The Yeast Gsk-3 Kinase Mck1 Is Necessary for Cell Wall Remodeling in Glucose-Starved and Cell Wall-Stressed Cells
Abstract
1. Introduction
2. Results
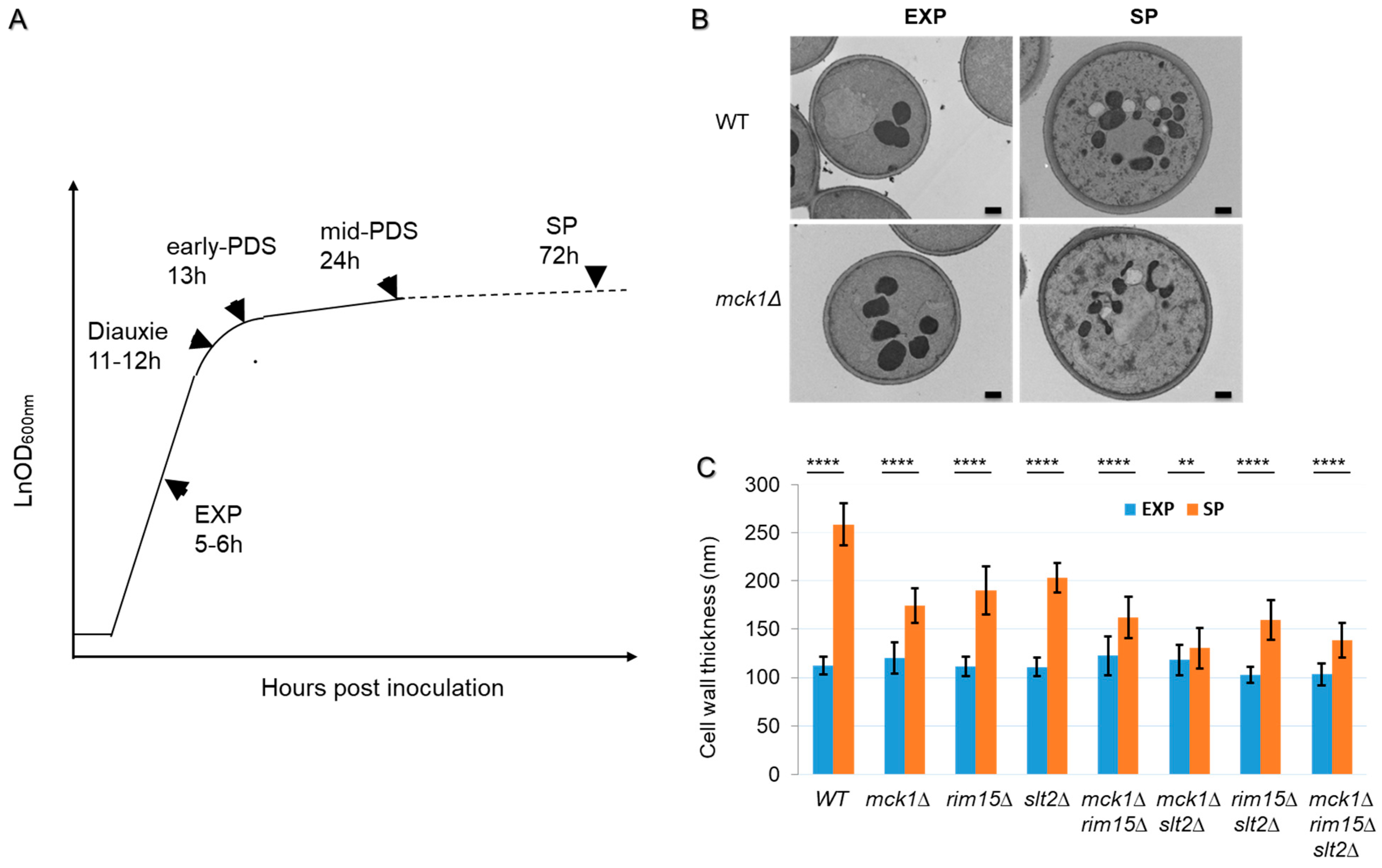
2.1. Mck1 Cooperates with Rim15 and Slt2 to Regulate Cell Wall Thickening in PDS Cells
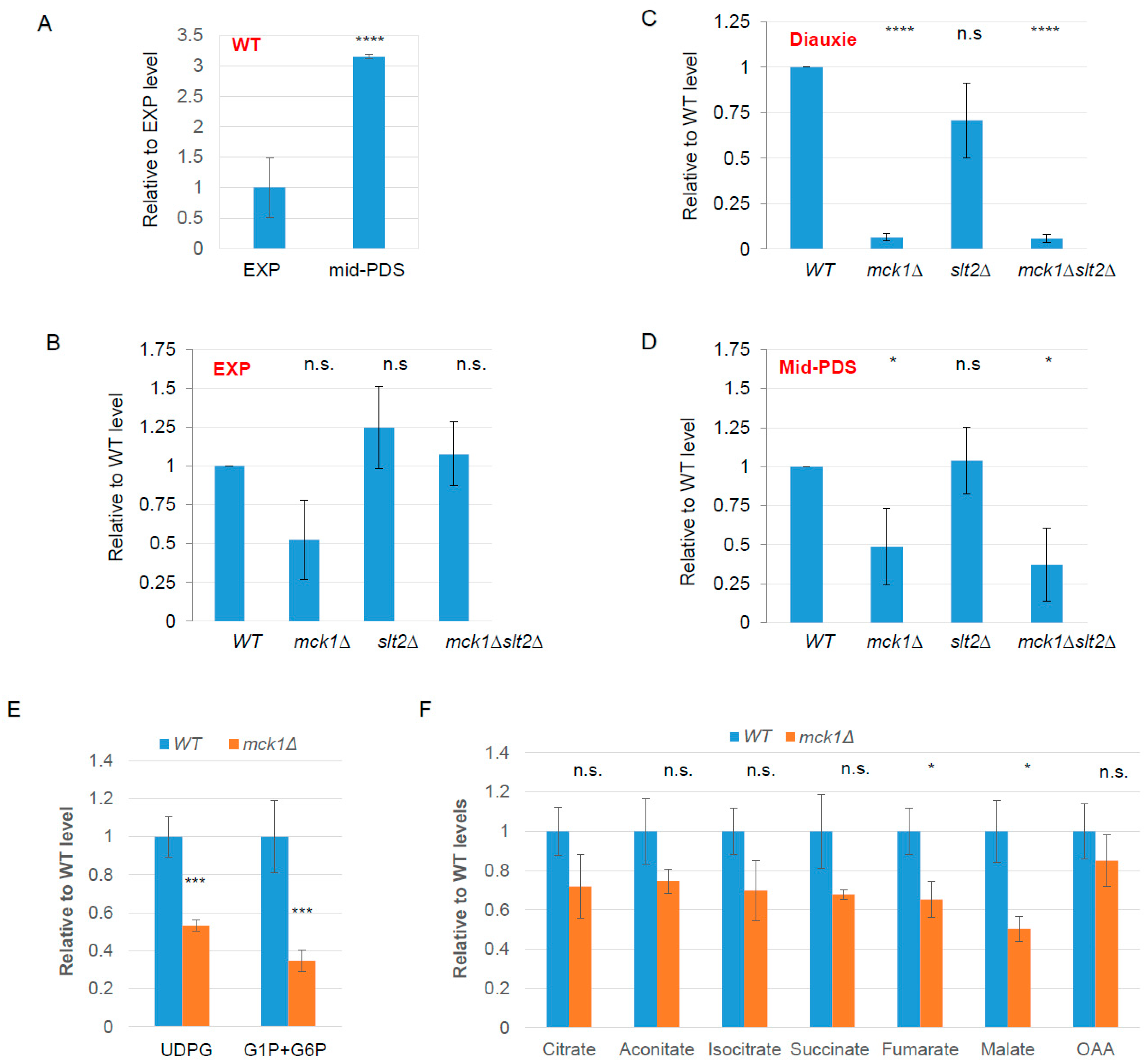
2.2. Mck1 Is Responsible for UDPG Accumulation in PDS Cells
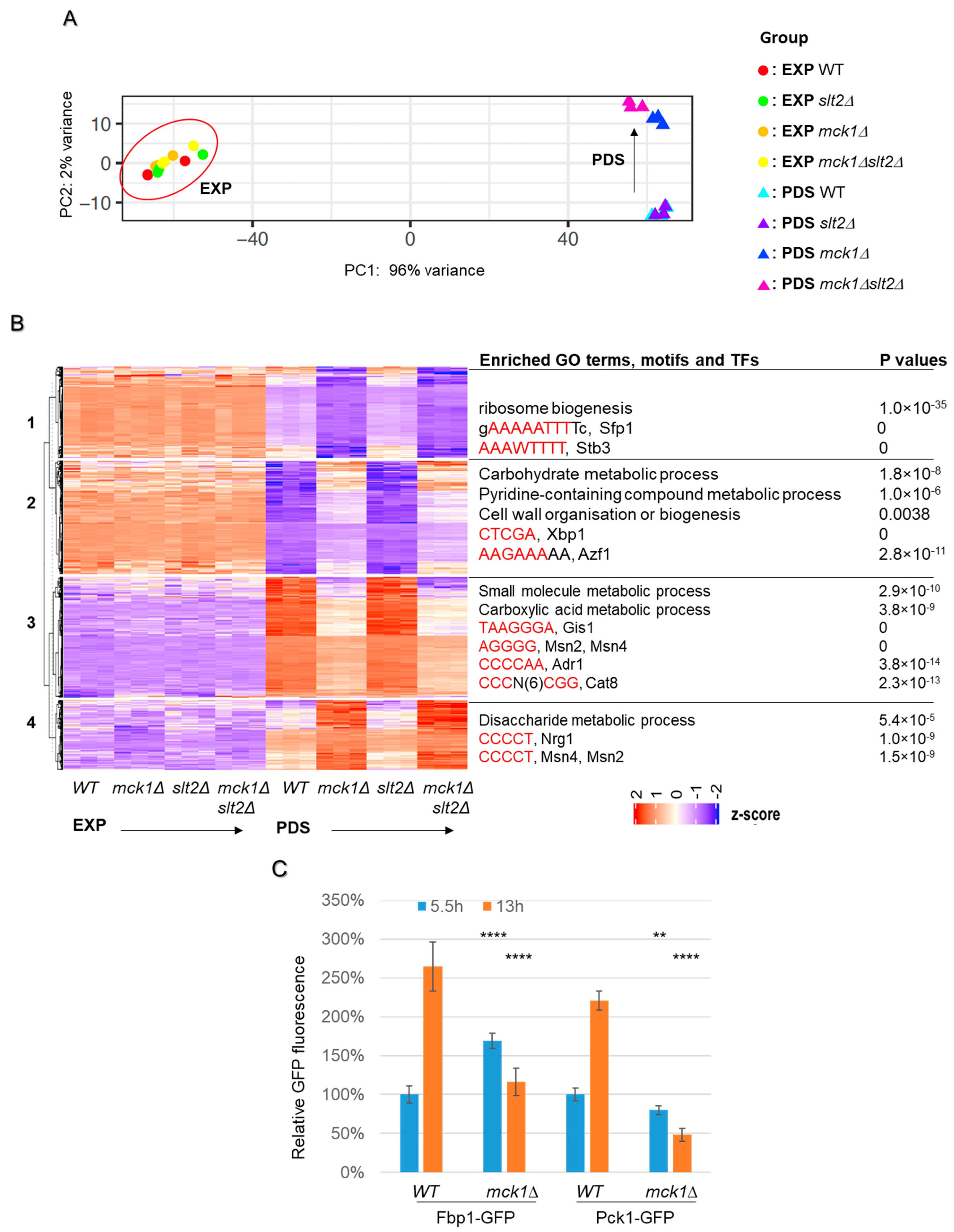
2.3. MCK1 Is Necessary to Promote Gene Expression Implicated in the Stress Response, TCA Cycle and Gluconeogenesis
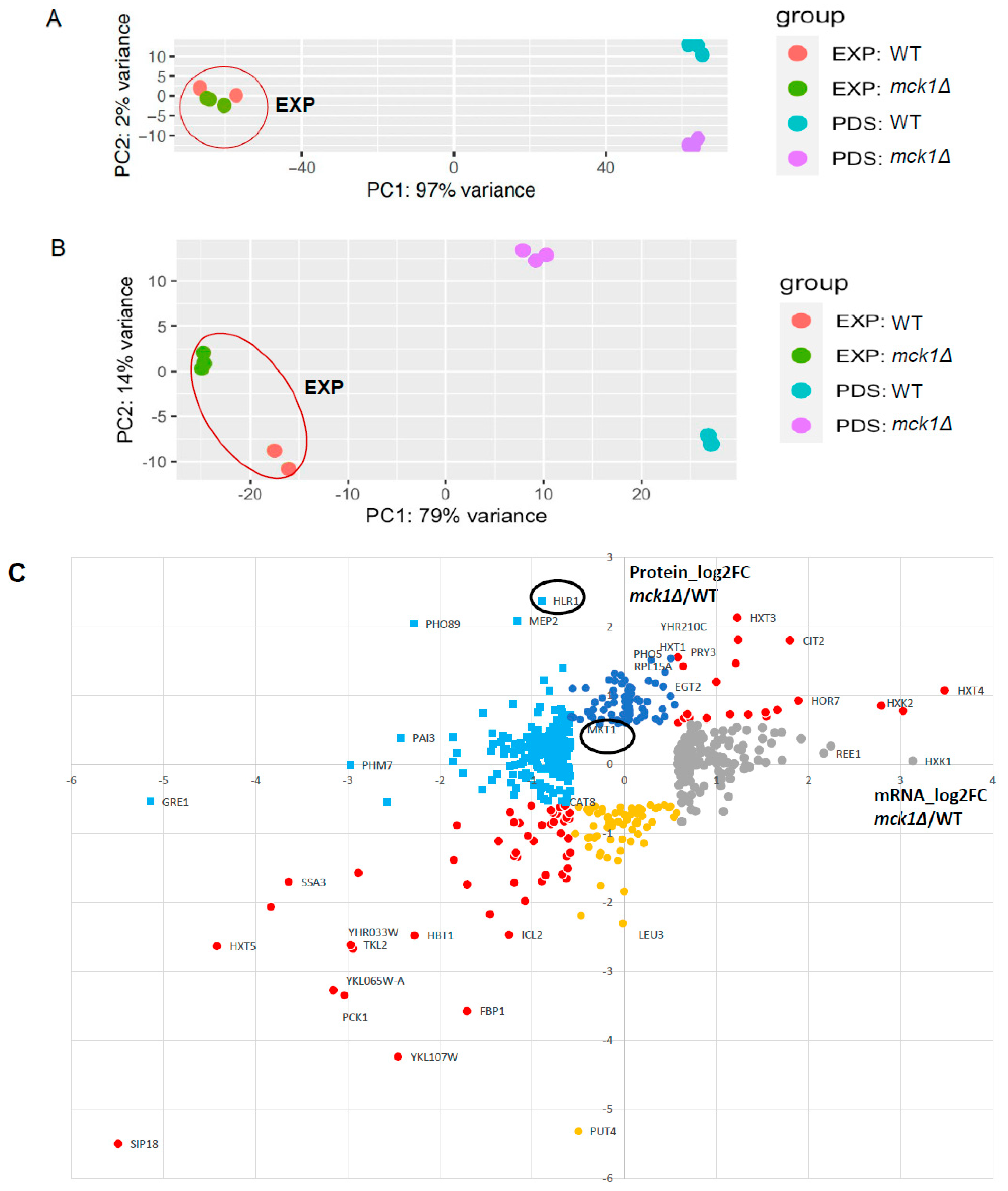
2.4. Integrative Analysis of the Proteome and the Transcriptome Confirms the Transcriptional Control of the Gluconeogenesis and Stress Response by Mck1
2.5. Analysis of the Mck1-Mediated Phosphoproteome Reveals the Conserved Sequence Motifs Targeted by the Gsk-3 Kinases
2.6. Mck1 May Be Involved in the Snf1-Directed Metabolic Reprogram Through the SAGA Coactivator and PKA Inhibition
2.7. Mck1 Phosphotargets Are Implicated in Polarized Growth and Cytokinesis
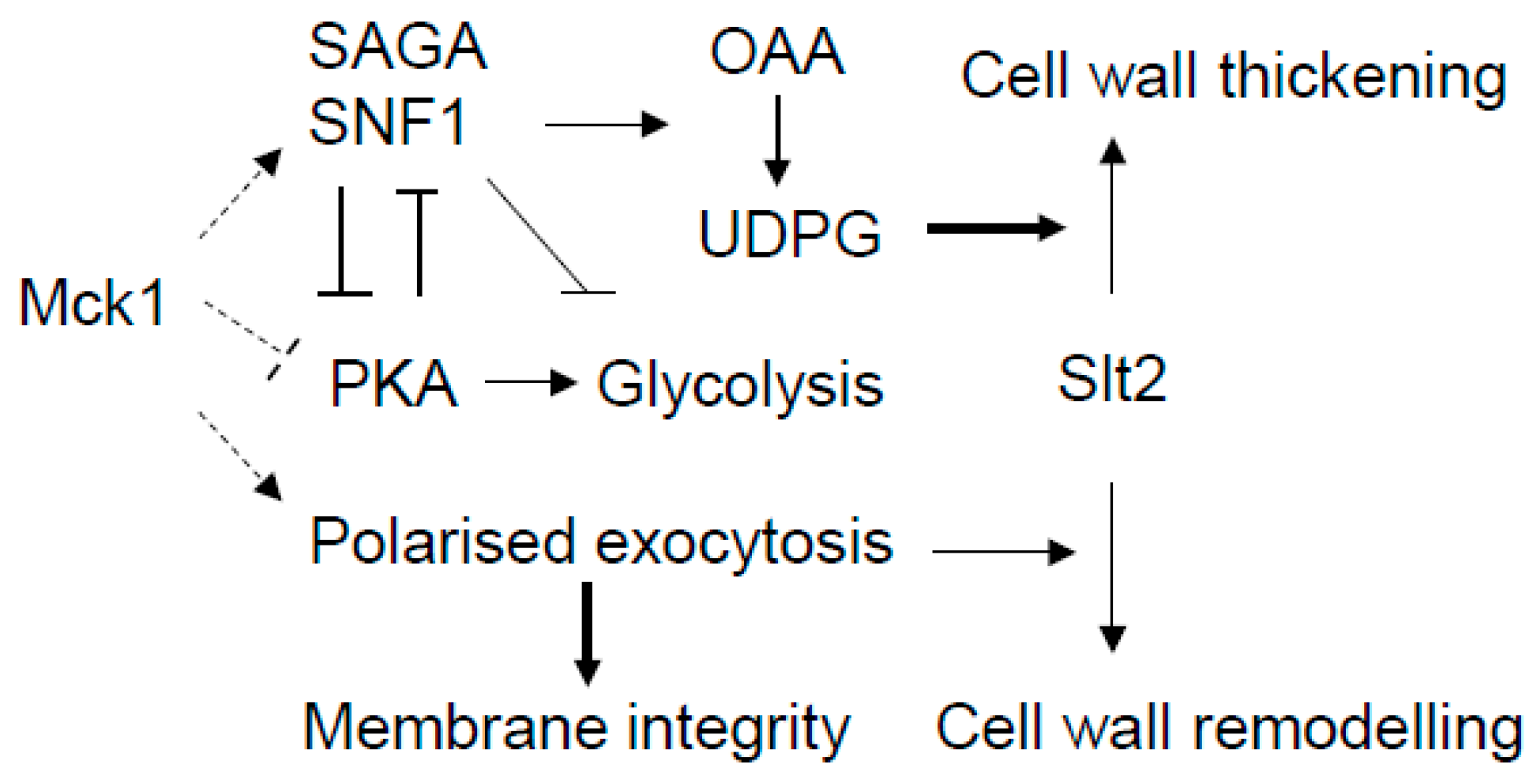
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Phenotypic Assays
4.3. Fluorescence Detection and Quantification
4.4. Transmission Electron Microscopy (TEM)
4.5. UDP-Glucose, TCA Cycle Metabolite Extraction and LC-MS/MS Analysis
4.6. Analyses of the Transcriptome Data
4.7. Total Proteome Isolation and Digestion
4.8. TMT Labeling and LC-MS/MS for Proteome and Phosphoproteome
4.9. Proteome and Phosphoproteome Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar] [CrossRef]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Pavón-Vergés, M.; Rodríguez-Peña, J.M.; Arroyo, J. Control of Gene Expression via the Yeast CWI Pathway. Int. J. Mol. Sci. 2022, 23, 1791. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI Pathway: Regulation of the Transcriptional Adaptive Response to Cell Wall Stress in Yeast. J. Fungi 2017, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Nombela, C.; Arroyo, J. Cooperation between SAGA and SWI/SNF complexes is required for efficient transcriptional responses regulated by the yeast MAPK Slt2. Nucleic Acids Res. 2016, 44, 7159–7172. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Nombela, C.; Arroyo, J. Slt2 MAPK association with chromatin is required for transcriptional activation of Rlm1 dependent genes upon cell wall stress. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 1029–1039. [Google Scholar] [CrossRef]
- Popolo, L.; Gualtieri, T.; Ragni, E. The yeast cell-wall salvage pathway. Med. Mycol. 2001, 39 (Suppl. S1), 111–121. [Google Scholar] [CrossRef]
- J Smits, G.; C Kapteyn, J.; van den Ende, H.; M Klis, F. Cell wall dynamics in yeast. Curr. Opin. Microbiol. 1999, 2, 348–352. [Google Scholar] [CrossRef]
- Smith, A.E.; Zhang, Z.; Thomas, C.R.; Moxham, K.E.; Middelberg, A.P.J. The mechanical properties of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 9871–9874. [Google Scholar] [CrossRef] [PubMed]
- De Virgilio, C. The essence of yeast quiescence. FEMS Microbiol. Rev. 2012, 36, 306–339. [Google Scholar] [CrossRef]
- Gray, J.V.; Petsko, G.A.; Johnston, G.C.; Ringe, D.; Singer, R.A.; Werner-Washburne, M. “Sleeping beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004, 68, 187–206. [Google Scholar] [CrossRef]
