Role and Potential of Artificial Intelligence in Biomarker Discovery and Development of Treatment Strategies for Amyotrophic Lateral Sclerosis
Abstract
1. Introduction
2. Methodology
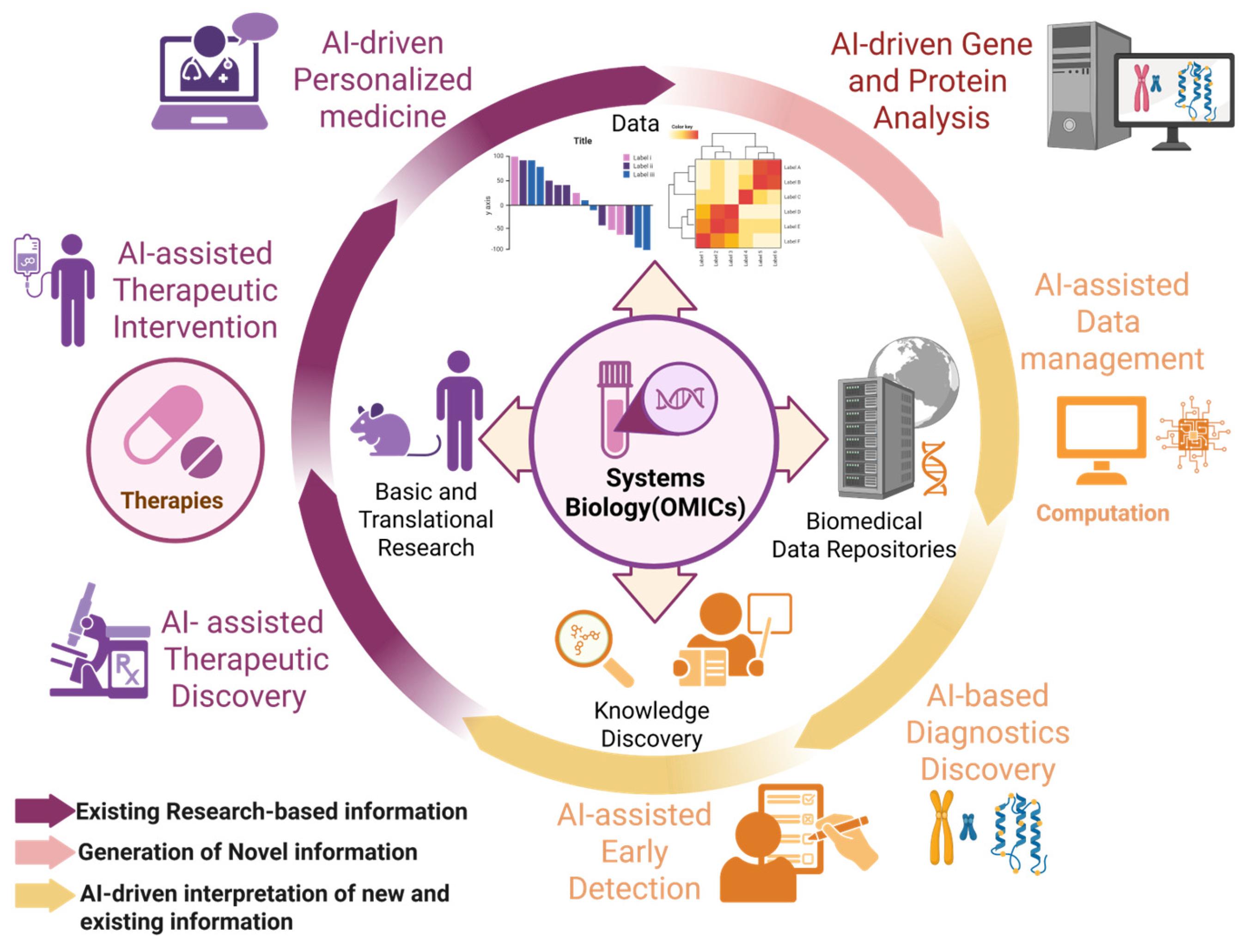
3. AI for Screening and Diagnosis of ALS
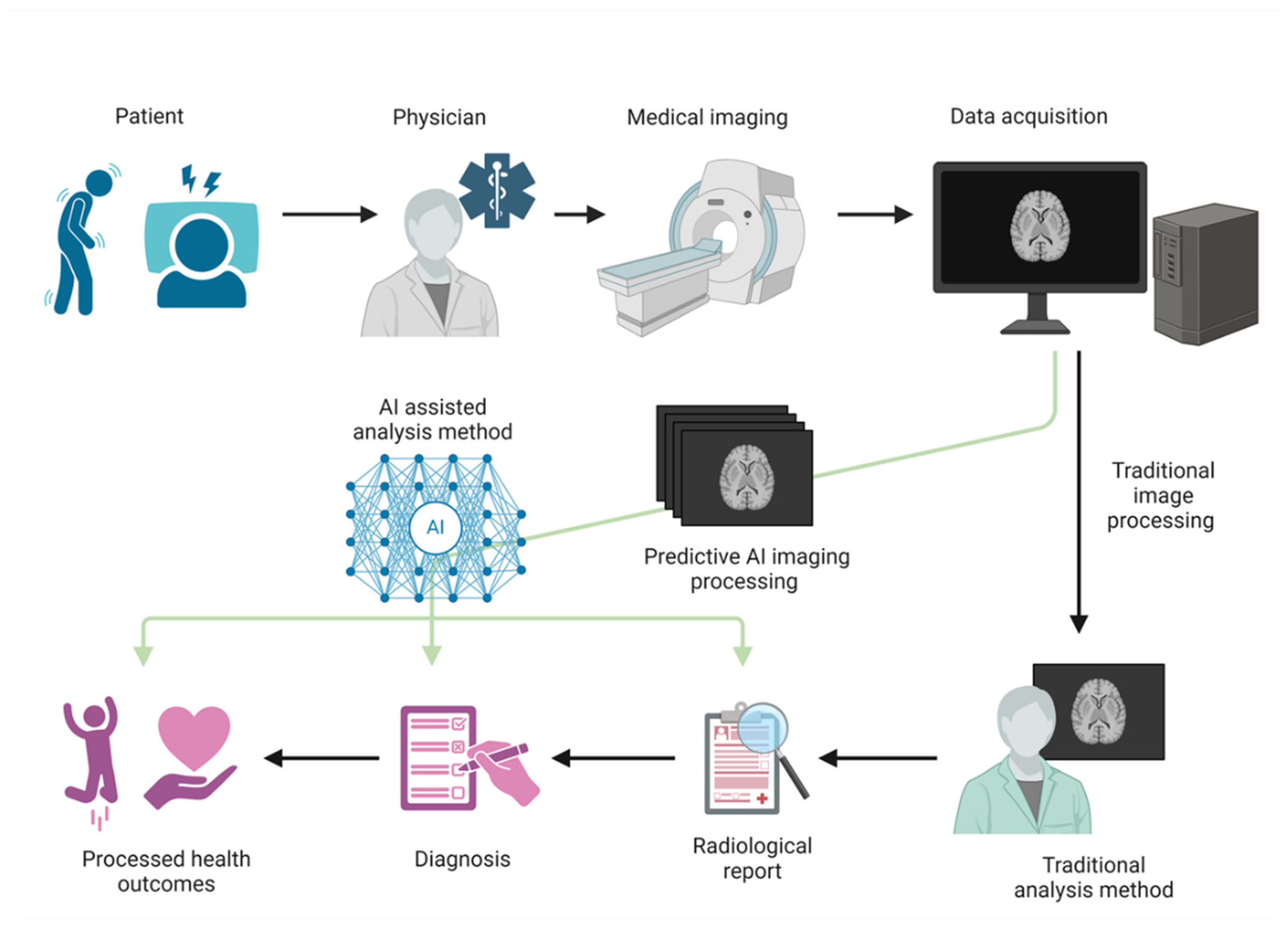
3.1. Diagnosis of Neurodegenerative Diseases
3.2. Integration and Impact of AI in ALS Diagnosis
4. Application of AI in ALS Biomarkers
4.1. Role of Biomarkers in Neurodegenerative Diseases
4.2. Role of Biomarkers in ALS
4.3. Research on the Development of Biomarkers for ALS Using AI
5. New Approaches to ALS Treatment
5.1. Development of Future ALS Treatment Methods
5.2. Development of Therapeutic Strategies Using AI
5.3. Progress in Personalized Medicine
5.4. Advantages and Limitations of AI Technology
5.5. Future Impact of AI on ALS Treatment Strategies
5.6. Issues in Translational Research Using AI
5.7. Future Prospects for AI Technology in ALS
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Menken, M.; Munsat, T.L.; Toole, J.F. The global burden of disease study: Implications for neurology. Arch. Neurol. 2000, 57, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.-M.; Hong, J.-S. Why neurodegenerative diseases are progressive: Uncontrolled inflammation drives disease progression. Trends Immunol. 2008, 29, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Fakhoury, M. Role of immunity and inflammation in the pathophysiology of neurodegenerative diseases. Neurodegener. Dis. 2015, 15, 63–69. [Google Scholar] [CrossRef]
- Taylor, J.P.; Brown, R.H.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Seki, S.; Kitaoka, Y.; Kawata, S.; Nishiura, A.; Uchihashi, T.; Hiraoka, S.-I.; Yokota, Y.; Isomura, E.T.; Kogo, M.; Tanaka, S. Characteristics of sensory neuron dysfunction in amyotrophic lateral sclerosis (ALS): Potential for ALS therapy. Biomedicines 2023, 11, 2967. [Google Scholar] [CrossRef]
- Kitaoka, Y.; Seki, S.; Kawata, S.; Nishiura, A.; Kawamura, K.; Hiraoka, S.-I.; Kogo, M.; Tanaka, S. Analysis of feeding behavior characteristics in the Cu/Zn Superoxide Dismutase 1 (SOD1) SOD1G93A Mice Model for Amyotrophic Lateral Sclerosis (ALS). Nutrients 2023, 15, 1651. [Google Scholar] [CrossRef]
- Doroszkiewicz, J.; Groblewska, M.; Mroczko, B. Molecular biomarkers and their implications for the early diagnosis of selected neurodegenerative diseases. Int. J. Mol. Sci. 2022, 23, 4610. [Google Scholar] [CrossRef]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef]
- Allegra, A.; Mirabile, G.; Tonacci, A.; Genovese, S.; Pioggia, G.; Gangemi, S. Machine learning approaches in diagnosis, prognosis and treatment selection of cardiac amyloidosis. Int. J. Mol. Sci. 2023, 24, 5680. [Google Scholar] [CrossRef]
- Gligorijevic, J.; Gligorijevic, D.; Pavlovski, M.; Milkovits, E.; Glass, L.; Grier, K.; Vankireddy, P.; Obradovic, Z. Optimizing clinical trials recruitment via deep learning. J. Am. Med. Inform. Assoc. 2019, 26, 1195–1202. [Google Scholar] [CrossRef]
- Wu, K.; Wu, E.; Dandrea, M.; Chitale, N.; Lim, M.; Dąbrowski, M.; Kantor, K.; Rangi, H.; Liu, R.; Garmhausen, M.; et al. Machine learning prediction of clinical trial operational efficiency. AAPS J. 2022, 24, 57. [Google Scholar] [CrossRef] [PubMed]
- Vora, L.K.; Gholap, A.D.; Jetha, K.; Thakur, R.R.S.; Solanki, H.K.; Chavda, V.P. Artificial intelligence in pharmaceutical technology and drug delivery design. Pharmaceutics 2023, 15, 1916. [Google Scholar] [CrossRef] [PubMed]
- Hessler, G.; Baringhaus, K.-H. Artificial intelligence in drug design. Molecules 2018, 23, 2520. [Google Scholar] [CrossRef]
- Gautam, R.; Sharma, M. Prevalence and diagnosis of neurological disorders using different deep learning techniques: A meta-analysis. J. Med. Syst. 2020, 44, 49. [Google Scholar] [CrossRef]
- Lan, A.Y.; Corces, M.R. Deep learning approaches for noncoding variant prioritization in neurodegenerative diseases. Front. Aging Neurosci. 2022, 14, 1027224. [Google Scholar] [CrossRef]
- Selvi, K.; Kalaivani, K.S.; Ramesh, H. Alzheimer’s Disease Prediction Using Deep Learning. In Proceedings of the International Conference on Circuit Power and Computing Technologies (ICCPCT), Kollam, India, 10–11 August 2023; Volume 2023, pp. 1161–1164. [Google Scholar] [CrossRef]
- Antonelli, L.; Guarracino, M.R.; Maddalena, L.; Sangiovanni, M. Integrating imaging and omics data: A review. Biomed. Signal Process. Control 2019, 52, 264–280. [Google Scholar] [CrossRef]
- Mead, R.J.; Shan, N.; Reiser, H.J.; Marshall, F.; Shaw, P.J. Amyotrophic lateral sclerosis: A neurodegenerative disorder poised for successful therapeutic translation. Nat. Rev. Drug Discov. 2023, 22, 185–212. [Google Scholar] [CrossRef]
- Kaur, S.; Singla, J.; Nkenyereye, L.; Jha, S.; Prashar, D.; Joshi, G.P.; El-Sappagh, S.; Islam, M.S.; Islam, S.M.R. Medical diagnostic systems using artificial intelligence (AI) algorithms: Principles and perspectives. IEEE Access 2020, 8, 228049–228069. [Google Scholar] [CrossRef]
- Papi, M.; Caracciolo, G. Principal component analysis of personalized biomolecular corona data for early disease detection. Nano Today 2018, 21, 14–17. [Google Scholar] [CrossRef]
