Epigenetic Biomarkers in Temporomandibular Joint Osteoarthritis: An Emerging Target in Treatment
Abstract
1. Introduction
Osteoarthritis of the Temporomandibular Joint
2. Epigenetics and OA
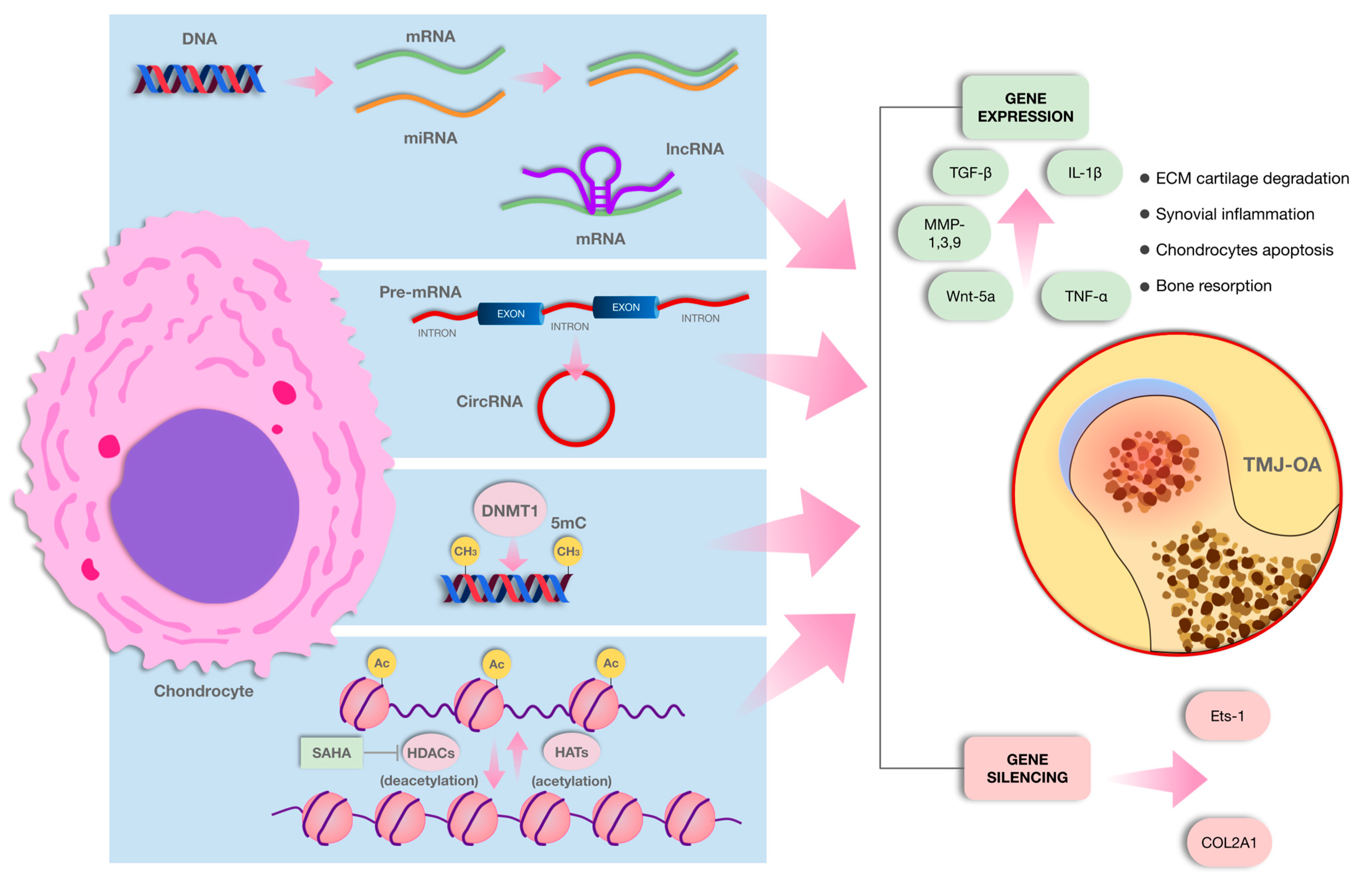
2.1. Post-Translational Histone Modification
Histone Modification and TMJ-OA
2.2. Non-Coding RNAs
2.2.1. MiRNAs
2.2.2. MiRNAs and TMJ-OA
2.2.3. Long Non-Coding RNAs
2.2.4. LncRNA and OA
2.2.5. LncRNA and TMJ-OA
2.2.6. Circular RNAs (circRNAs)
2.2.7. circRNAs and OA
2.2.8. circRNA and TMJ-OA
2.2.9. TMJ-OA and circRNA
2.3. DNA Methylation
2.3.1. DNA Methylation and OA
2.3.2. DNA Methylation and TMJ-OA
2.4. Knee/Hip OA
2.5. Scope and Limitations
2.6. Future Studies
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Su, N.; Liu, Y.; Yang, X.; Shen, J.; Wang, H. Association of malocclusion, self-reported bruxism and chewing-side preference with oral health-related quality of life in patients with temporomandibular joint osteoarthritis. Int. Dent. J. 2018, 68, 97–104. [Google Scholar] [CrossRef]
- Kang, S.C.; Lee, D.G.; Choi, J.H.; Kim, S.T.; Kim, Y.K.; Ahn, J. Association between estrogen receptor polymorphism and pain susceptibility in female temporomandibular joint osteoarthritis patients. Int. J. Oral Maxillofac. Surg. 2007, 36, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Zhang, J.N.; Gan, Y.H.; Zhou, Y.H. Current understanding of pathogenesis and treatment of TMJ osteoarthritis. J. Dent. Res. 2015, 94, 666. [Google Scholar] [CrossRef]
- Zhang, W.; Ouyang, H.; Dass, C.R.; Xu, J. Current research on pharmacologic and regenerative therapies for osteoarthritis. Bone Res. 2016, 4, 15040. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, H.; Bu, L.; He, D. Expression profile of circular RNAs in TMJ osteoarthritis synovial tissues and potential functions of hsa_circ_0000448 with specific back-spliced junction. Am. J. Transl. Res. 2019, 11, 5357–5374. [Google Scholar]
- Jeffries, M.A. Osteoarthritis year in review 2018: Genetics and epigenetics. Osteoarthr. Cartil. 2019, 27, 371–377. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Health Care Services; Board on Health Sciences Policy; Committee on Temporomandibular Disorders (TMDs): From Research Discoveries to Clinical Treatment; Yost, O.; Liverman, C.T.; English, R.; Mackey, S.; Bond, E.C. (Eds.) Temporomandibular Disorders: Priorities for Research and Care; National Academies Press: Washington, DC, USA, 2020. [Google Scholar]
- Lee, Y.H.; Park, H.K.; Auh, Q.S.; Nah, H.; Lee, J.S.; Moon, H.J.; Heo, D.N.; Kim, I.S.; Kwon, I.K. Emerging potential of exosomes in regenerative medicine for temporomandibular joint osteoarthritis. Int. J. Mol. Sci. 2020, 21, 1541. [Google Scholar] [CrossRef]
- Vos, L.M.; Kuijer, R.; Huddleston Slater, J.J.; Bulstra, S.K.; Stegenga, B. Inflammation is more distinct in temporomandibular joint osteoarthritis compared to the knee joint. J. Oral Maxillofac. Surg. 2014, 72, 35–40. [Google Scholar] [CrossRef]
- Farré-Guasch, E.; Aliberas, J.T.; Spada, N.F.; de Vries, R.; Schulten, E.A.J.M.; Lobbezoo, F. The role of inflammatory markers in temporomandibular myalgia: A systematic review. Jpn. Dent. Sci. Rev. 2023, 59, 281–288. [Google Scholar] [CrossRef]
- Qiao, Y.; Li, J.; Yuh, C.; Ko, F.; Mercuri, L.G.; Alkhudari, J.; Pourzal, R.; Oh, C.-d. Chemokine regulation in temporomandibular joint disease: A comprehensive review. Genes 2023, 14, 408. [Google Scholar] [CrossRef]
