m6A Methylation Mediated Autophagy and Nucleotide-Binding Oligomerization Domain-like Receptors Signaling Pathway Provides New Insight into the Mitigation of Oxidative Damage by Mulberry Leaf Polysaccharides
Abstract
1. Introduction
2. Results
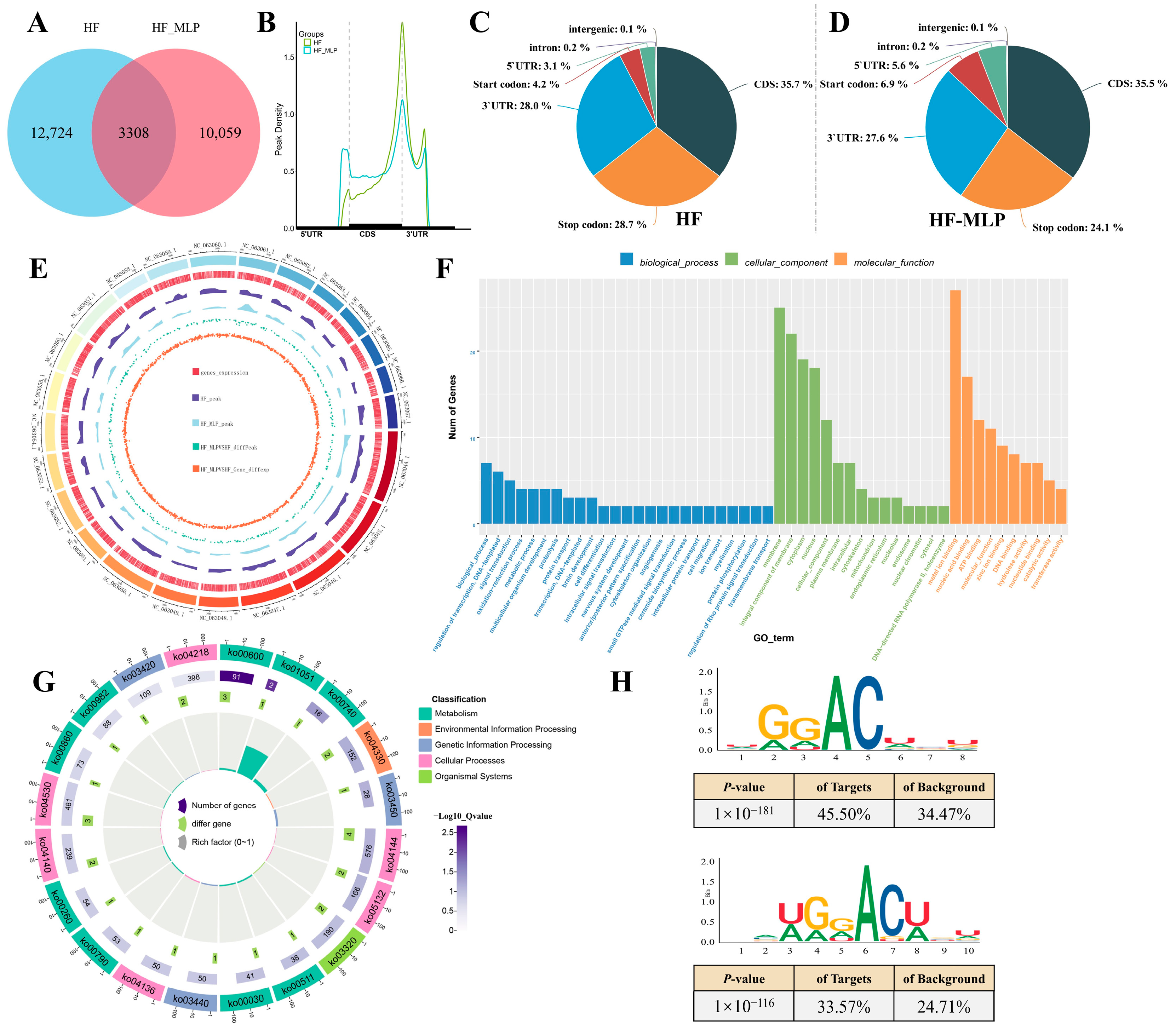
2.1. m6A Methylation Modification in Intestines of HF and HF-MLP Groups
2.2. Identification of DEGs in Intestinal of HF and HF-MLP Groups
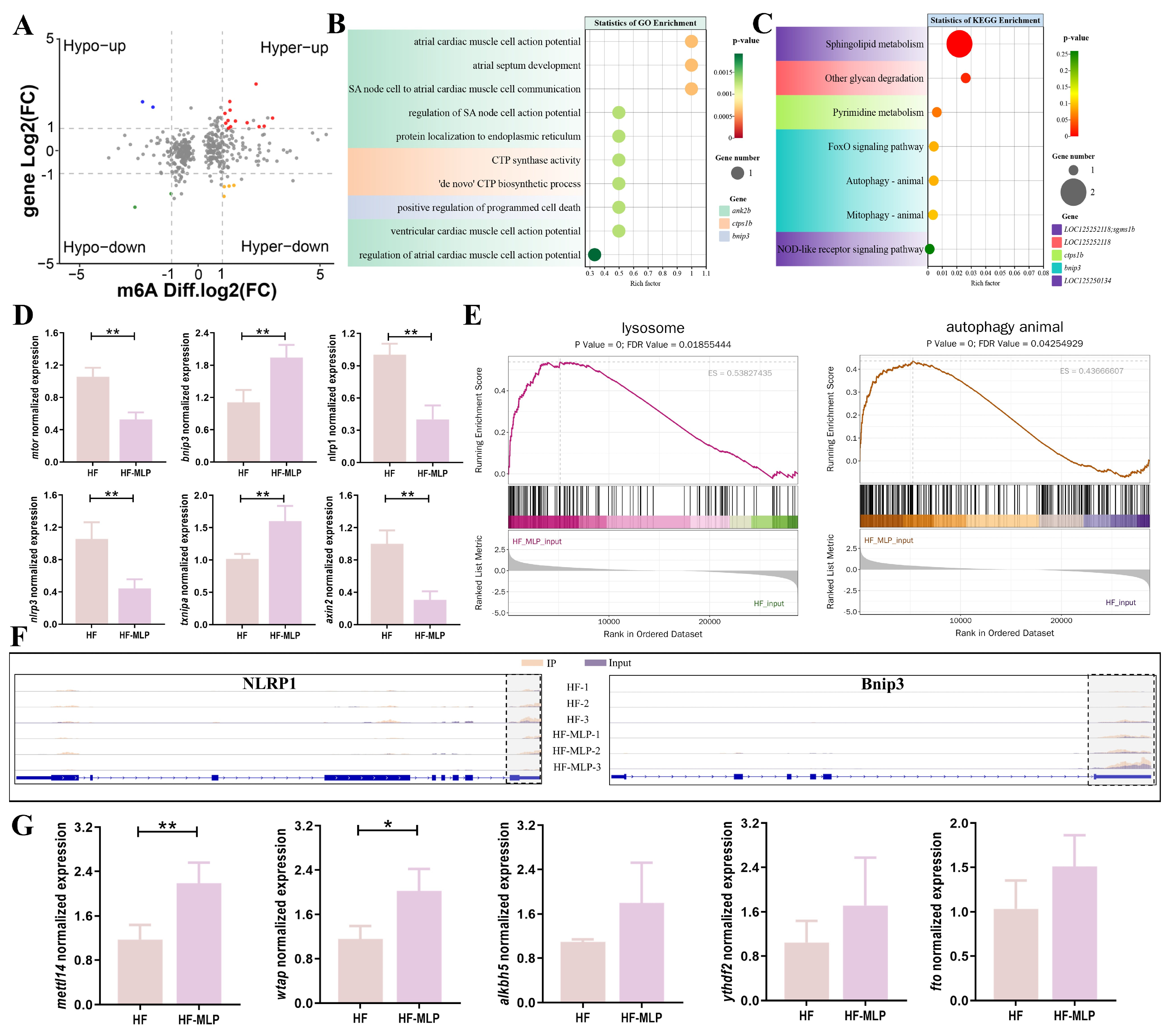
2.3. Assessment of the m6A-Modified Gene Transcription in Intestinal of HF and HF-MLP Groups
3. Discussion
4. Materials and Methods
4.1. MLPs
4.2. Diets
4.3. Fish and Feeding Trial
4.4. Sample Collection
4.5. RNA Isolation, Library Construction, and Sequencing
4.6. Bioinformatics Analysis
4.7. Real-Time PCR Analysis (qRT-PCR)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Bureau of Fisheries and Fishery Administration tMoA, National Fisheries Technology Extension Center, Fisheries. CSo. 2023 China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2023. [Google Scholar]
- Jiang, W.; Lin, Y.; Qian, L.; Lu, S.; Shen, H.; Ge, X.; Miao, L. Mulberry Leaf Polysaccharides Attenuate Oxidative Stress Injury in Peripheral Blood Leukocytes by Regulating Endoplasmic Reticulum Stress. Antioxidants 2024, 13, 136. [Google Scholar] [CrossRef]
- Ma, Q.; Li, L.Y.; Le, J.Y.; Lu, D.L.; Qiao, F.; Zhang, M.L.; Du, Z.Y.; Li, D.L. Dietary microencapsulated oil improves immune function and intestinal health in Nile tilapia fed with high-fat diet. Aquaculture 2018, 496, 19–29. [Google Scholar] [CrossRef]
- Tang, T.; Hu, Y.; Peng, M.; Chu, W.; Hu, Y.; Zhong, L. Effects of high-fat diet on growth performance, lipid accumulation and lipid metabolism-related MicroRNA/gene expression in the liver of grass carp (Ctenopharyngodon idella). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 234, 34–40. [Google Scholar] [CrossRef]
- Cao, X.F.; Dai, Y.J.; Liu, M.Y.; Yuan, X.Y.; Wang, C.C.; Huang, Y.Y.; Liu, W.B.; Jiang, G.Z. High-fat diet induces aberrant hepatic lipid secretion in blunt snout bream by activating endoplasmic reticulum stress-associated IRE1/XBP1 pathway. BBA-Mol. Cell Biol. Lipids 2019, 1864, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xia, T.; Yu, M.; Wang, L.; Song, K.; Zhang, C.; Lu, K. Hydroxytyrosol attenuates high-fat-diet-induced oxidative stress, apoptosis and inflammation of blunt snout bream (Megalobrama amblycephala) through its regulation of mitochondrial homeostasis. Fishes 2022, 7, 78. [Google Scholar] [CrossRef]
- Abasubong, K.P.; Jiang, G.Z.; Guo, H.X.; Wang, X.; Li, X.F.; Yan-Zou, D.; Liu, W.B.; Desouky, H.E. High-fat diet alters intestinal microbiota and induces endoplasmic reticulum stress via the activation of apoptosis and inflammation in blunt snout bream. Fish Physiol. Biochem. 2023, 49, 1079–1095. [Google Scholar] [CrossRef]
- Zhao, W.; Yao, R.; Wei, H.; Guo, Y.; Chen, A.; Chen, B. Astaxanthin, bile acid and chlorogenic acid attenuated the negative effects of high-fat diet on the growth, lipid deposition, and liver health of Oncorhynchus mykiss. Aquaculture 2023, 567, 739255. [Google Scholar] [CrossRef]