- Afzal, H.M.R.; Luo, S.; Ramadan, S.; Lechner-Scott, J. The emerging role of artificial intelligence in multiple sclerosis imaging. Mult. Scler. 2022, 28, 849–858. [Google Scholar] [CrossRef]
- de Jonge, S.; Potters, W.V.; Verhamme, C. Artificial intelligence for automatic classification of needle EMG signals: A scoping review. Clin. Neurophysiol. 2024, 159, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Kocar, T.D.; Müller, H.-P.; Ludolph, A.C.; Kassubek, J. Feature selection from magnetic resonance imaging data in ALS: A systematic review. Ther. Adv. Chronic Dis. 2021, 12, 20406223211051002. [Google Scholar] [CrossRef] [PubMed]
- Muhammed, S.; Upadhya, J.; Poudel, S.; Hasan, M.; Donthula, K.; Vargas, J.; Ranganathan, J.; Poudel, K. Improved Classification of Alzheimer’s Disease With Convolutional Neural Networks. In Proceedings of the 2023 IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 2 December 2023; pp. 1–7. [Google Scholar] [CrossRef]
- Sarica, A.; Cerasa, A.; Valentino, P.; Yeatman, J.; Trotta, M.; Barone, S.; Granata, A.; Nisticó, R.; Perrotta, P.; Pucci, F.; et al. The corticospinal tract profile in amyotrophic lateral sclerosis. Hum. Brain Mapp. 2017, 38, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
- Jiang, X.; Zhao, J.; Qian, W.; Song, W.; Lin, G.N. A generative adversarial network model for disease gene prediction with RNA-Seq data. IEEE Access 2020, 8, 37352–37360. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, J.-G.; Lin, D.; Zhang, L.; Shen, H.; Deng, H.-W. A systemic analysis of transcriptomic and epigenomic data to reveal regulation patterns for complex disease. G3 Genes Genomes Genet. 2017, 7, 2271–2279. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mubeen, H.; Masood, A.; Zafar, A.; Khan, Z.Q.; Khan, M.Q.; Nisa, A.U. Insights into AlphaFold’s Breakthrough in Neurodegenerative Diseases. Ir. J. Med. Sci. 2024, 193, 2577–2588. [Google Scholar] [CrossRef]
- Davis, S.A.; Itaman, S.; Khalid-Janney, C.M.; Sherard, J.A.; Dowell, J.A.; Cairns, N.J.; Gitcho, M.A. TDP-43 interacts with mitochondrial proteins critical for mitophagy and mitochondrial dynamics. Neurosci. Lett. 2018, 678, 8–15. [Google Scholar] [CrossRef]
- Van Deerlin, V.M.; Leverenz, J.B.; Bekris, L.M.; Bird, T.D.; Yuan, W.; Elman, L.B.; Clay, D.; Wood, E.M.; Chen-Plotkin, A.S.; Martinez-Lage, M.; et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: A genetic and histopathological analysis. Lancet Neurol. 2008, 7, 409–416. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhang, F.; Xu, L.; Yue, L.; Kon, O.L.; Zhu, Y.; Guo, T. High-throughput proteomics and AI for cancer biomarker discovery. Adv. Drug Deliv. Rev. 2021, 176, 113844. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.R.; Verstraete, E. What does imaging reveal about the pathology of amyotrophic lateral sclerosis? Curr. Neurol. Neurosci. Rep. 2015, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Barritt, A.W.; Gabel, M.C.; Cercignani, M.; Leigh, P.N. Emerging magnetic resonance imaging techniques and analysis methods in amyotrophic lateral sclerosis. Front. Neurol. 2018, 9, 1065. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.M.; Hildebrand, D.G.C.; Jeong, W.-K. FusionNet: A deep fully residual convolutional neural network for image segmentation in connectomics. Front. Comput. Sci. 2021, 3, 613981. [Google Scholar] [CrossRef]
- Fornito, A.; Zalesky, A.; Breakspear, M. The connectomics of brain disorders. Nat. Rev. Neurosci. 2015, 16, 159–172. [Google Scholar] [CrossRef]
- Umar, T.P.; Jain, N.; Papageorgakopoulou, M.; Shaheen, R.S.; Alsamhori, J.F.; Muzzamil, M.; Kostiks, A. Artificial intelligence for screening and diagnosis of amyotrophic lateral sclerosis: A systematic review and meta-analysis. Amyotroph. Lateral Scler. Front. Degener. 2024, 25, 425–436. [Google Scholar] [CrossRef]
- Tavazzi, E.; Longato, E.; Vettoretti, M.; Aidos, H.; Trescato, I.; Roversi, C.; Martins, A.S.; Castanho, E.N.; Branco, R.; Soares, D.F.; et al. Artificial intelligence and statistical methods for stratification and prediction of progression in amyotrophic lateral sclerosis: A systematic review. Artif. Intell. Med. 2023, 142, 102588. [Google Scholar] [CrossRef]
- Boyce, D.; Raymond, J.; Larson, T.C.; Kirkland, E.; Horton, D.K.; Mehta, P. What do you think caused your ALS? An analysis of the CDC national amyotrophic lateral sclerosis patient registry qualitative risk factor data using artificial intelligence and qualitative methodology. Amyotroph. Lateral Scler. Front. Degener. 2024, 25, 615–624. [Google Scholar] [CrossRef]
- Behler, A.; Müller, H.-P.; Ludolph, A.C.; Kassubek, J. Diffusion tensor imaging in amyotrophic lateral sclerosis: Machine learning for biomarker development. Int. J. Mol. Sci. 2023, 24, 1911. [Google Scholar] [CrossRef]
- McGown, A.; Stopford, M.J. High-throughput drug screens for amyotrophic lateral sclerosis drug discovery. Expert Opin. Drug Discov. 2018, 13, 1015–1025. [Google Scholar] [CrossRef]