- Herman, P.K. Stationary phase in yeast. Curr. Opin. Microbiol. 2002, 5, 602–607. [Google Scholar] [CrossRef]
- Carlson, M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999, 2, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Gancedo, J.M. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. MMBR 1998, 62, 334–361. [Google Scholar] [CrossRef]
- González, A.; Hall, M.N.; Lin, S.-C.; Hardie, D.G. AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab. 2020, 31, 472–492. [Google Scholar] [CrossRef] [PubMed]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov068. [Google Scholar] [CrossRef]
- Young, E.T.; Dombek, K.M.; Tachibana, C.; Ideker, T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 2003, 278, 26146–26158. [Google Scholar] [CrossRef]
- DeMille, D.; Badal, B.D.; Evans, J.B.; Mathis, A.D.; Anderson, J.F.; Grose, J.H. PAS kinase is activated by direct SNF1-dependent phosphorylation and mediates inhibition of TORC1 through the phosphorylation and activation of Pbp1. Mol. Biol. Cell 2015, 26, 569–582. [Google Scholar] [CrossRef]
- Grose, J.H.; Smith, T.L.; Sabic, H.; Rutter, J. Yeast PAS kinase coordinates glucose partitioning in response to metabolic and cell integrity signaling. EMBO J. 2007, 26, 4824–4830. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.L.; Rutter, J. Regulation of glucose partitioning by PAS kinase and Ugp1 phosphorylation. Mol. Cell 2007, 26, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Brauer, M.J.; Yuan, J.; Bennett, B.D.; Lu, W.; Kimball, E.; Botstein, D.; Rabinowitz, J.D. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA 2006, 103, 19302–19307. [Google Scholar] [CrossRef]
- Kassir, Y.; Rubin-Bejerano, I.; Mandel-Gutfreund, Y. The Saccharomyces cerevisiae GSK-3 beta homologs. Curr. Drug Targets 2006, 7, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Quan, Z.; Cao, L.; Tang, Y.; Yan, Y.; Oliver, S.G.; Zhang, N. The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan. PLoS Genet. 2015, 11, e1005282. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Dubouloz, F.; Cameroni, E.; Wanke, V.; Roosen, J.; Winderickx, J.; De Virgilio, C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 2003, 12, 1607–1613. [Google Scholar] [CrossRef]
- Reinders, A.; Bürckert, N.; Boller, T.; Wiemken, A.; De Virgilio, C. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 1998, 12, 2943–2955. [Google Scholar] [CrossRef]
- Aranda, S.; Laguna, A.; de la Luna, S. DYRK family of protein kinases: Evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011, 25, 449–462. [Google Scholar] [CrossRef]
- Cao, L.; Tang, Y.; Quan, Z.; Zhang, Z.; Oliver, S.G.; Zhang, N. Chronological Lifespan in Yeast Is Dependent on the Accumulation of Storage Carbohydrates Mediated by Yak1, Mck1 and Rim15 Kinases. PLoS Genet. 2016, 12, e1006458. [Google Scholar] [CrossRef]
- Zhang, N.; Cao, L. Starvation signals in yeast are integrated to coordinate metabolic reprogramming and stress response to ensure longevity. Curr. Genet. 2017, 63, 839–843. [Google Scholar] [CrossRef]
- Zhang, N.; Quan, Z.; Rash, B.; Oliver, S.G. Synergistic effects of TOR and proteasome pathways on the yeast transcriptome and cell growth. Open Biol. 2013, 3, 120137. [Google Scholar] [CrossRef]