- Nazet, U.; Neubert, P.; Schatz, V.; Grässel, S.; Proff, P.; Jantsch, J.; Schröder, A.; Kirschneck, C. Differential gene expression response of synovial fibroblasts from temporomandibular joints and knee joints to dynamic tensile stress. J. Orofac. Orthop. 2022, 83, 361–375. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cao, W.; Azeem, I.; Shao, Z. Epigenetics of osteoarthritis: Histones and TGF-β1. Clin. Chim. Acta 2020, 510, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lu, X.; Shen, B.; Zeng, Y. The therapeutic potential and role of miRNA, lncRNA, and circRNA in osteoarthritis. Curr. Gene Ther. 2019, 19, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Ratneswaran, A.; Kapoor, M. Osteoarthritis year in review: Genetics, genomics, epigenetics. Osteoarthr. Cartil. 2021, 29, 151–160. [Google Scholar] [CrossRef]
- Fraga, M.F.; Ballestar, E.; Paz, M.F.; Ropero, S.; Setien, F.; Ballestar, M.L.; Heine-Suñer, D.; Cigudosa, J.C.; Urioste, M.; Benitez, J.; et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA 2005, 102, 10604–10609. [Google Scholar] [CrossRef]
- Barter, M.J.; Young, D.A. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Curr. Rheumatol. Rep. 2013, 15, 353. [Google Scholar] [CrossRef]
- Hunter, D.J.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef]
- Zhang, Y.; Jordan, J.M. Epidemiology of osteoarthritis. Clin. Geriatr. Med. 2010, 26, 355–369, Erratum in Clin. Geriatr. Med. 2013, 29, ix. [Google Scholar] [CrossRef]
- Krakowski, P.; Rejniak, A.; Sobczyk, J.; Karpiński, R. Cartilage Integrity: A Review of Mechanical and Frictional Properties and Repair Approaches in Osteoarthritis. Healthcare 2024, 12, 1648. [Google Scholar] [CrossRef]
- Musumeci, G.; Aiello, F.; Szychlinska, M.; Di Rosa, M.; Castrogiovanni, P.; Mobasheri, A. Osteoarthritis in the XXIst Century: Risk Factors and Behaviours That Influence Disease Onset and Progression. Int. J. Mol. Sci. 2015, 16, 6093–6112. [Google Scholar] [CrossRef]
- Mobasheri, A.; Batt, M. An Update on the Pathophysiology of Osteoarthritis. Ann. Phys. Rehabil. Med. 2016, 59, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef]
- Lesseur, C.; Armstrong, D.A.; Murphy, M.A.; Appleton, A.A.; Koestler, D.C.; Paquette, A.G.; Lester, B.M.; Marsit, C.J. Sex-specific associations between placental leptin promoter DNA methylation and infant neurobehavior. Psychoneuroendocrinology 2014, 40, 1–9. [Google Scholar] [CrossRef][Green Version]
- Xiao, J.L.; Meng, J.H.; Gan, Y.H.; Li, Y.L.; Zhou, C.Y.; Ma, X.C. DNA methylation profiling in different phases of temporomandibular joint osteoarthritis in rats. Arch. Oral Biol. 2016, 68, 105–115. [Google Scholar] [CrossRef]
- Loughlin, J.; Reynard, L.N. Osteoarthritis: Epigenetics of articular cartilage in knee and hip OA. Nat. Rev. Rheumatol. 2015, 11, 6–7. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Parthun, M.R. Hat1: The emerging cellular roles of a type B histone acetyltransferase. Oncogene 2007, 26, 5319–5328. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Denu, J.M. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 2008, 18, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef]
- Dancy, B.M.; Cole, P.A. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015, 115, 2419–2452, Erratum in Chem. Rev. 2016, 116, 8314. [Google Scholar] [CrossRef]
- Sun, J.; Liao, W.; Su, K.; Jia, J.; Qin, L.; Liu, W.; He, Y.; Zhang, H.; Ou, F.; Zhang, Z.; et al. Suberoylanilide Hydroxamic Acid Attenuates Interleukin-1β-Induced Interleukin-6 Upregulation by Inhibiting the Microtubule Affinity-Regulating Kinase 4/Nuclear Factor-κB Pathway in Synovium-Derived Mesenchymal Stem Cells from the Temporomandibular Joint. Inflammation 2020, 43, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.W.; Watkins, G.; Le Good, N.; Roberts, S.; Murphy, C.L.; Brockbank, S.M.; Needham, M.R.; Read, S.J.; Newham, P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthr. Cartil. 2009, 17, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Sun, J.; Liao, W.; Qin, L.; Su, K.; He, Y.; Zhang, J.; Yang, R.; Zhang, Z.; Sun, Y. Knockdown of long non-coding RNA AK094629 attenuates the interleukin-1β-induced expression of interleukin-6 in synovium-derived mesenchymal stem cells from the temporomandibular joint. Mol. Med. Rep. 2020, 22, 1195–1204. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, R.; Wen, L.M. Long Non-coding RNA XIST Regulates Chondrogenic Differentiation of Synovium-derived Mesenchymal Stem Cells from Temporomandibular Joint via miR-27b-3p/ADAMTS-5 Axis. Cytokine 2021, 137, 155352. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Salzman, J. Detecting Circular RNAs: Bioinformatic and Experimental Challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, M.; O’Keefe, R.J.; Shen, J.; Li, Z.; Zhou, J.; Zhou, X.; Mao, J.J. Epigenetic and Therapeutic Implications of Dnmt3b in Temporomandibular Joint Osteoarthritis. Am. J. Transl. Res. 2019, 11, 1736–1747. [Google Scholar]
- Verza, F.A.; Das, U.; Fachin, A.L.; Dimmock, J.R.; Marins, M. Roles of histone deacetylases and inhibitors in anticancer therapy. Cancers 2020, 12, 1664. [Google Scholar] [CrossRef]
- Tang, J.; Yan, H.; Zhuang, S. Histone deacetylases as targets for treatment of multiple diseases. Clin. Sci. 2013, 124, 651–662. [Google Scholar] [CrossRef]
- Wang, X.; Song, Y.; Jacobi, J.L.; Tuan, R.S. Inhibition of histone deacetylases antagonized FGF2 and IL-1β effects on MMP expression in human articular chondrocytes. Growth Factors 2009, 27, 40–49. [Google Scholar] [CrossRef]
- Nasu, Y.; Nishida, K.; Miyazawa, S.; Komiyama, T.; Kadota, Y.; Abe, N.; Yoshida, A.; Hirohata, S.; Ohtsuka, A.; Ozaki, T. Trichostatin A, a histone deacetylase inhibitor, suppresses synovial inflammation and subsequent cartilage destruction in a collagen antibody-induced arthritis mouse model. Osteoarthr. Cartil. 2008, 16, 723–732. [Google Scholar] [CrossRef]
- Chen, W.P.; Bao, J.P.; Hu, P.F.; Feng, J.; Wu, L.D. Alleviation of osteoarthritis by Trichostatin A, a histone deacetylase inhibitor, in experimental osteoarthritis. Mol. Biol. Rep. 2010, 37, 3967–3972. [Google Scholar] [CrossRef]
- Qu, H.; Li, J.; Wu, L.D.; Chen, W.P. Trichostatin A increases the TIMP-1/MMP ratio to protect against osteoarthritis in an animal model of the disease. Mol. Med. Rep. 2016, 14, 2423–2430. [Google Scholar] [CrossRef] [PubMed]
- Chabane, N.; Zayed, N.; Afif, H.; Mfuna-Endam, L.; Benderdour, M.; Boileau, C.; Martel-Pelletier, J.; Pelletier, J.P.; Duval, N.; Fahmi, H. Histone deacetylase inhibitors suppress interleukin-1β-induced nitric oxide and prostaglandin E2 production in human chondrocytes. Osteoarthr. Cartil. 2008, 16, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.M.; Ding, Q.H.; Chen, W.P.; Luo, R.B. Vorinostat, a HDAC inhibitor, showed anti-osteoarthritic activities through inhibition of iNOS and MMP expression, p38 and ERK phosphorylation and blocking NF-κB nuclear translocation. Int. Immunopharmacol. 2013, 17, 329–335. [Google Scholar] [CrossRef]
- Higashiyama, R.; Miyaki, S.; Yamashita, S.; Yoshitaka, T.; Lindman, G.; Ito, Y.; Sasho, T.; Takahashi, K.; Lotz, M.; Asahara, H. Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Mod. Rheumatol. 2010, 20, 11–17. [Google Scholar] [CrossRef]
- Liao, W.T.; Sun, J.D.; Wang, Y.; He, Y.Q.; Su, K.; Lu, Y.Y.; Liao, G.; Sun, Y.P. Histone deacetylase inhibitors attenuated interleukin-1β-induced chondrogenesis inhibition in synovium-derived mesenchymal stem cells of the temporomandibular joint. Bone Joint Res. 2022, 11, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, P.; Hou, Y.; Chen, S.; Xiao, Z.; Zhan, J.; Luo, D.; Gu, M.; Lin, D. Berberine inhibits the interleukin-1β-induced inflammatory response via MAPK downregulation in rat articular chondrocytes. Drug Dev. Res. 2019, 80, 637–645. [Google Scholar] [CrossRef]