- Pinto, S.; Quintarelli, S.; Silani, V. New technologies and amyotrophic lateral sclerosis—Which step forward rushed by the COVID-19 pandemic? J. Neurol. Sci. 2020, 418, 117081. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, F.; Barbalho, I.; Barros, D.; Valentim, R.; Teixeira, C.; Henriques, J.; Gil, P.; Dourado Júnior, M. Biomedical signals and machine learning in amyotrophic lateral sclerosis: A systematic review. Biomed. Eng. Online 2021, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Cooper-Knock, J.; Harvey, C.; Zhang, S.; Moll, T.; Timpanaro, I.S.; Kenna, K.P.; Iacoangeli, A.; Veldink, J.H. Advances in the genetic classification of amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2021, 34, 756–764. [Google Scholar] [CrossRef]
- Johnson, K.A.; Minoshima, S.; Bohnen, N.I.; Donohoe, K.J.; Foster, N.L.; Herscovitch, P.; Karlawish, J.H.; Rowe, C.C.; Hedrick, S.; Pappas, V.; et al. Update on appropriate use criteria for amyloid PET imaging: Dementia experts, mild cognitive impairment, and education. Amyloid Imaging Task Force of the Alzheimer’s Association and Society for Nuclear Medicine and Molecular Imaging. Alzheimers Dement. 2013, 9, e106–e109. [Google Scholar] [CrossRef]
- Grimmer, T.; Wutz, C.; Alexopoulos, P.; Drzezga, A.; Förster, S.; Förstl, H.; Goldhardt, O.; Ortner, M.; Sorg, C.; Kurz, A. Visual versus fully automated analyses of 18F-FDG and amyloid PET for prediction of dementia due to Alzheimer disease in mild cognitive impairment. J. Nucl. Med. 2016, 57, 204–207. [Google Scholar] [CrossRef]
- Tai, Y.F.; Piccini, P. Applications of positron emission tomography (PET) in neurology. J. Neurol. Neurosurg. Psychiatry 2004, 75, 669–676. [Google Scholar] [CrossRef]
- Rocchi, L.; Niccolini, F.; Politis, M. Recent imaging advances in neurology. J. Neurol. 2015, 262, 2182–2194. [Google Scholar] [CrossRef]
- Rachakonda, V.; Pan, T.H.; Le, W.D. Biomarkers of neurodegenerative disorders: How good are they? Cell Res. 2004, 14, 347–358. [Google Scholar] [CrossRef]
- Ishii, K. PET approaches for diagnosis of dementia. AJNR Am. J. Neuroradiol. 2014, 35, 2030–2038. [Google Scholar] [CrossRef]
- Maschke, M.; Kastrup, O.; Forsting, M.; Diener, H.-C. Update on neuroimaging in infectious central nervous system disease. Curr. Opin. Neurol. 2004, 17, 475–480. [Google Scholar] [CrossRef]
- Cuddon, P.A. Electrophysiology in neuromuscular disease. Vet. Clin. N. Am. Small Anim. Pract. 2002, 32, 31–62. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Di Maria, G. Electrodiagnostic testing for disorders of peripheral nerves. Clin. Geriatr. Med. 2021, 37, 209–221. [Google Scholar] [CrossRef] [PubMed]
- DeBiasi, R.L.; Tyler, K.L. Molecular methods for diagnosis of viral encephalitis. Clin. Microbiol. Rev. 2004, 17, 903–925. [Google Scholar] [CrossRef] [PubMed]
- Mollenhauer, B.; Trenkwalder, C. Neurochemical biomarkers in the differential diagnosis of movement disorders. Mov. Disord. 2009, 24, 1411–1426. [Google Scholar] [CrossRef]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep learning applications in medical image analysis. IEEE Access 2018, 6, 9375–9389. [Google Scholar] [CrossRef]
- Kosvyra, A.; Filos, D.; Fotopoulos, D.; Olga, T.; Chouvarda, I. Towards data integration for AI in cancer research. In Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC 2021), Virtual, 1–5 November 2021; pp. 2054–2057. [Google Scholar] [CrossRef]
- Johnson, K.B.; Wei, W.-Q.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the future of personalized health care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef]
- Huber, R.G.; Pandey, S.; Chhangani, D.; Rincon-Limas, D.E.; Staff, N.P.; Yeo, C.J.J. Identification of potential pathways and biomarkers linked to progression in ALS. Ann. Clin. Transl. Neurol. 2023, 10, 150–165. [Google Scholar] [CrossRef]
- Imamura, K.; Izumi, Y.; Watanabe, A.; Tsukita, K.; Woltjen, K.; Yamamoto, T.; Hotta, A.; Kondo, T.; Kitaoka, S.; Ohta, A.; et al. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci. Transl. Med. 2017, 9, eaaf3962. [Google Scholar] [CrossRef]
- Gu, F.; Ma, S.; Wang, X.; Zhao, J.; Yu, Y.; Song, X. Evaluation of feature selection for Alzheimer’s disease diagnosis. Front. Aging Neurosci. 2022, 14, 924113. [Google Scholar] [CrossRef]
- Zhou, L.-Q.; Wang, J.-Y.; Yu, S.-Y.; Wu, G.-G.; Wei, Q.; Deng, Y.-B.; Wu, X.-L.; Cui, X.-W.; Dietrich, C.F. Artificial intelligence in medical imaging of the liver. World J. Gastroenterol. 2019, 25, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Khan, A. Transforming Healthcare through AI: Unleashing the Power of Personalized Medicine. Int. J. Multidiscip. Sci. Arts 2023, 2, 67–77. [Google Scholar] [CrossRef]
- Segura, T.; Medrano, I.H.; Collazo, S.; Maté, C.; Sguera, C.; Del Rio-Bermudez, C.; Casero, H.; Salcedo, I.; García-García, J.; Alcahut-Rodríguez, C. Symptoms timeline and outcomes in amyotrophic lateral sclerosis using artificial intelligence. Sci. Rep. 2023, 13, 702. [Google Scholar] [CrossRef] [PubMed]
- Bede, P.; Murad, A.; Hardiman, O. Pathological neural networks and artificial neural networks in ALS: Diagnostic classification based on pathognomonic neuroimaging features. J. Neurol. 2022, 269, 2440–2452. [Google Scholar] [CrossRef]