- Thomas-Chollier, M.; Sand, O.; Turatsinze, J.-V.; Janky, R.; Defrance, M.; Vervisch, E.; Brohée, S.; van Helden, J. RSAT: Regulatory sequence analysis tools. Nucleic Acids Res. 2008, 36, W119–W127. [Google Scholar] [CrossRef] [PubMed]
- McKnight, J.N.; Boerma, J.W.; Breeden, L.L.; Tsukiyama, T. Global Promoter Targeting of a Conserved Lysine Deacetylase for Transcriptional Shutoff during Quiescence Entry. Mol. Cell 2015, 59, 732–743. [Google Scholar] [CrossRef]
- Huber, A.; French, S.L.; Tekotte, H.; Yerlikaya, S.; Stahl, M.; Perepelkina, M.P.; Tyers, M.; Rougemont, J.; Beyer, A.L.; Loewith, R. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011, 30, 3052–3064. [Google Scholar] [CrossRef] [PubMed]
- Liko, D.; Conway, M.K.; Grunwald, D.S.; Heideman, W. Stb3 plays a role in the glucose-induced transition from quiescence to growth in Saccharomyces cerevisiae. Genetics 2010, 185, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Liko, D.; Slattery, M.G.; Heideman, W. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 26623–26628. [Google Scholar] [CrossRef]
- Albert, B.; Tomassetti, S.; Gloor, Y.; Dilg, D.; Mattarocci, S.; Kubik, S.; Hafner, L.; Shore, D. Sfp1 regulates transcriptional networks driving cell growth and division through multiple promoter-binding modes. Genes Dev. 2019, 33, 288–293. [Google Scholar] [CrossRef]
- Shore, D.; Zencir, S.; Albert, B. Transcriptional control of ribosome biogenesis in yeast: Links to growth and stress signals. Biochem. Soc. Trans. 2021, 49, 1589–1599. [Google Scholar] [CrossRef]
- Monteiro, P.T.; Oliveira, J.; Pais, P.; Antunes, M.; Palma, M.; Cavalheiro, M.; Galocha, M.; Godinho, C.P.; Martins, L.C.; Bourbon, N.; et al. YEASTRACT+: A portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res. 2020, 48, D642–D649. [Google Scholar] [CrossRef]
- Tachibana, C.; Yoo, J.Y.; Tagne, J.-B.; Kacherovsky, N.; Lee, T.I.; Young, E.T. Combined Global Localization Analysis and Transcriptome Data Identify Genes That Are Directly Coregulated by Adr1 and Cat8. Mol. Cell. Biol. 2005, 25, 2138–2146. [Google Scholar] [CrossRef]
- Ahuatzi, D.; Riera, A.; Pela Ez, R.; Herrero, P.; Moreno, F. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 2007, 282, 4485–4493. [Google Scholar] [CrossRef]
- Vega, M.; Riera, A.; Fernández-Cid, A.; Herrero, P.; Moreno, F. Hexokinase 2 Is an Intracellular Glucose Sensor of Yeast Cells That Maintains the Structure and Activity of Mig1 Protein Repressor Complex. J. Biol. Chem. 2016, 291, 7267–7285. [Google Scholar] [CrossRef] [PubMed]
- Lesko, M.A.; Chandrashekarappa, D.G.; Jordahl, E.M.; Oppenheimer, K.G.; Bowman, R.W.; Shang, C.; Durrant, J.D.; Schmidt, M.C.; O’Donnell, A.F. Changing course: Glucose starvation drives nuclear accumulation of Hexokinase 2 in S. cerevisiae. PLoS Genet. 2023, 19, e1010745. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.; Hayes, J.D.; Sutherland, C. A partnership with the proteasome; the destructive nature of GSK3. Biochem. Pharmacol. 2018, 147, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Doble, B.W.; Woodgett, J.R. GSK-3: Tricks of the trade for a multi-tasking kinase. J. Cell Sci. 2003, 116, 1175–1186. [Google Scholar] [CrossRef]