- Yin, P.; Xie, S.; Zhuang, Z.; He, X.; Tang, X.; Tian, L.; Liu, Y.; Niu, J. Dietary supplementation of bile acid attenuate adverse effects of high-fat diet on growth performance, antioxidant ability, lipid accumulation and intestinal health in juvenile largemouth bass (Micropterus salmoides). Aquaculture 2021, 531, 735864. [Google Scholar] [CrossRef]
- Cao, W.; Wu, J.; Zhao, X.; Li, Z.; Yu, J.; Shao, T.; Hou, X.; Zhou, L.; Wang, C.; Wang, G.; et al. Structural elucidation of an active polysaccharide from Radix Puerariae lobatae and its protection against acute alcoholic liver disease. Carbohydr. Polym. 2024, 325, 121565. [Google Scholar] [CrossRef]
- Lan, Y.; Wang, C.; Zhang, C.; Li, P.; Zhang, J.; Ji, H.; Yu, H. Dietary sea buckthorn polysaccharide reduced lipid accumulation, alleviated inflammation and oxidative stress, and normalized imbalance of intestinal microbiota that was induced by high-fat diet in zebrafish Danio rerio. Fish Physiol. Biochem. 2022, 48, 1717–1735. [Google Scholar] [CrossRef]
- Tan, X.; Sun, Z.; Ye, C. Dietary Lycium barbarum extract administration improved growth, meat quality and lipid metabolism in hybrid grouper (Epinephelus lanceolatus♂ × E. fuscoguttatus♀) fed high lipid diets. Aquaculture 2019, 504, 190–198. [Google Scholar] [CrossRef]
- Zou, C.; Fang, Y.; Lin, N.; Liu, H. Polysaccharide extract from pomelo fruitlet ameliorates diet-induced nonalcoholic fatty liver disease in hybrid grouper (Epinephelus lanceolatus♂ × Epinephelus fuscoguttatus♀). Fish Shellfish. Immunol. 2021, 119, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gu, W. Development Trends and Policy Recommendations for China’s Silkworm and Mulberry Industry in 2022. Chin. J. Anim. Sci. 2022, 58, 270–274. [Google Scholar]
- Li, L.; Zhang, S.; Xu, F.; Ma, H.; Zhuo, Y.; Qiao, Y. Research Progress of Mulberry Leaf Comprehensive Utilization. Sci. Technol. Food Ind. 2022, 43, 397–404. [Google Scholar]
- Yuan, Q.; Xie, Y.; Wang, W.; Yan, Y.; Ye, H.; Jabbar, S.; Zeng, X. Extraction optimization, characterization and antioxidant activity in vitro of polysaccharides from mulberry (Morus alba L.) leaves. Carbohydr. Polym. 2015, 128, 52–62. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, C.; Lu, G.; Cui, W.; Mu, Z.; Gao, H.; Wang, Y. Purification, characterization and anti-diabetic activity of a polysaccharide from mulberry leaf. Regul. Toxicol. Pharm. 2014, 70, 687–695. [Google Scholar] [CrossRef]
- Chen, X.; Sheng, Z.; Qiu, S.; Yang, H.; Jia, J.; Wang, J.; Jiang, C. Purification, characterization and in vitro and in vivo immune enhancement of polysaccharides from mulberry leaves. PLoS ONE 2019, 14, 0208611. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Cui, X.; Wang, D.; Hu, Y.; Wang, C. Structure-function relationship and biological activity of polysaccharides from mulberry leaves: A review. Int. J. Biol. Macromol. 2024, 268, 131701. [Google Scholar] [CrossRef]
- Chen, X.; Cai, B.; Wang, J.; Sheng, Z.; Yang, H.; Wang, D.; Chen, J.; Ning, Q. Mulberry leaf-derived polysaccharide modulates the immune response and gut microbiota composition in immunosuppressed mice. J. Funct. foods 2021, 83, 104545. [Google Scholar] [CrossRef]