- Olsson, B.; Zetterberg, H.; Hampel, H.; Blennow, K. Biomarker-based dissection of neurodegenerative diseases. Prog. Neurobiol. 2011, 95, 520–534. [Google Scholar] [CrossRef]
- Wang, J.; Bettegowda, C. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J. Mol. Diagn. 2017, 19, 24–34. [Google Scholar] [CrossRef]
- Shahim, P.; Månsson, J.-E.; Darín, N.; Zetterberg, H.; Mattsson, N. Cerebrospinal fluid biomarkers in neurological diseases in children. Eur. J. Paediatr. Neurol. 2013, 17, 7–13. [Google Scholar] [CrossRef]
- Kwiatkowski, T.J.; Bosco, D.A.; Leclerc, A.L.; Tamrazian, E.; Vanderburg, C.R.; Russ, C.; Davis, A.; Gilchrist, J.; Kasarskis, E.J.; Munsat, T.; et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009, 323, 1205–1208. [Google Scholar] [CrossRef]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.X.; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef]
- Millecamps, S.; Boillée, S.; Le Ber, I.; Seilhean, D.; Teyssou, E.; Giraudeau, M.; Moigneu, C.; Vandenberghe, N.; Danel-Brunaud, V.; Corcia, P.; et al. Phenotype difference between ALS patients with expanded repeats in C9ORF72 and patients with mutations in other ALS-related genes. J. Med. Genet. 2012, 49, 258–263. [Google Scholar] [CrossRef]
- Magen, I.; Yacovzada, N.S.; Yanowski, E.; Coenen-Stass, A.; Grosskreutz, J.; Lu, C.-H.; Greensmith, L.; Malaspina, A.; Fratta, P.; Hornstein, E. Circulating miR-181 is a prognostic biomarker for amyotrophic lateral sclerosis. Nat. Neurosci. 2021, 24, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Abu-Rumeileh, S.; Vacchiano, V.; Zenesini, C.; Polischi, B.; de Pasqua, S.; Fileccia, E.; Mammana, A.; Di Stasi, V.; Capellari, S.; Salvi, F.; et al. Diagnostic-prognostic value and electrophysiological correlates of CSF biomarkers of neurodegeneration and neuroinflammation in amyotrophic lateral sclerosis. J. Neurol. 2020, 267, 1699–1708. [Google Scholar] [CrossRef] [PubMed]
- Verde, F.; Steinacker, P.; Weishaupt, J.H.; Kassubek, J.; Oeckl, P.; Halbgebauer, S.; Tumani, H.; von Arnim, C.A.F.; Dorst, J.; Feneberg, E.; et al. Neurofilament light chain in serum for the diagnosis of amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2019, 90, 157–164. [Google Scholar] [CrossRef]
- Lilo, E.; Wald-Altman, S.; Solmesky, L.J.; Ben Yaakov, K.; Gershoni-Emek, N.; Bulvik, S.; Kassis, I.; Karussis, D.; Perlson, E.; Weil, M. Characterization of human sporadic ALS biomarkers in the familial ALS transgenic mSOD1(G93A) mouse model. Hum. Mol. Genet. 2013, 22, 4720–4725. [Google Scholar] [CrossRef]
- Thal, L.J.; Kantarci, K.; Reiman, E.M.; Klunk, W.E.; Weiner, M.W.; Zetterberg, H.; Galasko, D.; Praticò, D.; Griffin, S.; Schenk, D.; et al. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2006, 20, 6–15. [Google Scholar] [CrossRef]
- Raghunathan, R.; Turajane, K.; Wong, L.C. Biomarkers in neurodegenerative diseases: Proteomics spotlight on ALS and Parkinson’s disease. Int. J. Mol. Sci. 2022, 23, 9299. [Google Scholar] [CrossRef]
- Banack, S.A.; Dunlop, R.A.; Mehta, P.; Mitsumoto, H.; Wood, S.P.; Han, M.; Cox, P.A. A microRNA diagnostic biomarker for amyotrophic lateral sclerosis. Brain Commun. 2024, 6, fcae268. [Google Scholar] [CrossRef]
- Wiersema, A.F.; Rennenberg, A.; Smith, G.; Varderidou-Minasian, S.; Pasterkamp, R.J. Shared and distinct changes in the molecular cargo of extracellular vesicles in different neurodegenerative diseases. Cell. Mol. Life Sci. 2024, 81, 479. [Google Scholar] [CrossRef]
- Carata, E.; Muci, M.; Di Giulio, S.; Di Giulio, T.; Mariano, S.; Panzarini, E. The neuromuscular disorder mediated by extracellular vesicles in amyotrophic lateral sclerosis. Curr. Issues Mol. Biol. 2024, 46, 5999–6017. [Google Scholar] [CrossRef]
- Seki, S.; Yamamoto, T.; Quinn, K.; Spigelman, I.; Pantazis, A.; Olcese, R.; Wiedau-Pazos, M.; Chandler, S.H.; Venugopal, S. Circuit-specific early impairment of proprioceptive sensory neurons in the SOD1G93A mouse model for ALS. J. Neurosci. 2019, 39, 8798–8815. [Google Scholar] [CrossRef]
- Statland, J.M.; Barohn, R.J.; Mcvey, A.L.; Katz, J.S.; Dimachkie, M.M. Patterns of weakness, classification of motor neuron disease, and clinical diagnosis of sporadic amyotrophic lateral sclerosis. Neurol. Clin. 2015, 33, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; Corr, E.M.; Logroscino, G.; Robberecht, W.; Shaw, P.J.; Simmons, Z.; van den Berg, L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Kawata, S.; Seki, S.; Nishiura, A.; Kitaoka, Y.; Iwamori, K.; Fukada, S.-I.; Kogo, M.; Tanaka, S. Preservation of masseter muscle until the end stage in the SOD1G93A mouse model for ALS. Sci. Rep. 2024, 14, 24279. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A. The spectrum of cognitive dysfunction in amyotrophic lateral sclerosis: An update. Int. J. Mol. Sci. 2023, 24, 14647. [Google Scholar] [CrossRef]
- Balendra, R.; Isaacs, A.M. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 2018, 14, 544–558. [Google Scholar] [CrossRef]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef]
- Bachiller, S.; Jiménez-Ferrer, I.; Paulus, A.; Yang, Y.; Swanberg, M.; Deierborg, T.; Boza-Serrano, A. Microglia in neurological diseases: A road map to brain-disease dependent-inflammatory response. Front. Cell. Neurosci. 2018, 12, 488. [Google Scholar] [CrossRef]