- Patel, P.; Woodgett, J.R. Glycogen Synthase Kinase 3: A Kinase for All Pathways? Curr. Top. Dev. Biol. 2017, 123, 277–302. [Google Scholar]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Mok, J.; Kim, P.M.; Lam, H.Y.K.; Piccirillo, S.; Zhou, X.; Jeschke, G.R.; Sheridan, D.L.; Parker, S.A.; Desai, V.; Jwa, M.; et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 2010, 3, ra12. [Google Scholar] [CrossRef]
- Valk, E.; Örd, M.; Faustova, I.; Loog, M. CDK signaling via nonconventional CDK phosphorylation sites. Mol. Biol. Cell 2023, 34, pe5. [Google Scholar] [CrossRef]
- Kishi, T.; Ikeda, A.; Nagao, R.; Koyama, N. The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc. Natl. Acad. Sci. USA 2007, 104, 17418–17423. [Google Scholar] [CrossRef]
- Lee, J.; Moir, R.D.; McIntosh, K.B.; Willis, I.M. TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol. Cell 2012, 45, 836–843. [Google Scholar] [CrossRef]
- Zimmermann, C.; Santos, A.; Gable, K.; Epstein, S.; Gururaj, C.; Chymkowitch, P.; Pultz, D.; Rødkær, S.V.; Clay, L.; Bjørås, M.; et al. TORC1 inhibits GSK3-mediated Elo2 phosphorylation to regulate very long chain fatty acid synthesis and autophagy. Cell Rep. 2013, 5, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Mizunuma, M.; Hirata, D.; Miyaoka, R.; Miyakawa, T. GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J. 2001, 20, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Bodenmiller, B.; Wanka, S.; Kraft, C.; Urban, J.; Campbell, D.; Pedrioli, P.G.; Gerrits, B.; Picotti, P.; Lam, H.; Vitek, O.; et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 2010, 3, rs4. [Google Scholar] [CrossRef]
- Gutin, J.; Joseph-Strauss, D.; Sadeh, A.; Shalom, E.; Friedman, N. Genetic screen of the yeast environmental stress response dynamics uncovers distinct regulatory phases. Mol. Syst. Biol. 2019, 15, e8939. [Google Scholar] [CrossRef]
- Hirata, Y.; Andoh, T.; Asahara, T.; Kikuchi, A. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol. Biol. Cell 2003, 14, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.C. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000, 19, 4936–4943. [Google Scholar] [CrossRef]
- Vincent, O.; Townley, R.; Kuchin, S.; Carlson, M. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 2001, 15, 1104–1114. [Google Scholar] [CrossRef]
- Hedbacker, K.; Carlson, M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008, 13, 2408–2420. [Google Scholar] [CrossRef]
- Fischer, V.; Schumacher, K.; Tora, L.; Devys, D. Global role for coactivator complexes in RNA polymerase II transcription. Transcription 2019, 10, 29–36. [Google Scholar] [CrossRef]
- Timmers, H.T.M. SAGA and TFIID: Friends of TBP drifting apart. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194604. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Mocciaro, G.; Griffin, J.L.; Zhang, N. The Saccharomyces cerevisiae acetyltransferase Gcn5 exerts antagonistic pleiotropic effects on chronological ageing. Aging 2023, 15, 10915–10937. [Google Scholar] [CrossRef] [PubMed]
- Abate, G.; Bastonini, E.; Braun, K.A.; Verdone, L.; Young, E.T.; Caserta, M. Snf1/AMPK regulates Gcn5 occupancy, H3 acetylation and chromatin remodelling at S. cerevisiae ADY2 promoter. Biochim Biophys Acta 2012, 1819, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Agricola, E.; Verdone, L.; Di Mauro, E.; Caserta, M. H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes. Mol. Microbiol. 2006, 62, 1433–1446. [Google Scholar] [CrossRef]