- Li, R.; Xue, Z.; Li, S.; Zhou, J.; Liu, J.; Zhang, M.; Panichayupakaranant, P.; Chen, H. Mulberry leaf polysaccharides ameliorate obesity through activation of brown adipose tissue and modulation of the gut microbiota in high-fat diet fed mice. Food Funct. 2022, 13, 561–573. [Google Scholar] [CrossRef]
- Li, R.; Xue, Z.; Jia, Y.; Wang, Y.; Li, S.; Zhou, J.; Liu, J.; Zhang, M.; He, C.; Chen, H. Polysaccharides from mulberry (Morus alba L.) leaf prevents obesity by inhibiting pancreatic lipase in high-fat diet induced mice. Int. J. Biol. Macromol. 2021, 192, 452–460. [Google Scholar] [CrossRef]
- Zhao, X.; Fu, Z.; Yao, M.; Cao, Y.; Zhu, T.; Mao, R.; Huang, M.; Pang, Y.; Meng, X.; Li, L.; et al. Mulberry (Morus alba L.) leaf polysaccharide ameliorates insulin resistance-and adipose deposition-associated gut microbiota and lipid metabolites in high-fat diet-induced obese mice. Food. Sci. Nutr. 2022, 10, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Ichimiya, T.; Yamakawa, T.; Hirano, T.; Yokoyama, Y.; Hayashi, Y.; Hirayama, D.; Wagatsuma, K.; Itoi, T.; Nakase, H. Autophagy and autophagy-related diseases: A review. Int. J. Mol. Sci. 2020, 21, 8974. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, M.S. Autophagy as a crosstalk mediator of metabolic organs in regulation of energy metabolism. Rev. Endocr. Metab. Disord. 2014, 15, 11–20. [Google Scholar] [CrossRef]
- Painter, J.D.; Galle-Treger, L.; Akbari, O. Role of autophagy in lung inflammation. Front. Immunol. 2020, 11, 1337. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wei, C.; Yang, S.; Zhao, T.; Luo, Z. Effects of fat and fatty acids on the formation of autolysosomes in the livers from yellow catfish Pelteobagrus fulvidraco. Genes 2019, 10, 751. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xia, T.; Lin, J.; Wang, L.; Song, K.; Zhang, C. Quercetin attenuates high-fat diet-induced excessive fat deposition of spotted seabass (Lateolabrax maculatus) through the regulatory for mitochondria and endoplasmic reticulum. Front. Mar. Sci. 2021, 8, 746811. [Google Scholar] [CrossRef]
- Liu, X.C.; Huang, M.; Huang, X.P.; Guan, J.F.; Li, X.F.; Xie, D.Z.; Xu, C. NLRP3 is a promising target for regulating high glucose-induced inflammatory response in Megalobrama amblycephala. Aquaculture 2022, 555, 738220. [Google Scholar] [CrossRef]
- Su, S.; Wu, Y.; Lin, Q.; Wang, D.; Hai, J. URB597 protects against NLRP3 inflammasome activation by inhibiting autophagy dysfunction in a rat model of chronic cerebral hypoperfusion. J. Neuroinflammation 2019, 16, 260. [Google Scholar] [CrossRef]
- Lin, Q.; Li, S.; Jiang, N.; Shao, X.; Zhang, M.; Xi, H.; Zhang, Z.; Shen, J.; Zhou, Y.; Zhou, W.; et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019, 26, 101254. [Google Scholar] [CrossRef]
- Hou, Y.; Lin, S.; Qiu, J.; Sun, W.; Dong, M.; Xiang, Y.; Wang, L.; Du, P. NLRP3 inflammasome negatively regulates podocyte autophagy in diabetic nephropathy. Biochem. Biophys. Res. Commun. 2020, 521, 791–798. [Google Scholar] [CrossRef]