- Emde, A.; Eitan, C.; Liou, L.-L.; Libby, R.T.; Rivkin, N.; Magen, I.; Reichenstein, I.; Oppenheim, H.; Eilam, R.; Silvestroni, A.; et al. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: A new mechanism for ALS. EMBO J. 2015, 34, 2633–2651. [Google Scholar] [CrossRef]
- Gonzalez-Garza, M.T.; Martinez, H.R.; Cruz-Vega, D.E.; Hernandez-Torre, M.; Moreno-Cuevas, J.E. Adipsin, MIP-1b, and IL-8 as CSF biomarker panels for ALS diagnosis. Dis. Markers 2018, 2018, 3023826. [Google Scholar] [CrossRef]
- Frank, R.; Hargreaves, R. Clinical biomarkers in drug discovery and development. Nat. Rev. Drug Discov. 2003, 2, 566–580. [Google Scholar] [CrossRef]
- Török, N.; Tanaka, M.; Vécsei, L. Searching for peripheral biomarkers in neurodegenerative diseases: The tryptophan-kynurenine metabolic pathway. Int. J. Mol. Sci. 2020, 21, 9338. [Google Scholar] [CrossRef] [PubMed]
- van den Bos, M.A.J.; Geevasinga, N.; Higashihara, M.; Menon, P.; Vucic, S. Pathophysiology and diagnosis of ALS: Insights from advances in neurophysiological techniques. Int. J. Mol. Sci. 2019, 20, 2818. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; von Arnim, C.A.F.; Burnie, N.; Bozeat, S.; Cummings, J. Biomarkers in Alzheimer’s disease: Role in early and differential diagnosis and recognition of atypical variants. Alzheimer’s Res. Ther. 2023, 15, 175. [Google Scholar] [CrossRef] [PubMed]
- owser, R.; Turner, M.R.; Shefner, J. Biomarkers in amyotrophic lateral sclerosis: Opportunities and limitations. Nat. Rev. Neurol. 2011, 7, 631–638. [Google Scholar] [CrossRef]
- Borrebaeck, C.A.K. Precision diagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 2017, 17, 199–204. [Google Scholar] [CrossRef]
- Liu, A.; Schisterman, E.F.; Zhu, Y. On linear combinations of biomarkers to improve diagnostic accuracy. Stat. Med. 2005, 24, 37–47. [Google Scholar] [CrossRef]
- Nakamori, M.; Ishikawa, R.; Watanabe, T.; Toko, M.; Naito, H.; Takahashi, T.; Simizu, Y.; Yamazaki, Y.; Maruyama, H. Swallowing sound evaluation using an electronic stethoscope and artificial intelligence analysis for patients with amyotrophic lateral sclerosis. Front. Neurol. 2023, 14, 1212024. [Google Scholar] [CrossRef]
- Pancotti, C.; Birolo, G.; Rollo, C.; Sanavia, T.; Di Camillo, B.; Manera, U.; Chiò, A.; Fariselli, P. Deep learning methods to predict amyotrophic lateral sclerosis disease progression. Sci. Rep. 2022, 12, 13738. [Google Scholar] [CrossRef]
- Din Abdul Jabbar, M.A.; Guo, L.; Nag, S.; Guo, Y.; Simmons, Z.; Pioro, E.P.; Ramasamy, S.; Yeo, C.J.J. Predicting amyotrophic lateral sclerosis (ALS) progression with machine learning. Amyotroph. Lateral Scler. Front. Degener. 2024, 25, 242–255. [Google Scholar] [CrossRef]
- Awwalu, J.; Garba, A.G.; Ghazvini, A.; Atuah, R. Artificial intelligence in personalized medicine application of AI algorithms in solving personalized medicine problems. Int. J. Comput. Theor. Eng. 2015, 7, 439–443. [Google Scholar] [CrossRef]
- Schork, N.J. Artificial intelligence and personalized medicine. Cancer Treat. Res. 2019, 178, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhu, S.; Lu, W.; Liu, Z.; Huang, J.; Zhou, Y.; Fang, J.; Huang, Y.; Guo, H.; Li, L.; et al. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 2020, 11, 1775–1797. [Google Scholar] [CrossRef] [PubMed]
- Parekh, A.E.; Shaikh, O.A.; Simran, F.; Manan, S.; Hasibuzzaman, M.A. Artificial intelligence (AI) in personalized medicine: AI-generated personalized therapy regimens based on genetic and medical history: Short communication. Ann. Med. Surg. 2023, 85, 5831–5833. [Google Scholar] [CrossRef] [PubMed]
- Harwin, W.S.; Patton, J.L.; Edgerton, V.R. Challenges and opportunities for robot-mediated neurorehabilitation. Proc. IEEE 2006, 94, 1717–1726. [Google Scholar] [CrossRef]
- Sidyakina, I.V.; Voronova, M.V.; Ivanov, V.V.; Snopkov, P.S.; Epifanov, V.A. Questions of neurorehabilitation. Innovative technologies of neurorehabilitation. Technol. Neurorehab. Fizioter. 2020, 4, 61–65. [Google Scholar] [CrossRef]
- Himabindhu, G. Recent advances in neurorehabilitation an editorial. Int. J. Neurorehab. 2020, 7, 4. [Google Scholar] [CrossRef]
- Jaiswal, M.K. Riluzole and Edaravone: A tale of two amyotrophic lateral sclerosis drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef]
- Writing Group; Edaravone. Safety and efficacy of Edaravone in well defined patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017, 16, 505–512. [Google Scholar] [CrossRef]
- Nizzardo, M.; Simone, C.; Rizzo, F.; Ruggieri, M.; Salani, S.; Riboldi, G.; Faravelli, I.; Zanetta, C.; Bresolin, N.; Comi, G.P.; et al. Minimally invasive transplantation of iPSC-derived ALDHhiSSCloVLA4+ neural stem cells effectively improves the phenotype of an amyotrophic lateral sclerosis model. Hum. Mol. Genet. 2014, 23, 342–354. [Google Scholar] [CrossRef]
- Pun, F.W.; Liu, B.H.M.; Long, X.; Leung, H.W.; Leung, G.H.D.; Mewborne, Q.T.; Gao, J.; Shneyderman, A.; Ozerov, I.V.; Wang, J.; et al. Identification of therapeutic targets for amyotrophic lateral sclerosis using PandaOmics—An AI-enabled biological target discovery platform. Front. Aging Neurosci. 2022, 14, 914017. [Google Scholar] [CrossRef]