- Agricola, E.; Verdone, L.; Xella, B.; Di Mauro, E.; Caserta, M. Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae. Biochemistry 2004, 43, 8878–8884. [Google Scholar] [CrossRef]
- Biddick, R.K.; Law, G.L.; Young, E.T. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS ONE 2008, 3, e1436. [Google Scholar] [CrossRef]
- Biddick, R.K.; Law, G.L.; Chin, K.K.B.; Young, E.T. The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. J. Biol. Chem. 2008, 283, 33101–33109. [Google Scholar] [CrossRef]
- Kassem, S.; Villanyi, Z.; Collart, M.A. Not5-dependent co-translational assembly of Ada2 and Spt20 is essential for functional integrity of SAGA. Nucleic Acids Res. 2017, 45, 1186–1199. [Google Scholar] [CrossRef]
- Cui, Y.; Ramnarain, D.B.; Chiang, Y.-C.; Ding, L.-H.; McMahon, J.S.; Denis, C.L. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genom. 2008, 279, 323–337. [Google Scholar] [CrossRef]
- Venters, B.J.; Wachi, S.; Mavrich, T.N.; Andersen, B.E.; Jena, P.; Sinnamon, A.J.; Jain, P.; Rolleri, N.S.; Jiang, C.; Hemeryck-Walsh, C.; et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 2011, 41, 480–492. [Google Scholar] [CrossRef]
- Peng, W.; Togawa, C.; Zhang, K.; Kurdistani, S.K. Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics 2008, 179, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Griffioen, G.; Swinnen, S.; Thevelein, J.M. Feedback inhibition on cell wall integrity signaling by Zds1 involves Gsk3 phosphorylation of a cAMP-dependent protein kinase regulatory subunit. J. Biol. Chem. 2003, 278, 23460–23471. [Google Scholar] [CrossRef] [PubMed]
- Rayner, T.F.; Gray, J.V.; Thorner, J.W. Direct and novel regulation of cAMP-dependent protein kinase by Mck1p, a yeast glycogen synthase kinase-3. J. Biol. Chem. 2002, 277, 16814–16822. [Google Scholar] [CrossRef]
- Barrett, L.; Orlova, M.; Maziarz, M.; Kuchin, S. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell 2012, 11, 119–128. [Google Scholar] [CrossRef]
- Hedbacker, K.; Townley, R.; Carlson, M. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 2004, 24, 1836–1843. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.R.; Johnson, T.R.; Dollard, C.; Shuster, J.R.; Denis, C.L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell 1989, 56, 409–419. [Google Scholar] [CrossRef]
- Ratnakumar, S.; Kacherovsky, N.; Arms, E.; Young, E.T. Snf1 controls the activity of adr1 through dephosphorylation of Ser230. Genetics 2009, 182, 735–745. [Google Scholar] [CrossRef]
- Bi, E.; Park, H.-O. Cell polarization and cytokinesis in budding yeast. Genetics 2012, 191, 347–387. [Google Scholar] [CrossRef]
- Etienne-Manneville, S.; Hall, A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr. Opin. Cell Biol. 2003, 15, 67–72. [Google Scholar] [CrossRef]
- Wu, C.-F.; Lew, D.J. Beyond symmetry-breaking: Competition and negative feedback in GTPase regulation. Trends Cell Biol. 2013, 23, 476–483. [Google Scholar] [CrossRef]
- Chiou, J.-G.; Balasubramanian, M.K.; Lew, D.J. Cell Polarity in Yeast. Annu. Rev. Cell Dev. Biol. 2017, 33, 77–101. [Google Scholar] [CrossRef] [PubMed]