- Vrentas, C.E.; Boggiatto, P.M.; Olsen, S.C.; Leppla, S.H.; Moayeri, M. Characterization of the NLRP1 inflammasome response in bovine species. Innate Immun. 2020, 26, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.R.; Xing, R.L.; Wang, P.M.; Zhang, N.S.; Yin, S.J.; Li, X.C.; Zhang, L. NLRP1 and NLRP3 inflammasomes mediate LPS/ATP-induced pyroptosis in knee osteoarthritis. Mol. Med. Rep. 2018, 17, 5463–5469. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Sun, J.; Zhang, W.; Wang, X.; Xu, Y.; Peng, Y.; Zhang, L.; Xiong, W.; Liu, Y.; Liu, H. Novel insights into the roles of N6-methyladenosine (m6A) modification and autophagy in human diseases. Int. J. Biol. Sci. 2023, 19, 705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, X.; Huang, X. m6A-related bioinformatics analysis and functional characterization reveals that METTL3-mediated NPC1L1 mRNA hypermethylation facilitates progression of atherosclerosis via inactivation of the MAPK pathway. Inflamm. Res. 2023, 72, 429–442. [Google Scholar] [CrossRef]
- Panserat, S.; Marandel, L.; Geurden, I.; Veron, V.; Dias, K.; Plagnes-Juan, E.; Pegourié, G.; Arbenoits, E.; Santigosa, E.; Weber, G.; et al. Muscle catabolic capacities and global hepatic epigenome are modified in juvenile rainbow trout fed different vitamin levels at first feeding. Aquaculture 2017, 468, 515–523. [Google Scholar] [CrossRef]
- Xie, N.; Tian, J.; Meng, X.; Dong, L.; Jiang, M.; Wen, H.; Lu, X. DNA methylation profiling and transcriptome sequencing reveal the molecular mechanism of the high-carbohydrate diet on muscle growth of grass carp (Ctenopharyngodon idella). Aquac. Rep. 2023, 30, 101545. [Google Scholar] [CrossRef]
- Terova, G.; Díaz, N.; Rimoldi, S.; Ceccotti, C.; Gliozheni, E.; Piferrer, F. Effects of sodium butyrate treatment on histone modifications and the expression of genes related to epigenetic regulatory mechanisms and immune response in European sea bass (Dicentrarchus labrax) fed a plant-based diet. PLoS ONE 2016, 11, 0160332. [Google Scholar] [CrossRef]
- Jacobsen, S.C.; Brøns, C.; Bork-Jensen, J.; Ribel-Madsen, R.; Yang, B.; Lara, E.; Hall, E.; Calvanese, V.; Nilsson, E.; Jørgensen, S.W.; et al. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia 2012, 55, 3341–3349. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Zhou, D.; Moody, L.; Lezmi, S.; Chen, H.; Pan, Y.X. High-fat diet caused widespread epigenomic differences on hepatic methylome in rat. Physiol. Genom. 2015, 47, 514–523. [Google Scholar] [CrossRef]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef]
- Sun, J.; Li, M.; Xiong, Y.; Zhai, L.; Zhao, J. Oxidative stress mediated by n6-methyladenosine methylation contributes to high-fat diet induced male reproductive dysfunction. Mol. Nutr. Food Res. 2023, 67, e2101052. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, D.; Cao, S.; Qiao, M.; Zhang, P.; Zhao, Q.; Shen, Y.; Huang, X.; Song, L. The effects of folic acid on RNA m6A methylation in hippocampus as well as learning and memory ability of rats with acute lead exposure. J. Funct. Foods 2021, 76, 104276. [Google Scholar] [CrossRef]
- Fu, R.; Chen, D.; Tian, G.; Zheng, P.; He, J.; Yu, J.; Mao, X.; Huang, Z.; Pu, J.; Yu, B. Betaine affects abdominal flare fat metabolism via regulating m6A RNA methylation in finishing pigs fed a low-energy diet. J. Funct. Foods 2023, 107, 105620. [Google Scholar] [CrossRef]