- Kaushik, A.; Jayant, R.D.; Bhardwaj, V.; Nair, M. Personalized nanomedicine for CNS diseases. Drug Discov. Today 2018, 23, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Todo, T.; Ito, H.; Ino, Y.; Ohtsu, H.; Ota, Y.; Shibahara, J.; Tanaka, M. Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: A phase 2 trial. Nat. Med. 2022, 28, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jou, T.H.T.; Hsin, J.; Wang, Z.; Huang, K.; Ye, J.; Yin, H.; Xing, Y. Talimogene Laherparepvec (T-VEC): A Review of the Recent Advances in Cancer Therapy. J. Clin. Med. 2023, 12, 1098. [Google Scholar] [CrossRef] [PubMed]
- Emani, S.; Rui, A.; Rocha, H.A.L.; Rizvi, R.F.; Juaçaba, S.F.; Jackson, G.P.; Bates, D.W. Physicians’ perceptions of and satisfaction with artificial intelligence in cancer treatment: A clinical decision support system experience and implications for low-middle-income countries. JMIR Cancer 2022, 8, e31461. [Google Scholar] [CrossRef]
- Founta, K.; Dafou, D.; Kanata, E.; Sklaviadis, T.; Zanos, T.P.; Gounaris, A.; Xanthopoulos, K. Gene targeting in amyotrophic lateral sclerosis using causality-based feature selection and machine learning. Mol. Med. 2023, 29, 12. [Google Scholar] [CrossRef]
- Doxakis, E. Therapeutic antisense oligonucleotides for movement disorders. Med. Res. Rev. 2021, 41, 2656–2688. [Google Scholar] [CrossRef]
- ElMallah, M.K.; Keeler-Klunk, A.; Zieger, M.; McCall, A.L.; Pucci, L.; Brown, R.H.; Mueller, C. Respiratory directed gene therapy prolongs survival in an ALS mouse model. FASEB J. 2019, 33, 843.11. [Google Scholar] [CrossRef]
- Bakkar, N.; Kovalik, T.; Lorenzini, I.; Spangler, S.; Lacoste, A.; Sponaugle, K.; Ferrante, P.; Argentinis, E.; Sattler, R.; Bowser, R. Artificial intelligence in neurodegenerative disease research: Use of IBM Watson to identify additional RNA-binding proteins altered in amyotrophic lateral sclerosis. Acta Neuropathol. 2018, 135, 227–247. [Google Scholar] [CrossRef]
- Seki, S.; Tanaka, S.; Yamada, S.; Tsuji, T.; Enomoto, A.; Ono, Y.; Chandler, S.H.; Kogo, M. Neuropeptide Y modulates membrane excitability in neonatal rat mesencephalic V neurons. J. Neurosci. Res. 2020, 98, 921–935. [Google Scholar] [CrossRef]
- Tanaka, S.; Tomita, I.; Seki, S.; Yamada, S.; Kogo, M.; Furusawa, K. Serotonergic modulation of slow inward rectification in mesencephalic trigeminal neurons. Brain Res. 2019, 1718, 126–136. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, F.; Tang, J.; Nussinov, R.; Cheng, F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health 2020, 2, e667–e676. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, K.; Chen, C.Z.; Marshall, R.E.; Mihatov, N.; Choi, Y.; Nguyen, D.-T.; Southall, N.; Chen, K.G.; Park, J.K.; Zheng, W. Advancing precision medicine with personalized drug screening. Drug Discov. Today 2019, 24, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Smalley, E. AI-powered drug discovery captures pharma Interest. Nat. Biotechnol. 2017, 35, 604–605. [Google Scholar] [CrossRef]
- Harrold, J.M.; Ramanathan, M.; Mager, D.E. Network-based approaches in drug discovery and early development. Clin. Pharmacol. Ther. 2013, 94, 651–658. [Google Scholar] [CrossRef]
- Aliyeva, A. Transhumanism: Integrating cochlear implants with artificial intelligence and the brain-machine interface. Cureus 2023, 15, e50378. [Google Scholar] [CrossRef]
- Migliorelli, L.; Scoppolini Massini, L.; Coccia, M.; Villani, L.; Frontoni, E.; Squartini, S. A deep learning-based Telemonitoring application to automatically assess oral diadochokinesis in patients with bulbar amyotrophic lateral sclerosis. Comput. Methods Programs Biomed. 2023, 242, 107840. [Google Scholar] [CrossRef]
- Card, N.S.; Wairagkar, M.; Iacobacci, C.; Hou, X.; Singer-Clark, T.; Willett, F.R.; Kunz, E.M.; Fan, C.; Vahdati Nia, M.; Deo, D.R.; et al. An accurate and rapidly calibrating speech neuroprosthesis. N. Engl. J. Med. 2024, 391, 609–618. [Google Scholar] [CrossRef]
- Azzaz, F.; Yahi, N.; Chahinian, H.; Fantini, J. The Epigenetic Dimension of Protein Structure is an intrinsic weakness of the AlphaFold program. Biomolecules 2022, 12, 1527. [Google Scholar] [CrossRef]
- Fantini, J.; Azzaz, F.; Di Scala, C.; Aulas, A.; Chahinian, H.; Yahi, N. Conformationally adaptive therapeutic peptides for diseases caused by intrinsically disordered proteins (IDPs). New paradigm for drug discovery: Target the target, not the arrow. Pharmacol. Therapeutics. 2025, 267, 108797. [Google Scholar] [CrossRef]
- Zhang, L.; Hong, H. Genomic discoveries and personalized medicine in neurological diseases. Pharmaceutics 2015, 7, 542–553. [Google Scholar] [CrossRef]
- Luo, M.; Lee, L.K.C.; Peng, B.; Choi, C.H.J.; Tong, W.Y.; Voelcker, N.H. Delivering the promise of gene therapy with nanomedicines in treating central nervous system diseases. Adv. Sci. 2022, 9, e2201740. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Hamad, S.; Shedeed, H.A.; Hussein, A.S. Enhanced deep learning model for personalized cancer treatment. IEEE Access 2022, 10, 106050–106058. [Google Scholar] [CrossRef]