- Pruyne, D.; Bretscher, A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000, 113 Pt 3, 365–375. [Google Scholar] [CrossRef]
- Xie, Y.; Miao, Y. Polarisome assembly mediates actin remodeling during polarized yeast and fungal growth. J. Cell Sci. 2021, 134, jcs247916. [Google Scholar] [CrossRef] [PubMed]
- Kang, P.J.; Lee, M.E.; Park, H.-O. Bud3 activates Cdc42 to establish a proper growth site in budding yeast. J. Cell Biol. 2014, 206, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Takaku, T.; Ogura, K.; Kumeta, H.; Yoshida, N.; Inagaki, F. Solution structure of a novel Cdc42 binding module of Bem1 and its interaction with Ste20 and Cdc42. J. Biol. Chem. 2010, 285, 19346–19353. [Google Scholar] [CrossRef]
- Lamson, R.E.; Winters, M.J.; Pryciak, P.M. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 2002, 22, 2939–2951. [Google Scholar] [CrossRef]
- Mösch, H.U.; Roberts, R.L.; Fink, G.R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5352–5356. [Google Scholar] [CrossRef]
- Bretscher, A. Polarized growth and organelle segregation in yeast: The tracks, motors, and receptors. J. Cell Biol. 2003, 160, 811–816. [Google Scholar] [CrossRef]
- Buxbaum, A.R.; Haimovich, G.; Singer, R.H. In the right place at the right time: Visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015, 16, 95–109. [Google Scholar] [CrossRef]
- Lwin, K.M.; Li, D.; Bretscher, A. Kinesin-related Smy1 enhances the Rab-dependent association of myosin-V with secretory cargo. Mol. Biol. Cell 2016, 27, 2450–2462. [Google Scholar] [CrossRef]
- Glomb, O.; Wu, Y.; Rieger, L.; Rüthnick, D.; Mulaw, M.A.; Johnsson, N. The cell polarity proteins Boi1 and Boi2 direct an actin nucleation complex to sites of exocytosis in Saccharomyces cerevisiae. J. Cell Sci. 2020, 133, jcs237982. [Google Scholar] [CrossRef] [PubMed]
- Masgrau, A.; Battola, A.; Sanmartin, T.; Pryszcz, L.P.; Gabaldón, T.; Mendoza, M. Distinct roles of the polarity factors Boi1 and Boi2 in the control of exocytosis and abscission in budding yeast. Mol. Biol. Cell 2017, 28, 3082–3094. [Google Scholar] [CrossRef]
- Luo, G.; Gruhler, A.; Liu, Y.; Jensen, O.N.; Dickson, R.C. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 2008, 283, 10433–10444. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Aguilar, P.S.; Fröhlich, F.; Chu, F.; Moreira, K.; Burlingame, A.L.; Walter, P. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007, 26, 4946–4955. [Google Scholar] [CrossRef]
- Smythe, E.; Ayscough, K.R. The Ark1/Prk1 family of protein kinases. Regulators of endocytosis and the actin skeleton. EMBO Rep. 2003, 4, 246–251. [Google Scholar] [CrossRef]
- Roelants, F.M.; Leskoske, K.L.; Pedersen, R.T.A.; Muir, A.; Liu, J.M.-H.; Finnigan, G.C.; Thorner, J. TOR Complex 2-Regulated Protein Kinase Fpk1 Stimulates Endocytosis via Inhibition of Ark1/Prk1-Related Protein Kinase Akl1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2017, 37, e00627-16. [Google Scholar] [CrossRef]
- Bourgoint, C.; Rispal, D.; Berti, M.; Filipuzzi, I.; Helliwell, S.B.; Prouteau, M.; Loewith, R. Target of rapamycin complex 2-dependent phosphorylation of the coat protein Pan1 by Akl1 controls endocytosis dynamics in Saccharomyces cerevisiae. J. Biol. Chem. 2018, 293, 12043–12053. [Google Scholar] [CrossRef]
