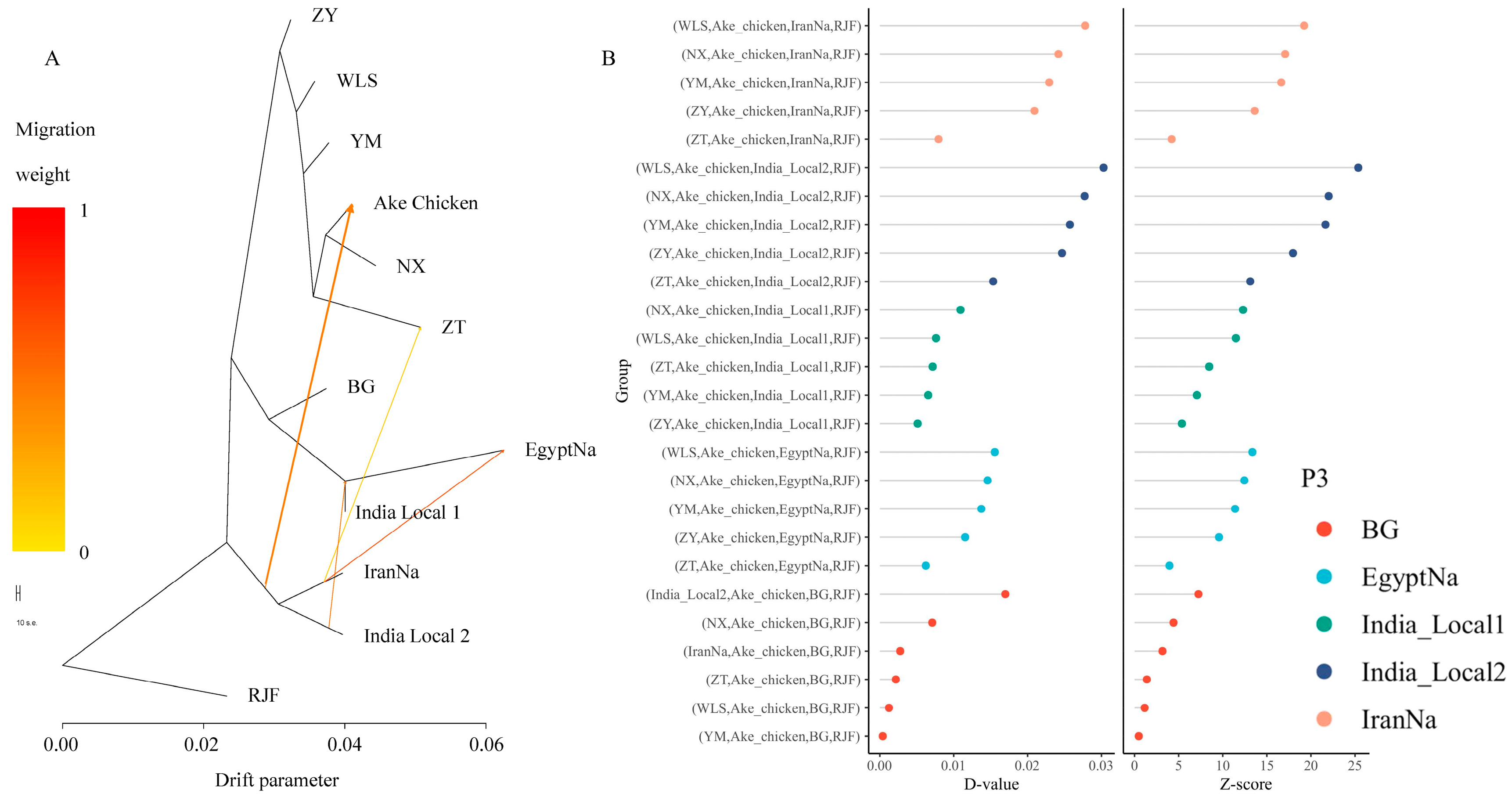
2.4. Gene Flow Analysis
2.5. Effective Population Size and Population Separation History Estimation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Ethical Statement
5.2. Sample and Datasets
5.3. DNA Extraction
5.4. Whole-Genome Sequencing, Reads Mapping, and Single-Nucleotide Polymorphism (SNP) Detection
5.5. Population Phylogenetic and Structure Analysis
5.6. Detection of Selection Signatures
5.7. Migration Events and Admixture Analysis
5.8. Effective Population Size and Population Separation History Estimation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brugaletta, G.; Teyssier, J.R.; Rochell, S.J.; Dridi, S.; Sirri, F. A review of heat stress in chickens. Part I: Insights into physiology and gut health. Front. Physiol. 2022, 13, 934381. [Google Scholar] [CrossRef] [PubMed]
- Apalowo, O.O.; Ekunseitan, D.A.; Fasina, Y.O. Impact of Heat Stress on Broiler Chicken Production. Poultry 2024, 3, 107–128. [Google Scholar] [CrossRef]
- Cahaner, A.; Leenstra, F. Effects of High-Temperature on Growth and Efficiency of Male and Female Broilers from Lines Selected for High Weight-Gain, Favorable Feed Conversion, and High or Low Fat-Content. Poult. Sci. 1992, 71, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Merat, P. Potential Usefulness of the Na (Naked-Neck) Gene in Poultry Production. World Poult. Sci. J. 1986, 42, 124–142. [Google Scholar] [CrossRef]
- Ladjali, K.; Tixier-Boichard, M.; Bordas, A.; Merat, P. Cytogenetic study of early chicken embryos: Effect of naked neck gene and high ambient temperature. Poult. Sci. 1995, 74, 903–909. [Google Scholar] [CrossRef]
- Cai, R.; Yin, Z.; Gu, S.; Deng, X.; Darwish, H.Y.A.; Sun, C.; Yang, N. Research note: Genetic basis of the naked-neck trait in ake chickens revealed by whole-genome sequencing. Poult. Sci. 2025, 104, 104934. [Google Scholar] [CrossRef] [PubMed]
- Schiffels, S.; Durbin, R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 2014, 46, 919–925. [Google Scholar] [CrossRef]
- Peng, Y.; Cai, X.; Wang, Y.; Liu, Z.; Zhao, Y. Genome-wide analysis suggests multiple domestication events of Chinese local pigs. Anim. Genet. 2022, 53, 293–306. [Google Scholar] [CrossRef]
- Hlongwane, N.L.; Hadebe, K.; Dzomba, E.F.; Muchadeyi, F.C. Genome Wide Assessment of Genetic Variation and Population Distinctiveness of the Pig Family in South Africa. Front. Genet. 2020, 11, 344. [Google Scholar] [CrossRef]
- Zhong, H.A.; Kong, X.Y.; Zhang, Y.W.; Su, Y.K.; Zhang, B.; Zhu, L.; Chen, H.; Gou, X.; Zhang, H. Microevolutionary mechanism of high-altitude adaptation in Tibetan chicken populations from an elevation gradient. Evol. Appl. 2022, 15, 2100–2112. [Google Scholar] [CrossRef]