- Kale, M.; Wankhede, N.; Pawar, R.; Ballal, S.; Kumawat, R.; Goswami, M.; Khalid, M.; Taksande, B.; Upaganlawar, A.; Umekar, M.; et al. AI-driven innovations in Alzheimer’s disease: Integrating early diagnosis, personalized treatment, and prognostic modeling. Aging Res. Rev. 2024, 101, 102497. [Google Scholar] [CrossRef] [PubMed]
- Marriott, H.; Kabiljo, R.; Hunt, G.P.; Khleifat, A.A.; Jones, A.; Troakes, C.; Project MinE ALS Sequencing Consortium; TargetALS Sequencing Consortium; Pfaff, A.L.; Quinn, J.P.; et al. Unsupervised Machine Learning Identifies Distinct ALS Molecular Subtypes in Post-Mortem Motor Cortex and Blood Expression Data. Acta Neuropathol. Commun. 2023, 11, 208. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef]
- van der Velden, B.H.M.; Kuijf, H.J.; Gilhuijs, K.G.A.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef]
- Sheu, R.-K.; Pardeshi, M.S. A survey on medical explainable AI (XAI): Recent progress, explainability approach, human interaction and scoring system. Sensors 2022, 22, 8068. [Google Scholar] [CrossRef]
- Pierce, R.; Sterckx, S.; Van Biesen, W. A riddle, wrapped in a mystery, inside an enigma: How semantic black boxes and opaque artificial intelligence confuse medical decision-making. Bioethics 2022, 36, 113–120. [Google Scholar] [CrossRef]
- Anderson, M.; Anderson, S.L. How Should AI Be developed, validated, and implemented in Patient Care? AMA J. Ethics 2019, 21, E125–E130. [Google Scholar] [CrossRef]
- Yu, F.; Moehring, A.; Banerjee, O.; Salz, T.; Agarwal, N.; Rajpurkar, P. Heterogeneity and predictors of the effects of AI assistance on radiologists. Nat. Med. 2024, 30, 837–849. [Google Scholar] [CrossRef]
- Lauraitis, A.; Maskeliūnas, R.; Damaševičius, R.; Połap, D.; Woźniak, M. A smartphone application for automated decision support in cognitive task based evaluation of central nervous system motor disorders. IEEE J. Biomed. Health Inform. 2019, 23, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- Elm, J.J.; Daeschler, M.; Bataille, L.; Schneider, R.; Amara, A.; Espay, A.J.; Afek, M.; Admati, C.; Teklehaimanot, A.; Simuni, T. Feasibility and utility of a clinician dashboard from wearable and mobile application Parkinson’s disease data. npj Digit. Med. 2019, 2, 95. [Google Scholar] [CrossRef] [PubMed]
- Papaiz, F.; Dourado, M.E.T.; Valentim, R.A.M.; de Morais, A.H.F.; Arrais, J.P. Machine learning solutions applied to amyotrophic lateral sclerosis prognosis: A review. Front. Comput. Sci. 2022, 4, 869140. [Google Scholar] [CrossRef]
- Musk, E.; Neuralink. An integrated brain-machine interface platform with thousands of channels. J. Med. Internet Res. 2019, 21, e16194. [Google Scholar] [CrossRef]
- Bartus, R.T.; Weinberg, M.S.; Samulski, R.J. Parkinson’s disease gene therapy: Success by design meets failure by efficacy. Mol. Ther. 2014, 22, 487–497. [Google Scholar] [CrossRef]
- Oliveira, A.A.; Haeser, A.; Pranke, P. Stem cell grafts as therapeutic tools for central nervous system disorders. Psychol. Neurosci. 2008, 1, 47–54. [Google Scholar] [CrossRef]
- Liyao, R. Design innovation driven by artificial intelligence AI multifunctional wheelchair design based on the needs of patients with ALS. J. Phys. Conf. Ser. 2021, 1880, 012022. [Google Scholar] [CrossRef]
- Haglin, J.M.; Jimenez, G.; Eltorai, A.E.M. Artificial neural networks in medicine. Health Technol. 2019, 9, 1–6. [Google Scholar] [CrossRef]

| Year | Authors | Title | Journal | Focus | Reference |
|---|---|---|---|---|---|
| 2018 | McGown & Stopford | High-Throughput Drug Screens (HTDS) or ALS Drug Discovery | Expert Opinion on Drug Discovery | Overview of HTDS methods and artificial intelligence (AI) applications in ALS drug discovery. | [42] |
| 2020 | Pinto et al. | New Technologies and Amyotrophic Lateral Sclerosis | Journal of Neurological Sciences | AI, telemedicine and assistive technologies accelerated by COVID-19. | [43] |
| 2021 | Fernandes et al. | Biomedical Signals and Machine Learning (ML) in Amyotrophic Lateral Sclerosis | BioMed Eng Online | ML applications in ALS diagnosis, communication, and survival prediction. | [44] |
| 2021 | Cooper-Knock et al. | Advances in the Genetic Classification of Amyotrophic Lateral Sclerosis | Current Opinion in Neurology | Genetic classification and ML models for understanding ALS. | [45] |
| 2022 | Behler et al. | Diffusion Tensor Imaging in ALS: Machine Learning for Biomarker Development | International Journal of Molecular Sciences | Use of diffusion tensor imaging (DTI) and ML for ALS biomarker discovery and stratification. | [41] |
| 2023 | Tavazzi et al. | AI and Statistical Methods for Stratification and Prediction of ALS | AI in Medicine | AI methods for stratification and prediction of ALS progression. | [39] |
| 2024 | Boyce et al. | What Do You Think Caused Your ALS? | ALS and Frontotemporal Degeneration | AI and qualitative methods to analyze patient-reported causes of ALS. | [40] |
| 2024 | Umar et al. | AI for Screening and Diagnosis of ALS | ALS and Frontotemporal Degeneration | Meta-analysis of AI tools for ALS screening and diagnosis. | [38] |
| Biomarker Type | Examples | Diagnostic Relevance | Associated Diseases | Details and Clinical Applications | References |