- Liu, W.; Sun, Y.; He, Y.; Zhang, H.; Zheng, Y.; Yao, Y.; Zhang, Z. IL-1β impedes the chondrogenic differentiation of synovial fluid mesenchymal stem cells in the human temporomandibular joint. Int. J. Mol. Med. 2017, 39, 317–326. [Google Scholar] [CrossRef]
- Pujol, J.-P.; Chadjichristos, C.; Legendre, F.; Baugé, C.; Beauchef, G.; Andriamanalijaona, R.; Galéra, P.; Boumediene, K. Interleukin-1 and transforming growth factor-beta 1 as crucial factors in osteoarthritic cartilage metabolism. Connect Tissue Res. 2008, 49, 293–297. [Google Scholar] [CrossRef]
- Makki, M.S.; Haqqi, T.M. Histone deacetylase inhibitor vorinostat (SAHA, MK0683) perturbs miR-9-MCPIP1 axis to block IL-1β-induced IL-6 expression in human OA chondrocytes. Connect. Tissue Res. 2017, 58, 64–75. [Google Scholar] [CrossRef]
- Shen, J.; Abu-Amer, Y.; O’Keefe, R.J.; McAlinden, A. Inflammation and epigenetic regulation in osteoarthritis. Connect Tissue Res. 2017, 58, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol. 2004, 14, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Murchison, E.P.; Hannon, G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004, 16, 223–229. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Görlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F.; Fischer, S.E.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.W.; Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Fang, L.L.; Wang, X.H.; Sun, B.F.; Zhang, X.D.; Zhu, X.H.; Yu, Z.J.; Luo, H. Expression, regulation and mechanism of action of the miR-17-92 cluster in tumor cells (Review). Int. J. Mol. Med. 2017, 40, 1624–1630. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Y.; Deng, M.; Li, J.; Cai, H.; Meng, Q.; Fang, W.; Long, X.; Ke, J. MicroRNA221-3p modulates Ets-1 expression in synovial fibroblasts from patients with osteoarthritis of the temporomandibular joint. Osteoarthr. Cartil. 2016, 24, 2003–2011. [Google Scholar] [CrossRef]
- Wu, W.; Xuan, Y.; Ge, Y.; Mu, S.; Hu, C.; Fan, R. Plasma miR-146a and miR-365 expression and inflammatory factors in patients with osteoarthritis. Malays. J. Pathol. 2021, 43, 311–317. [Google Scholar]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Shan, Y.; Pan, Y.; Ma, J.; Jia, L. Long non-coding RNA HOTAIR promotes osteoarthritis progression via miR-17-5p/FUT2/β-catenin axis. Cell Death Dis. 2018, 9, 711. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Cheon, E.J.; Lee, M.H.; Kim, H.A. MicroRNA-127-5p regulates matrix metalloproteinase 13 expression and interleukin-1β-induced catabolic effects in human chondrocytes. Arthritis Rheum. 2014, 66, 1394. [Google Scholar] [CrossRef]
- Mehana, E.E.; Khafaga, A.F.; El-Blehi, S.S. The role of matrix metalloproteinases in osteoarthritis pathogenesis: An updated review. Life Sci. 2019, 234, 116786. [Google Scholar] [CrossRef] [PubMed]
- Van der Kraan, P.M.; Goumans, M.J.; Blaney Davidson, E.; ten Dijke, P. Age-dependent alteration of TGF-beta signalling in osteoarthritis. Cell Tissue Res. 2012, 347, 257–265. [Google Scholar] [CrossRef]
- Cao, Y.; Tang, S.; Nie, X.; Zhou, Z.; Ruan, G.; Han, W.; Zhu, Z.; Ding, C. Decreased miR-214-3p activates NF-κB pathway and aggravates osteoarthritis progression. EBioMedicine 2021, 65, 103283. [Google Scholar] [CrossRef]
- Li, W.; Zhao, S.; Yang, H.; Zhang, C.; Kang, Q.; Deng, J.; Xu, Y.; Ding, Y.; Li, S. Potential Novel Prediction of TMJ-OA: MiR-140-5p Regulates Inflammation Through Smad/TGF-β Signaling. Front. Pharmacol. 2019, 10, 15. [Google Scholar] [CrossRef]
- Peters, C.L.; Morris, C.J.; Mapp, P.I.; Blake, D.R.; Lewis, C.E.; Winrow, V.R. The transcription factors hypoxia-inducible factor 1alpha and Ets-1 colocalize in the hypoxic synovium of inflamed joints in adjuvant-induced arthritis. Arthritis Rheum. 2004, 50, 291–296. [Google Scholar] [CrossRef]