- Bai, H.; Zhao, N.; Li, X.; Ding, Y.; Guo, Q.; Chen, G.; Chang, G. Whole-genome resequencing identifies candidate genes associated with heat adaptation in chickens. Poult. Sci. 2024, 103, 104139. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Lan, F.R.; Chen, X.M.; Yan, Y.Y.; Li, G.Q.; Wu, G.Q.; Sun, C.J.; Yang, N. Runs of homozygosity and selection signature analyses reveal putative genomic regions for artificial selection in layer breeding. BMC Genom. 2024, 25, 638. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Z.; Zhang, M.; Wang, S.; Gao, T.; Huang, H.; Zhang, T.; Cai, H.; Liu, X.; Fu, T.; et al. Population Structure and Selection Signal Analysis of Nanyang Cattle Based on Whole-Genome Sequencing Data. Genes 2024, 15, 351. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, L.; Gao, L.; Wei, Z.; Dang, D.; Yang, L. Population Structure and Genetic Diversity of Yunling Cattle Determined by Whole-Genome Resequencing. Genes 2023, 14, 2141. [Google Scholar] [CrossRef]
- Lv, F.H.; Cao, Y.H.; Liu, G.J.; Luo, L.Y.; Lu, R.; Liu, M.J.; Li, W.R.; Zhou, P.; Wang, X.H.; Shen, M.; et al. Whole-Genome Resequencing of Worldwide Wild and Domestic Sheep Elucidates Genetic Diversity, Introgression, and Agronomically Important Loci. Mol. Biol. Evol. 2022, 39, msab353. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.R.; Lv, F.H.; He, S.G.; Tian, S.L.; Peng, W.F.; Sun, Y.W.; Zhao, Y.X.; Tu, X.L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef]
- Wang, M.S.; Thakur, M.; Peng, M.S.; Jiang, Y.; Frantz, L.A.F.; Li, M.; Zhang, J.J.; Wang, S.; Peters, J.; Otecko, N.O.; et al. 863 genomes reveal the origin and domestication of chicken. Cell Res. 2020, 30, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Lin, D.; Wang, Y.Z.; Peng, D.Z.; Li, H.F.; Fei, J.; Chen, K.W.; Yang, N.; Hu, X.X.; Zhao, Y.Q.; et al. Widespread introgression in Chinese indigenous chicken breeds from commercial broiler. Evol. Appl. 2019, 12, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.; Richter, K.K.; Wilkin, S.; Stark, S.; Mir-Makhamad, B.; Fernandes, R.; Maksudov, F.; Mirzaakhmedov, S.; Rahmonov, H.; Schirmer, S.; et al. Archaeological and molecular evidence for ancient chickens in Central Asia. Nat. Commun. 2024, 15, 2697. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Proc, G.P.D. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics. 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data. PLoS Genet. 2012, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.Y.; Patterson, N.; Reich, D.; Slatkin, M. Testing for Ancient Admixture between Closely Related Populations. Mol. Biol. Evol. 2011, 28, 2239–2252. [Google Scholar] [CrossRef]
- Malinsky, M.; Matschiner, M.; Svardal, H. Dsuite—Fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 2021, 21, 584–595. [Google Scholar] [CrossRef]