- Shao, Y.A.; Duan, X.M.; Zhao, X.L.; Lv, Z.M.; Li, C.H. Global N6-methyladenosine methylation analysis reveals the positive correlation between m6A modification and mRNA abundance during Apostichopus japonicus disease development. Dev. Comp. Immunol. 2022, 133, 104434. [Google Scholar] [CrossRef]
- Ji, Q.; Xie, Z.L.; Li, L.Z.; Han, X.L.; Song, W. A Characterization of the RNA modification response to starvation under low temperatures in large yellow croaker (Larimichthys crocea). Fishes 2024, 9, 41. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Su, T.; Chen, J.; Huo, X.; Kuang, L.; Yan, T.; Gao, F.; Wu, D. Transcriptome-wide m6A methylation and metabolomic analysis reveal regulatory networks in rice roots under manganese stress. Environ. Exp. Bot. 2024, 226, 105906. [Google Scholar] [CrossRef]
- Chen, J.; Cao, H.; Chen, D.; Kuang, L.; Wu, D. Transcriptome-wide analysis of m6A methylation reveals genetic responses to cadmium stress at germination stage in rice. Environ. Exp. Bot. 2023, 205, 105130. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef]
- Kramer, M.C.; Janssen, K.A.; Palos, K.; Nelson, A.D.; Vandivier, L.E.; Garcia, B.A.; Lyons, E.; Beilstein, M.A.; Gregory, B.D. N6-methyladenosine and RNA secondary structure affect transcript stability and protein abundance during systemic salt stress in Arabidopsis. Plant Direct 2020, 4, e00239. [Google Scholar] [CrossRef] [PubMed]
- Clancy, M.J.; Shambaugh, M.E.; Timpte, C.S.; Bokar, J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002, 30, 4509–4518. [Google Scholar] [CrossRef]
- Harper, J.E.; Miceli, S.M.; Roberts, R.J.; Manley, J.L. Sequence specificity of the human messenger-RNA n6-adenosine methylase invitro. Nucleic Acids Res. 1990, 18, 5735–5741. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Ynag, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- He, M.; Lei, H.; He, X.; Liu, Y.; Wang, A.; Ren, Z.; Liu, X.; Yan, G.; Wang, W.; Wang, Y.; et al. METTL14 regulates osteogenesis of bone marrow mesenchymal stem cells via inducing autophagy through m6A/IGF2BPs/Beclin-1 signal axis. Stem Cells Transl. Med. 2022, 11, 987–1001. [Google Scholar] [CrossRef]
- Huang, M.; He, J.; He, X. Mechanism of METTL14 affecting IR injury of cardiomyocytes by m6A methylation regulation sirt6-mediated mitochondrial autophagy. Lab. Med. Clin. 2024, 21, 3669–3674+3681. [Google Scholar]
- Liu, X.K. Mechanism Study on the Restorative Effect of Notoginsenoside R1 to RNA Methylation Mediated Skin Ultraviolet Damage. Ph.D. Thesis, Changchun University of Chinese Medicine, Changchun, China, June 2024. [Google Scholar]
- Luo, G.Z.; MacQueen, A.; Zheng, G.; Duan, H.; Dore, L.C.; Lu, Z.; Liu, J.; Chen, K.; Jia, G.; Bergelson, J.; et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef]
- Niu, Y.; Lin, Z.Y.; Wan, A.; Chen, H.L.; Liang, H.; Sun, L.; Wang, G. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression updates through inhibiting BNIP3. Mol. Cancer 2019, 18, 46. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Wang, F.; Zeng, X.J.; Huang, Z.; Dong, K.F. Astragalus polysaccharide ameliorates steroid-induced osteonecrosis of femoral head through miR-206/HIF-1α/BNIP3 axis. Kaohsiung J. Med. Sci. 2021, 37, 1089–1100. [Google Scholar] [CrossRef]