- Li, X.; Huang, T.L.; Zhang, G.D.; Jiang, J.T.; Guo, P.Y. LncRNA ANRIL Impacts the Progress of Osteoarthritis via Regulating Proliferation and Apoptosis of Osteoarthritis Synoviocytes. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9729–9737. [Google Scholar] [CrossRef]
- Lu, Z.; Luo, M.; Huang, Y. lncRNA-CIR Regulates Cell Apoptosis of Chondrocytes in Osteoarthritis. J. Cell. Biochem. 2018, 120, 7229–7237. [Google Scholar] [CrossRef]
- Li, Y.F.; Li, S.H.; Liu, Y.; Luo, Y.T. Long Noncoding RNA CIR Promotes Chondrocyte Extracellular Matrix Degradation in Osteoarthritis by Acting as a Sponge for Mir-27b. Cell Physiol. Biochem. 2018, 43, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Sun, M.L.; Zhang, X.A.; Wang, X.Q. Crosstalk Among circRNA/lncRNA, miRNA, and mRNA in Osteoarthritis. Front. Cell Dev. Biol. 2021, 9, 774370. [Google Scholar] [CrossRef] [PubMed]
- Cen, X.; Huang, X.Q.; Sun, W.T.; Liu, Q.; Liu, J. Long noncoding RNAs: A new regulatory code in osteoarthritis. Am. J. Transl. Res. 2017, 9, 4747–4755. [Google Scholar] [PubMed]
- Tan, F.; Wang, D.; Yuan, Z. The Fibroblast-Like Synoviocyte Derived Exosomal Long Non-coding RNA H19 Alleviates Osteoarthritis Progression Through the miR-106b-5p/TIMP2 Axis. Inflammation 2020, 43, 1498–1509. [Google Scholar] [CrossRef]
- Zhang, S.; Yap, A.U.; Toh, W.S. Stem Cells for Temporomandibular Joint Repair and Regeneration. Stem Cell Rev. Rep. 2015, 11, 728–742. [Google Scholar] [CrossRef]
- Saurenmann, R.K.; Kellenberger, C.J. Assessing arthritis in the temporomandibular joint. J. Rheumatol. 2015, 42, 2000–2002. [Google Scholar] [CrossRef][Green Version]
- Zhang, C.; Wang, P.; Jiang, P.; Lv, Y.; Dong, C.; Dai, X. Upregulation of lncRNA HOTAIR contributes to IL-1β-induced MMP overexpression and chondrocytes apoptosis in temporomandibular joint osteoarthritis. Gene 2016, 586, 248–253. [Google Scholar] [CrossRef]
- Jiang, M.; Liu, J.; Luo, T.; Chen, Q.; Lu, M.; Meng, D. LncRNA PACER is down-regulated in osteoarthritis and regulates chondrocyte apoptosis and lncRNA HOTAIR expression. Biosci. Rep. 2019, 39, BSR20190404. [Google Scholar] [CrossRef]
- Liang, Q.; Asila, A.; Deng, Y.; Liao, J.; Liu, Z.; Fang, R. Osteopontin-induced lncRNA HOTAIR expression is involved in osteoarthritis by regulating cell proliferation. BMC Geriatr. 2021, 21, 57. [Google Scholar] [CrossRef]
- Xu, K.; Meng, Z.; Xian, X.M.; Deng, M.H.; Meng, Q.G.; Fang, W.; Zhang, D.; Long, X. LncRNA PVT1 induces chondrocyte apoptosis through upregulation of TNF-α in synoviocytes by sponging miR-211-3p. Mol. Cell. Probes 2020, 52, 101560. [Google Scholar] [CrossRef]
- Loda, A.; Heard, E. Xist RNA in Action: Past, Present, and Future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [PubMed]
- Pintacuda, G.; Young, A.N.; Cerase, A. Function by Structure: Spotlights on Xist Long Non-coding RNA. Front. Mol. Biosci. 2017, 4, 90. [Google Scholar] [CrossRef]
- Hu, S.; Chang, J.; Li, Y.; Wang, W.; Guo, M.; Zou, E.C.; Wang, Y.; Yang, Y. Long Non-coding RNA XIST as a Potential Prognostic Biomarker in Human Cancers: A Meta-analysis. Oncotarget 2018, 9, 13911–13919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sharma, N.; Drobinski, P.; Kayed, A.; Chen, Z.; Kjelgaard-Petersen, C.F.; Gantzel, T.; Thudium, C.S. Inflammation and Joint Destruction May Be Linked to the Generation of Cartilage Metabolites of ADAMTS-5 Through Activation of Toll-like Receptors. Osteoarthr. Cartil. 2020, 28, 658–668. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, X.; Hu, X.; Dai, L.; Fu, X.; Zhang, J.; Ao, Y. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a miR-136 ‘Sponge’ in Human Cartilage Degradation. Sci. Rep. 2016, 6, 22572. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Zhang, Y.; Wang, J.J. CircRNA hsa_circ_0005105 Upregulates NAMPT Expression and Promotes Chondrocyte Extracellular Matrix Degradation by Sponging miR-26a. Cell Biol. Int. 2017, 41, 1283–1289. [Google Scholar] [CrossRef]