- Terhorst, J.; Kamm, J.A.; Song, Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2017, 49, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.; Mugal, C.; Nabholz, B.; Schielzeth, H.; Wolf, J.B.; Backstrom, N.; Kunstner, A.; Balakrishnan, C.N.; Heger, A.; Ponting, C.P.; et al. Molecular evolution of genes in avian genomes. Genome Biol. 2010, 11, R68. [Google Scholar] [CrossRef] [PubMed]



| Regional Classification of Breeds | Breeds | Number | |
|---|---|---|---|
| Southwest China | Native breeds from Yunnan Province | Ake chicken | 30 |
| Nixi chicken (NX) | 10 | ||
| Zhaotong chicken (ZT) | 10 | ||
| Yimen chicken (YM) | 10 | ||
| Zhenyuan chicken (ZY) | 10 | ||
| Wuliangshan chicken (WLS) | 10 | ||
| Banna game chicken (BG) | 10 | ||
| Chahua chicken (CH) | 25 | ||
| Red jungle fowl (RJF) | 10 | ||
| Native breeds from Tibet Autonomous Region | Shannan chicken (TSN) | 10 | |
| Linzhi chicken (LZ) | 9 | ||
| Gongbujiangda chicken (GB) | 10 | ||
| Native breeds from Guangxi Province | Yao chicken (Yao) | 14 | |
| Longsheng Feng Chicken (LSFC) | 13 | ||
| South China | Native breeds from Guangdong Province | Huiyang beard chicken (HYB) | 14 |
| Huaixiang chicken (HX) | 10 | ||
| Xinghua chicken (XH) | 10 | ||
| Native breed from Hainan Province | Wenchang chicken (WC) | 14 | |
| Other countries | Foreign breeds | Indian local chicken 1 | 13 |
| Indian local chicken 2 | 22 | ||
| Iranian Naked-neck chicken (IranNa) | 3 | ||
| Egyptian Naked-neck chicken (EgyptNa) | 30 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, R.; Gu, S.; Zhang, B.; Deng, X.; Abdelfattah, M.G.; Yang, N.; Darwish, H.Y.A.; Sun, C. Genetic and Evolutionary Analysis of Ake Chicken: New Insights into China’s Sole Indigenous Naked-Neck Chicken Breed. Int. J. Mol. Sci. 2025, 26, 4399. https://doi.org/10.3390/ijms26094399
Cai R, Gu S, Zhang B, Deng X, Abdelfattah MG, Yang N, Darwish HYA, Sun C. Genetic and Evolutionary Analysis of Ake Chicken: New Insights into China’s Sole Indigenous Naked-Neck Chicken Breed. International Journal of Molecular Sciences. 2025; 26(9):4399. https://doi.org/10.3390/ijms26094399
Chicago/Turabian StyleCai, Ronglang, Shuang Gu, Boxuan Zhang, Xuemei Deng, Mostafa Galal Abdelfattah, Ning Yang, Hesham Y. A. Darwish, and Congjiao Sun. 2025. "Genetic and Evolutionary Analysis of Ake Chicken: New Insights into China’s Sole Indigenous Naked-Neck Chicken Breed" International Journal of Molecular Sciences 26, no. 9: 4399. https://doi.org/10.3390/ijms26094399
APA StyleCai, R., Gu, S., Zhang, B., Deng, X., Abdelfattah, M. G., Yang, N., Darwish, H. Y. A., & Sun, C. (2025). Genetic and Evolutionary Analysis of Ake Chicken: New Insights into China’s Sole Indigenous Naked-Neck Chicken Breed. International Journal of Molecular Sciences, 26(9), 4399. https://doi.org/10.3390/ijms26094399