- Tolsma, T.O.; Cuevas, L.M.; Di Pietro, S.M. The Sla1 adaptor-clathrin interaction regulates coat formation and progression of endocytosis. Traffic 2018, 19, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Tolsma, T.O.; Febvre, H.P.; Olson, D.M.; Di Pietro, S.M. Cargo-mediated recruitment of the endocytic adaptor protein Sla1 in S. cerevisiae. J. Cell Sci. 2020, 133, jcs247684. [Google Scholar] [CrossRef]
- Feliciano, D.; Di Pietro, S.M. SLAC, a complex between Sla1 and Las17, regulates actin polymerization during clathrin-mediated endocytosis. Mol. Biol. Cell 2012, 23, 4256–4272. [Google Scholar] [CrossRef]
- Sun, Y.; Leong, N.T.; Wong, T.; Drubin, D.G. A Pan1/End3/Sla1 complex links Arp2/3-mediated actin assembly to sites of clathrin-mediated endocytosis. Mol. Biol. Cell 2015, 26, 3841–3856. [Google Scholar] [CrossRef] [PubMed]
- de León, N.; Valdivieso, M.-H. The long life of an endocytic patch that misses AP-2. Curr. Genet. 2016, 62, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.S.; Allwood, E.G.; Smith, A.P.C.; Gardiner, F.C.; Costa, R.; Winder, S.J.; Ayscough, K.R. The WASP homologue Las17 activates the novel actin-regulatory activity of Ysc84 to promote endocytosis in yeast. Mol. Biol. Cell 2009, 20, 1618–1628. [Google Scholar] [CrossRef]
- Tonikian, R.; Xin, X.; Toret, C.P.; Gfeller, D.; Landgraf, C.; Panni, S.; Paoluzi, S.; Castagnoli, L.; Currell, B.; Seshagiri, S.; et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009, 7, e1000218. [Google Scholar] [CrossRef]
- Farrell, K.B.; McDonald, S.; Lamb, A.K.; Worcester, C.; Peersen, O.B.; Di Pietro, S.M. Novel function of a dynein light chain in actin assembly during clathrin-mediated endocytosis. J. Cell Biol. 2017, 216, 2565–2580. [Google Scholar] [CrossRef]
- Lamb, A.K.; Fernandez, A.N.; Peersen, O.B.; Di Pietro, S.M. The dynein light chain protein Tda2 functions as a dimerization engine to regulate actin capping protein during endocytosis. Mol. Biol. Cell 2021, 32, 1459–1473. [Google Scholar] [CrossRef] [PubMed]
- Lamb, A.K.; Fernandez, A.N.; Eadaim, A.; Johnson, K.; Di Pietro, S.M. Mechanism of actin capping protein recruitment and turnover during clathrin-mediated endocytosis. J. Cell Biol. 2024, 223, e202306154. [Google Scholar] [CrossRef]
- Shin, M.; van Leeuwen, J.; Boone, C.; Bretscher, A. Yeast Aim21/Tda2 both regulates free actin by reducing barbed end assembly and forms a complex with Cap1/Cap2 to balance actin assembly between patches and cables. Mol. Biol. Cell 2018, 29, 923–936. [Google Scholar] [CrossRef]
- Marquardt, J.; Chen, X.; Bi, E. Reciprocal regulation by Elm1 and Gin4 controls septin hourglass assembly and remodeling. J. Cell Biol. 2024, 223, e202308143. [Google Scholar] [CrossRef]
- Okada, H.; Chen, X.; Wang, K.; Marquardt, J.; Bi, E. Bni5 tethers myosin-II to septins to enhance retrograde actin flow and the robustness of cytokinesis. BioRxiv, 2023; BioRxiv:2023.11.07.566094. [Google Scholar]
- Cassani, C.; Raspelli, E.; Chiroli, E.; Fraschini, R. Vhs2 is a novel regulator of septin dynamics in budding yeast. Cell Cycle 2014, 13, 1590–1601. [Google Scholar] [CrossRef]
- Devrekanli, A.; Foltman, M.; Roncero, C.; Sanchez-Diaz, A.; Labib, K. Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J. Cell Sci. 2012, 125, 5453–5466. [Google Scholar] [CrossRef] [PubMed]