- Wu, Y.; Hong, Z.; Xu, W.; Chen, J.; Wang, Q.; Chen, J.; Ni, W.; Mei, Z.; Xie, Z.; Ma, Y.; et al. Circular RNA circPDE4D Protects Against Osteoarthritis by Binding to miR-103a-3p and Regulating FGF18. Mol. Ther. 2021, 29, 308–323. [Google Scholar] [CrossRef] [PubMed]
- Tachmazidou, I.; Hatzikotoulas, K.; Southam, L.; Esparza-Gordillo, J.; Haberland, V.; Zheng, J.; Johnson, T.; Koprulu, M.; Zengini, E.; Steinberg, J. Identification of New Therapeutic Targets for Osteoarthritis Through Genome-wide Analyses of UK Biobank Data. Nat. Genet. 2019, 51, 230–236. [Google Scholar] [CrossRef]
- Yao, X.; Zhang, J.; Jing, X.; Ye, Y.; Guo, J.; Sun, K.; Guo, F. Fibroblast Growth Factor 18 Exerts Anti-osteoarthritic Effects Through PI3K-AKT Signaling and Mitochondrial Fusion and Fission. Pharmacol. Res. 2019, 139, 314–324. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, Y.; Wang, C.; Zhang, X.; He, D. CircGCN1L1 Promotes Synoviocyte Proliferation and Chondrocyte Apoptosis by Targeting miR-330-3p and TNF-α in TMJ Osteoarthritis. Cell Death Dis. 2020, 11, 284. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Tajes, J.; Soto-Hermida, A.; Vazquez-Mosquera, M.E.; Cortes-Pereira, E.; Mosquera, A.; Fernandez-Moreno, M.; Oreiro, N.; Fernandez-Lopez, C.; Fernandez, J.L.; Rego-Perez, I.; et al. Genome-wide DNA Methylation Analysis of Articular Chondrocytes Reveals a Cluster of Osteoarthritic Patients. Ann. Rheum. Dis. 2014, 73, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Rushton, M.D.; Young, D.A.; Loughlin, J.; Reynard, L.N. Differential DNA Methylation and Expression of Inflammatory and Zinc Transporter Genes Defines Subgroups of Osteoarthritic Hip Patients. Ann. Rheum. Dis. 2015, 74, 1778–1782. [Google Scholar] [CrossRef]
- Alvarez-Garcia, O.; Fisch, K.M.; Wineinger, N.E.; Akagi, R.; Saito, M.; Sasho, T.; Su, A.I.; Lotz, M.K. Increased DNA Methylation and Reduced Expression of Transcription Factors in Human Osteoarthritis Cartilage. Arthritis Rheumatol. 2016, 68, 1876–1886. [Google Scholar] [CrossRef] [PubMed]
- Izda, V.; Martin, J.; Sturdy, C.; Jeffries, M.A. DNA Methylation and Noncoding RNA in OA: Recent Findings and Methodological Advances. Osteoarthr. Cartil. Open 2021, 3, 100208. [Google Scholar] [CrossRef]
- Reynard, L.N. Analysis of Genetics and DNA Methylation in Osteoarthritis: What Have We Learnt About the Disease? Semin. Cell Dev. Biol. 2017, 62, 57–66. [Google Scholar] [CrossRef]
- Ramasamy, S.K.; Kusumbe, A.P.; Wang, L.; Adams, R.H. Endothelial Notch Activity Promotes Angiogenesis and Osteogenesis in Bone. Nature 2014, 507, 376–380. [Google Scholar] [CrossRef]
- Wu, Q.; Zhu, M.; Rosier, R.N.; Zuscik, M.J.; O’Keefe, R.J.; Chen, D. Beta-Catenin, Cartilage, and Osteoarthritis. Ann. N. Y. Acad. Sci. 2010, 1192, 344–350. [Google Scholar] [CrossRef]
- Kawaguchi, H. Regulation of Osteoarthritis Development by Wnt-Beta-Catenin Signaling Through the Endochondral Ossification Process. J. Bone Miner. Res. 2009, 24, 8–11. [Google Scholar] [CrossRef]
- Wang, M.; Li, S.; Xie, W.; Shen, J.; Im, H.J.; Holz, J.D.; Wang, M.; Diekwisch, T.G.; Chen, D. Activation of Beta-Catenin Signalling Leads to Temporomandibular Joint Defects. Eur. Cell Mater. 2014, 28, 223–235. [Google Scholar] [CrossRef]