- Chen, G.; Shaw, M.H.; Kim, Y.G.; Nuñez, G. NOD-Like Receptors: Role in Innate Immunity and Inflammatory Disease. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 365–398. [Google Scholar] [CrossRef]
- Li, J.Y.; Gao, K.; Shao, T.; Fan, D.D.; Hu, C.B.; Sun, C.C.; Dong, W.R.; Lin, A.F.; Xiang, L.X.; Shao, J.Z. Characterization of an NLRP1 Inflammasome from Zebrafish Reveals a Unique Sequential Activation Mechanism Underlying Inflammatory Caspases in Ancient Vertebrates. J. Immunol. 2018, 201, 1946–1966. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Guang, Q.; Hao, Y.; Yu, X.; Wang, Y.; Zhai, L. Antrodia camphorata polysaccharide alleviates inflammatory damage of cortical neurons through ROS-NOX2-NLRP1 signal pathway. Zhejiang Med. J. 2021, 43, 244–249+346. [Google Scholar]
- Zhao, H.; Wang, H.; Liu, R.; Liang, Y.; Li, K.; Shan, S.; An, L.; Yang, G.; Li, H. Activation of the NLRP1 inflammasome and its ligand recognition in the antibacterial immune response of common carp (Cyprinus carpio). Fish Shellfish. Immunol. 2022, 125, 238–246. [Google Scholar] [CrossRef]
- Xu, W.N.; Chen, D.H.; Liu, W.B.; Xu, J.X.; Yang, S.S. Molecular characterization of microtubule-associated protein 1-light chain 3B in Megalobrama amblycephala fed with high fat/berberine diets. J. Appl. Genet. 2018, 59, 345–355. [Google Scholar] [CrossRef]
- Meng, X.H.; Chen, B.; Zhang, J.P. Intracellular insulin and impaired autophagy in a zebrafish model and a cell model of Type 2 diabetes. Int. J. Biol. Sci. 2017, 13, 985–995. [Google Scholar] [CrossRef]
- Liu, S.Y.; Chen, L.; Li, X.C.; Hu, Q.K.; He, L.J. Lycium barbarum polysaccharide protects diabetic peripheral neuropathy by enhancing autophagy via mTOR/p70S6K inhibition in Streptozotocin-induced diabetic rats. J. Chem. Neuroanat. 2018, 89, 37–42. [Google Scholar] [CrossRef]
- Chen, D.D.; Xu, R.; Zhou, J.Y.; Chen, J.Q.; Wang, L.; Liu, X.S.; Liang, C.L.; Liu, B.H.; Lu, R.R.; Wu, J.B.; et al. Cordyceps militaris polysaccharides exerted protective effects on diabetic nephropathy in mice via regulation of autophagy. Food Funct. 2019, 10, 5102–5114. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gong, H.; Yang, S.; Yang, L.; Fan, Y.; Zhou, Y. Pectic bee pollen polysaccharide from Rosa rugosa alleviates diet-induced hepatic steatosis and insulin resistance via induction of AMPK/mTOR-mediated autophagy. Molecules 2017, 22, 699. [Google Scholar] [CrossRef]
- Li, A.; Shuai, X.; Jia, Z.; Li, H.; Liang, X.; Su, D.; Guo, W. Ganoderma lucidum polysaccharide extract inhibits hepatocellular carcinoma growth by downregulating regulatory T cells accumulation and function by inducing microRNA-125b. J. Transl. Med. 2015, 13, 100. [Google Scholar] [CrossRef]
- Chen, H.L.; Tan, H.L.; Yang, J.; Wei, Y.Y.; Hu, T.J. Sargassum polysaccharide inhibits inflammatory response in PCV2 infected-RAW264. 7 cells by regulating histone acetylation. Carbohydr. Polym. 2018, 200, 633–640. [Google Scholar] [CrossRef]