|---|---|---|---|---|---|
| Protein Biomarkers | Neurofilament light chain (NFL) | Indicator of axonal damage, correlates with disease severity | ALS, Alzheimer’s | Used in CSF and blood tests; prognostic marker for disease progression | [74,75,77] |
| Genetic Markers | TDP-43 | Linked to neuronal degeneration, found in cytoplasmic inclusions | ALS, Frontotemporal dementia | Identifies TDP-43 proteinopathies; aids in differential diagnosis | [31,76] |
| Mutations in SOD1 | Common genetic cause of familial ALS | ALS | Screening in at-risk populations; genetic counseling | [70,71] | |
| C9ORF72 expansions | Most common genetic variation in familial ALS and FTD | ALS, FTD | Helps in confirming familial cases; guides prognosis and management | [72] | |
| TDP-43 mutations | Implicated in ALS pathology, affects RNA processing | ALS | Useful for familial ALS cases; potential targets for therapy | [76] | |
| Molecular Biomarkers | Plasma cell-free miRNA | Non-invasive markers that reflect gene expression changes | ALS | Potential for early diagnosis and monitoring of disease progression | [73] |
| The cargo content of extracellular vesicles (EVs) | ALS | [79,80,81] | |||
| Electrophysiological Biomarkers | Motor Unit Number Estimation (MUNE) | Quantifies the number of functional motor units | ALS | Assesses disease progression and response to treatment in ALS | [78] |
| AI Application | Techniques Used | Description and Use Cases | Impact and Clinical Relevance | Associated Diseases | References |
|---|---|---|---|---|---|
| Diagnostic Imaging | Deep Learning, Convolutional Neural Networks | AI algorithms analyze MRI, PET scans to detect and quantify pathological changes | Enhances accuracy and speed of diagnosis | ALS, Alzheimer’s, Parkinson’s | [23,24,25] |
| Automated measurement of brain atrophy and detection of specific protein accumulations | Provides early-detection capabilities | ||||
| Drug Discovery | Machine Learning, Network Analysis | Identification of new drug targets and repurposing of existing drugs | Speeds up drug-discovery process, reduces costs | ALS, Alzheimer’s, Parkinson’s | [12,104] |
| AI-driven simulations predict drug interactions and effectiveness | Improves safety and efficacy of new drugs | ||||
| Clinical Trials | Deep Learning, Predictive Analytics | Optimization of clinical-trial design and participant selection | Increases efficiency and efficacy of trials | Neurodegenerative diseases | [10,11] |
| Real-time data analysis predicts treatment outcomes | Facilitates faster regulatory approvals | ||||
| Personalized Medicine | Machine Learning, Genomic Data Analysis | Customization of treatment plans based on patient genetic profiles | Enhances treatment effectiveness and reduces adverse effects | Neurodegenerative diseases | [102,103,105] |
| AI models predict disease progression and treatment responses | Allows timely adjustments to therapy | ||||
| Neurorehabilitation | AI-driven Robotics, Neurofeedback | AI algorithms control robotic devices for physical therapy | Improves motor function and recovery rates | Stroke, ALS, Parkinson’s | [106,107,108] |
| Neurofeedback techniques train patients to modify brain activity | Enhances cognitive rehabilitation | ||||
| Predictive Analytics | Machine Learning, Big Data Analysis | Analysis of large-scale health data to predict disease trends | Aids in public-health planning and resource allocation | Neurodegenerative diseases | [19,20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitaoka, Y.; Uchihashi, T.; Kawata, S.; Nishiura, A.; Yamamoto, T.; Hiraoka, S.-i.; Yokota, Y.; Isomura, E.T.; Kogo, M.; Tanaka, S.; et al. Role and Potential of Artificial Intelligence in Biomarker Discovery and Development of Treatment Strategies for Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2025, 26, 4346. https://doi.org/10.3390/ijms26094346
Kitaoka Y, Uchihashi T, Kawata S, Nishiura A, Yamamoto T, Hiraoka S-i, Yokota Y, Isomura ET, Kogo M, Tanaka S, et al. Role and Potential of Artificial Intelligence in Biomarker Discovery and Development of Treatment Strategies for Amyotrophic Lateral Sclerosis. International Journal of Molecular Sciences. 2025; 26(9):4346. https://doi.org/10.3390/ijms26094346
Chicago/Turabian StyleKitaoka, Yoshihiro, Toshihiro Uchihashi, So Kawata, Akira Nishiura, Toru Yamamoto, Shin-ichiro Hiraoka, Yusuke Yokota, Emiko Tanaka Isomura, Mikihiko Kogo, Susumu Tanaka, and et al. 2025. "Role and Potential of Artificial Intelligence in Biomarker Discovery and Development of Treatment Strategies for Amyotrophic Lateral Sclerosis" International Journal of Molecular Sciences 26, no. 9: 4346. https://doi.org/10.3390/ijms26094346
APA StyleKitaoka, Y., Uchihashi, T., Kawata, S., Nishiura, A., Yamamoto, T., Hiraoka, S.-i., Yokota, Y., Isomura, E. T., Kogo, M., Tanaka, S., Spigelman, I., & Seki, S. (2025). Role and Potential of Artificial Intelligence in Biomarker Discovery and Development of Treatment Strategies for Amyotrophic Lateral Sclerosis. International Journal of Molecular Sciences, 26(9), 4346. https://doi.org/10.3390/ijms26094346