- Zhu, M.; Tang, D.; Wu, Q.; Hao, S.; Chen, M.; Xie, C.; Rosier, R.N.; O’Keefe, R.J.; Zuscik, M.; Chen, D. Activation of β-Catenin Signaling in Articular Chondrocytes Leads to an Osteoarthritis-Like Phenotype in Adult β-Catenin Conditional Activation Mice. J. Bone Miner. Res. 2009, 24, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Challen, G.A.; Sun, D.; Mayle, A.; Jeong, M.; Luo, M.; Rodriguez, B.; Mallaney, C.; Celik, H.; Yang, L.; Xia, Z.; et al. Dnmt3a and Dnmt3b Have Overlapping and Distinct Functions in Hematopoietic Stem Cells. Cell Stem Cell 2014, 15, 350–364. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.-D.; Maity, S.N.; Lu, J.-F.; Zhang, J.; Liang, S.; Coustry, F.; de Crombrugghe, B.; Yasuda, H. Identification of SOX9 interaction sites in the genome of chondrocytes. PLoS ONE 2010, 5, e10113. [Google Scholar] [CrossRef]
- Tardif, G.; Hum, D.; Pelletier, J.P.; Duval, N.; Martel-Pelletier, J. Regulation of the IGFBP-5 and MMP-13 Genes by the MicroRNAs MiR-140 and MiR-27a in Human Osteoarthritic Chondrocytes. BMC Musculoskelet. Disord. 2009, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Szala, D.; Kopańska, M.; Trojniak, J.; Jabłoński, J.; Hanf-Osetek, D.; Snela, S.; Zawlik, I. The Role of MicroRNAs in the Pathophysiology of Osteoarthritis. Int. J. Mol. Sci. 2024, 25, 6352. [Google Scholar] [CrossRef]
- Duan, L.; Liang, Y.; Xu, X.; Xiao, Y.; Wang, D. Recent progress on the role of miR-140 in cartilage matrix remodelling and its implications for osteoarthritis treatment. Arthritis Res. Ther. 2020, 22, 194. [Google Scholar] [CrossRef]
- Cai, Z.; Long, T.; Zhao, Y.; Lin, R.; Wang, Y. Epigenetic Regulation in Knee Osteoarthritis. Front. Genet. 2022, 13, 942982. [Google Scholar] [CrossRef]
- Miranda-Duarte, A. DNA Methylation in Osteoarthritis: Current Status and Therapeutic Implications. Open Rheumatol. J. 2018, 12, 37–49. [Google Scholar] [CrossRef]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef]
- Cömert Kiliç, S.; Kiliç, N.; Sümbüllü, M.A. Temporomandibular joint osteoarthritis: Cone beam computed tomography findings, clinical features, and correlations. Int. J. Oral Maxillofac. Surg. 2015, 44, 1268–1274. [Google Scholar] [CrossRef] [PubMed]
- Günther, K.P.; Sun, Y. Reliability of radiographic assessment in hip and knee osteoarthritis. Osteoarthr. Cartil. 1999, 7, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Shen, J.; Hui, T. Epigenetic and microRNA regulation during osteoarthritis development. F1000Res. 2015, 4, F1000 Faculty Rev-1092. [Google Scholar] [CrossRef] [PubMed][Green Version]
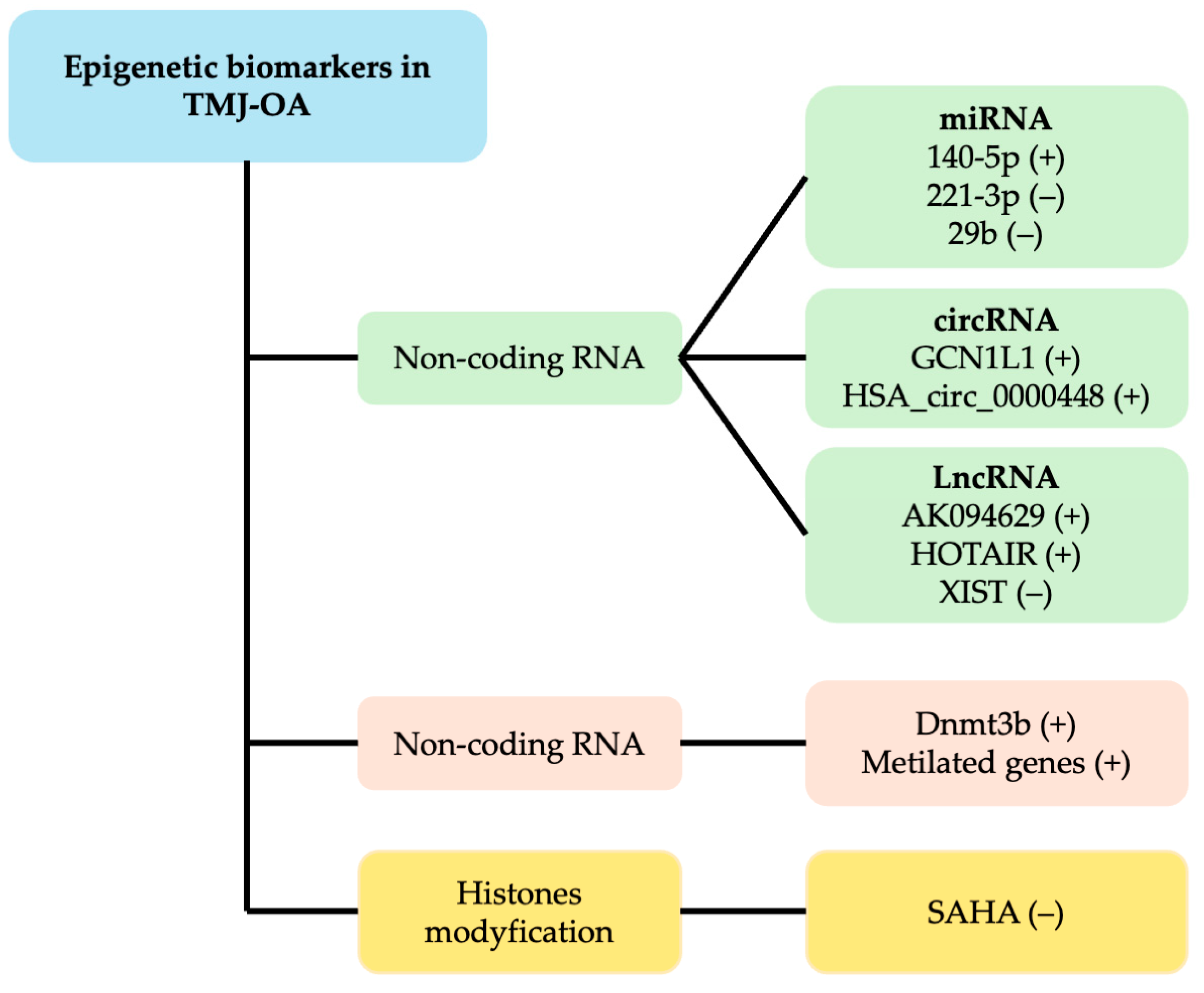
| Reference | TMJ Sample | Sample Characteristics | Epigenetic Biomarkers | Main Results |
|---|---|---|---|---|
| [5] | Synovial tissues | TMJ-OA patients undergoing TMJ disk surgery. | circRNA | circRNA_0000448 upregulation is related to the TNF, IL-1, and IFN-signaling pathways, and downregulated circRNAs are related to myogenesis. |
| [25] | Articular cartilage | Surgically induced TMJ-OA rats. | DNA methylation | There was significant differential methylation of several genes involved in the pathogenesis of TMJ-OA. In the early stage, this resulted in the methylation of genes of the TNF family, such as Adamts5 and Runx. In late stage, this included genes of the VEGFA, CTGF, MEPE, and OMD families. |
| [32] | SMSCs | Patients with TMJ-OA undergoing TMJ debridement surgery. | Histone deacetylation | SAHA attenuated IL-6 secretion in IL-1β-induced SMSCs through the inhibition of the MARK4/NF-κB pathway. |
| [32] | BMSCs of subchondral bones | A mouse model with an OA-like change in the TMJ induced by an experimentally UAC. | miRNA | miR-29b was markedly lower in BMSCs from subchondral bones of TMJ-OA. |
| [33] | Synovial tissues | TMJ-OA patients | miRNA | IL-1β reduced miRNA221-3p expression in a time- and dose-dependent manner in TMJOA synovial fibroblasts. The expression of Ets-1 was induced. |
| [34] | SMSCs | Patients with TMJ-OA were treated surgically. | lncRNA | The downregulation of lncRNA AK094629 attenuated IL-1β-regulated IL-6 expression in TMJ -OA SMSCs by inhibiting MAP3K4. |