- Ruan, Y.; Li, H.; Pu, L.; Shen, T. Tremella fuciformis Polysaccharides Attenuate Oxidative Stress and Inflammation in Macrophages through miR-155. Anal. Cell. Pathol. 2018, 2018, 5762371. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Yoon, S.O.; Lee, H.J.; Oh, J.Y. Rapamycin regulates macrophage activation by inhibiting NLRP3 inflammasome-p38 MAPK-NFκB pathways in autophagy- and p62-dependent manners. Oncotarget 2017, 8, 40817–40831. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.M.; Li, J.; Shen, R.; Li, J.; Yu, Y.; Li, L.; Deng, S.H.; Liu, T.; Zhang, T.; Xu, Y.; et al. Autophagy induced by micheliolide alleviates acute irradiation-induced intestinal injury via inhibition of the NLRP3 inflammasome. Front. Pharmacol. 2022, 12, 773150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Z.; Feng, D.; Zu, G.; Li, Y.; Zhao, Y.; Wang, G.; Ning, S.; Zhu, J.; Zhang, F.; et al. Autophagy Induction Ameliorates Inflammatory Responses in Intestinal Ischemia—Reperfusion Through Inhibiting NLRP3 Inflammasome Activation. Shock 2019, 52, 387–395. [Google Scholar] [CrossRef]


| Predict Function | Gene Name | Transcript ID | Regulation | Log2FC | p-Value |
|---|---|---|---|---|---|
| Autophagy–animal | ddit4 | XM_048171097.1 | Up | 4.33 | 0.00 |
| hif1al2 | XM_048166513.1 | Up | 2.17 | 0.01 | |
| bnip3 | XM_048170148.1 | Up | 1.32 | 0.02 | |
| mtor | XM_048210663.1 | Down | −1.56 | 0.02 | |
| ulk2 | XM_048161082.1 | Up | 3.47 | 0.00 | |
| ulk1a | XM_048209404.1 | Up | 1.23 | 0.00 | |
| irs2b | XM_048193118.1 | Up | 4.79 | 0.00 | |
| irs2a | XM_048197081.1 | Up | 3.62 | 0.00 | |
| zfyve1 | XM_048206589.1 | Up | 1.26 | 0.02 | |
| cflara | XM_048173454.1 | Down | −1.13 | 0.01 | |
| ctsbb | XM_048207873.1 | Up | 2.91 | 0.00 | |
| ctsl.1 | XM_048191905.1 | Up | 3.14 | 0.00 | |
| ctsla | XM_048189060.1 | Up | 2.11 | 0.00 | |
| NOD-like receptor signaling pathway | mapk12b | XM_048157097.1 | Up | 2.13 | 0.03 |
| traf3 | XM_048191482.1 | Down | −1.05 | 0.01 | |
| nlrc3 | XM_048175695.1 | Up | 1.94 | 0.01 | |
| nlrp3 | XM_048210961.1 | Down | −3.50 | 0.00 | |
| nlrp12 | XM_048190599.1 | Down | −3.12 | 0.00 | |
| nlrp1 | XM_048162530.1 | Down | −1.59 | 0.00 |
| GeneName | MeRIP-Seq | RNA−Seq | Expression Pattern | |||
|---|---|---|---|---|---|---|
| Peak Annotation | Diffpeak. Log2(Fc) | m6A Regulation | Diffgene. Log2(Fc) | Gene Regulation | ||
| wnk1a | UTR3 | −1.02 | down | −1.90 | down | Hypo-down |
| loc125252147 | exonic | −2.44 | down | −2.50 | down | |
| ank2b | UTR3 | −2.14 | down | 2.18 | up | Hypo-up |
| znhit6 | UTR3 | −1.73 | down | 1.94 | up | |
| znf420 | UTR3 | 1.26 | up | −1.56 | down | Hyper-down |
| loc125275941 | UTR3 | 1.06 | up | −2.02 | down | |
| nlrp1 | UTR3 | 1.07 | up | −1.59 | down | |
| map4 | exonic | 1.45 | up | −1.53 | down | |
| ubald2 | UTR3 | 2.32 | up | 2.97 | up | Hyper-up |
| mmadhca | UTR3 | 1.29 | up | 2.19 | up | |
| zgc:110699 | UTR3 | 1.32 | up | 1.07 | up | |
| tmem119b | UTR3 | 1.24 | up | 1.01 | up | |
| zranb1b | exonic | 2.44 | up | 1.07 | up | |
| sh3d19 | exonic | 2.64 | up | 1.10 | up | |
| ctps1b | UTR3 | 1.10 | up | 1.66 | up | |
| dock4b | UTR3 | 1.30 | up | 1.81 | up | |