| [35] | SMSCs | Patients with TMJ-OA under- going TMJ surgery. | lncRNA | XIST decreased During the chondrogenic differentiation of SMSCs from TMJ. XIST knockdown promoted the chondrogenic differentiation of SMSCs. XIST directly bound to miR-27b-3p and regulated the expression of ADAMTS-5. |
| [36] | MCCs | MCCs from mice were induced by IL-1β (A TMJ-OA model in vitro). | miRNA | MMP13, miR-140-5p, and NF-kB were significantly increased in IL-1β inflammatory responses in MCCs. miR-140-5p regulates TMJ-OA pathogenesis through TGF-β/Smad. |
| [37] | TMJ tissues | TMJ rabbits with surgically induced AL. TMJ from rats with MIA-induced OA. | DNA methylation | The overexpression of Dnmt3b in TMJ stem/progenitor cells led to elevated levels of collagen type II and a reduction in collagen type X, while Dnmt3b knockdown produced the reverse pattern—diminished expression of collagen type II alongside an increase in collagen type X. |
| Feature | Temporomandibular Joint Osteoarthritis (TMJ-OA) | Knee/Hip Osteoarthritis (OA) | References |
|---|---|---|---|
| Joint Structure | Synovial joint with fibrocartilage lining | Synovial joints with hyaline cartilage | [3,9] |
| Primary Function | Mastication, speech; low-load, high-precision | Load-bearing (locomotion and posture) | [121] |
| Mechanical Stress Type | Shear and compressive (chewing) | Predominantly compressive and torsional | [12] |
| Cartilage Regenerative Capacity | Higher due to fibrocartilage | Lower hyaline cartilage is poorly regenerative | [3] |
| Common Age Group | Often affects younger adults (20–40s), especially females | Typically affects older adults (50+ years) | [2] |
| Symptoms | Jaw pain, joint sounds (clicking/crepitus), and limited motion | Pain, stiffness, swelling, and decreased mobility | [1] |
| Imaging Findings | Condylar erosion, osteophytes, and joint space narrowing (MRI/CBCT) | Joint space narrowing, osteophytes, and subchondral sclerosis (X-ray/MRI) | [122,123] |
| Inflammatory Profile | Inflammation may be more prominent in the TMJ | Chronic low-grade inflammation typical | [9] |
| Epigenetic Regulation | TMJ-specific: altered DNA methylation (e.g., DNMT3B, ADAMTS, TGF-β, and Wnt/β-catenin); distinct miRNA and lncRNA expression (e.g., miR-140-5p, HOTAIR, and XIST) | Knee/hip: methylation of SOX9, RUNX2, and COL2A1; miR-140 downregulation; and global DNA methylation alterations | [25,119,124] |
| Therapeutic Research | Focus on epigenetic therapies, e.g., HDAC inhibitors (SAHA and TSA) | Focus on biologicals (e.g., anti-NGF) and joint replacements in advanced cases | [32,51] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, S.; Santander, J.; Barria, D.; Salazar, L.A.; Sandoval, C.; Arias, C.; Iturriaga, V. Epigenetic Biomarkers in Temporomandibular Joint Osteoarthritis: An Emerging Target in Treatment. Int. J. Mol. Sci. 2025, 26, 3668. https://doi.org/10.3390/ijms26083668
Wen S, Santander J, Barria D, Salazar LA, Sandoval C, Arias C, Iturriaga V. Epigenetic Biomarkers in Temporomandibular Joint Osteoarthritis: An Emerging Target in Treatment. International Journal of Molecular Sciences. 2025; 26(8):3668. https://doi.org/10.3390/ijms26083668
Chicago/Turabian StyleWen, Schilin, Javiera Santander, Daniel Barria, Luis A. Salazar, Cristian Sandoval, Consuelo Arias, and Verónica Iturriaga. 2025. "Epigenetic Biomarkers in Temporomandibular Joint Osteoarthritis: An Emerging Target in Treatment" International Journal of Molecular Sciences 26, no. 8: 3668. https://doi.org/10.3390/ijms26083668
APA StyleWen, S., Santander, J., Barria, D., Salazar, L. A., Sandoval, C., Arias, C., & Iturriaga, V. (2025). Epigenetic Biomarkers in Temporomandibular Joint Osteoarthritis: An Emerging Target in Treatment. International Journal of Molecular Sciences, 26(8), 3668. https://doi.org/10.3390/ijms26083668