| mnta | UTR3 | 1.13 | up | 1.23 | up | |
| sialidase-4 | UTR5 | 2.97 | up | 1.46 | up | |
| mtss1la | UTR3 | 1.22 | up | 1.35 | up | |
| sgms1b | UTR5 | 1.97 | up | 1.25 | up | |
| bnip3 | UTR3 | 1.51 | up | 1.32 | up | |
| Genes | Primer Sequence (5′–3′) | Product Length (bps) | Accession No. | |
|---|---|---|---|---|
| mettl14 | Forward | TCGGCCGACATGGTACAAAT | 120 | XM_048197582.1 |
| Reverse | TGGTCTTGCCAGGGTTGTTT | |||
| wtap | Forward | AGAGCTCAAGAGCAGCCAAG | 200 | XM_048206845.1 |
| Reverse | GTTCAGAGGCCGTTGAAGGA | |||
| alkbh5 | Forward | TGCACACAGGCCTCGTATTT | 131 | XM_048197746.1 |
| Reverse | AGCCCGGCTCTCTATCTTCA | |||
| fto | Forward | ACGGCACAGGAGAACAGAAG | 107 | XM_048184628.1 |
| Reverse | GCCTGAAGGATTGTCCTGCT | |||
| ythdf2 | Forward | CAAAGGGCCCCTCTATCTGC | 221 | XM_048191909.1 |
| Reverse | TGGTCACCGGCTTATTCTCG | |||
| txnipa | Forward | GAGAACACCTGCTCTCGCAT | 168 | XM_048201110.1 |
| Reverse | CACACGAATGCTCTTCCCCT | |||
| nlrp1 | Forward | ACTCAGCAAAGCAGGAAAAGC | 161 | XM_048162530.1 |
| Reverse | AGGTCTCAACGAGGGAAATG | |||
| nlrp3 | Forward | TGGAGTTGTGTCTCTCCAACG | 163 | XM_048194926.1 |
| Reverse | CCTTCCGGACCAGTCCATTC | |||
| axin2 | Forward | GTCTGAAGCGGGAACAGGAA | 121 | XM_048190722.1 |
| Reverse | AAAGGCAGAGAGTGGGATGC | |||
| bnip3 | Forward | GAGGTGGCAGCAGTCCTAAA | 125 | XM_048170148.1 |
| Reverse | ATCACATGGCAGGCTTCCTC | |||
| mtor | Forward | GCCTCAAGTTATGCCCACCT | 91 | XM_048210663.1 |
| Reverse | CACAACCATCCCCATCTGCT | |||
| β-actin | Forward | TCGTCCACCGCAAATGCTTCTA | 152 | XM_048192430.1 |
| Reverse | CCGTCACCTTCACCGTTCCAGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, W.; Lin, Y.; Qian, L.; Lu, S.; Gu, Z.; Ge, X.; Miao, L. m6A Methylation Mediated Autophagy and Nucleotide-Binding Oligomerization Domain-like Receptors Signaling Pathway Provides New Insight into the Mitigation of Oxidative Damage by Mulberry Leaf Polysaccharides. Int. J. Mol. Sci. 2025, 26, 4345. https://doi.org/10.3390/ijms26094345
Jiang W, Lin Y, Qian L, Lu S, Gu Z, Ge X, Miao L. m6A Methylation Mediated Autophagy and Nucleotide-Binding Oligomerization Domain-like Receptors Signaling Pathway Provides New Insight into the Mitigation of Oxidative Damage by Mulberry Leaf Polysaccharides. International Journal of Molecular Sciences. 2025; 26(9):4345. https://doi.org/10.3390/ijms26094345
Chicago/Turabian StyleJiang, Wenqiang, Yan Lin, Linjie Qian, Siyue Lu, Zhengyan Gu, Xianping Ge, and Linghong Miao. 2025. "m6A Methylation Mediated Autophagy and Nucleotide-Binding Oligomerization Domain-like Receptors Signaling Pathway Provides New Insight into the Mitigation of Oxidative Damage by Mulberry Leaf Polysaccharides" International Journal of Molecular Sciences 26, no. 9: 4345. https://doi.org/10.3390/ijms26094345
APA StyleJiang, W., Lin, Y., Qian, L., Lu, S., Gu, Z., Ge, X., & Miao, L. (2025). m6A Methylation Mediated Autophagy and Nucleotide-Binding Oligomerization Domain-like Receptors Signaling Pathway Provides New Insight into the Mitigation of Oxidative Damage by Mulberry Leaf Polysaccharides. International Journal of Molecular Sciences, 26(9), 4345. https://doi.org/10.3390/ijms26094345