- Jendretzki, A.; Ciklic, I.; Rodicio, R.; Schmitz, H.-P.; Heinisch, J.J. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol. Genet. Genomics 2009, 282, 437–451. [Google Scholar] [CrossRef]
- Korinek, W.S.; Bi, E.; Epp, J.A.; Wang, L.; Ho, J.; Chant, J. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 2000, 10, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Nishihama, R.; Onishi, M.; Pringle, J.R. Role of the Hof1-Cyk3 interaction in cleavage-furrow ingression and primary-septum formation during yeast cytokinesis. Mol. Biol. Cell 2018, 29, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; MacTaggart, B.; Ohya, Y.; Bi, E. The kinetic landscape and interplay of protein networks in cytokinesis. iScience 2021, 24, 101917. [Google Scholar] [CrossRef]
- Kono, K.; Al-Zain, A.; Schroeder, L.; Nakanishi, M.; Ikui, A.E. Plasma membrane/cell wall perturbation activates a novel cell cycle checkpoint during G1 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2016, 113, 6910–6915. [Google Scholar] [CrossRef]
- Herth, W. Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: Evidence for a gap between polymerization and microfibril formation. J. Cell Biol. 1980, 87, 442–450. [Google Scholar] [CrossRef]
- Martín, H.; Rodríguez-Pachón, J.M.; Ruiz, C.; Nombela, C.; Molina, M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 1511–1519. [Google Scholar] [CrossRef]
- Shimizu, J.; Yoda, K.; Yamasaki, M. The hypo-osmolarity-sensitive phenotype of the Saccharomyces cerevisiae hpo2 mutant is due to a mutation in PKC1, which regulates expression of beta-glucanase. Mol. Gen. Genet. 1994, 242, 641–648. [Google Scholar] [CrossRef]
- Martín, H.; Castellanos, M.C.; Cenamor, R.; Sánchez, M.; Molina, M.; Nombela, C. Molecular and functional characterization of a mutant allele of the mitogen-activated protein-kinase gene SLT2(MPK1) rescued from yeast autolytic mutants. Curr. Genet. 1996, 29, 516–522. [Google Scholar] [CrossRef]
- Brennwald, P.; Rossi, G. Spatial regulation of exocytosis and cell polarity: Yeast as a model for animal cells. FEBS Lett. 2007, 581, 2119–2124. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Feng, S.; Wu, B.; Guo, W. Polarized Exocytosis. Cold Spring Harb. Perspect. Biol. 2017, 9, a027870. [Google Scholar] [CrossRef]
- Park, H.-O.; Bi, E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007, 71, 48–96. [Google Scholar] [CrossRef] [PubMed]
- Palomino, A.; Herrero, P.; Moreno, F. Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae. Biochem. J. 2005, 388, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Jouandot, D.; Roy, A.; Kim, J.-H. Functional dissection of the glucose signaling pathways that regulate the yeast glucose transporter gene (HXT) repressor Rgt1. J. Cell. Biochem. 2011, 112, 3268–3275. [Google Scholar] [CrossRef]
- Kim, J.-H.; Johnston, M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 26144–26149. [Google Scholar] [CrossRef]
- Palomino, A.; Herrero, P.; Moreno, F. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 2006, 34, 1427–1438. [Google Scholar] [CrossRef]
- Roy, A.; Shin, Y.J.; Cho, K.H.; Kim, J.-H. Mth1 regulates the interaction between the Rgt1 repressor and the Ssn6-Tup1 corepressor complex by modulating PKA-dependent phosphorylation of Rgt1. Mol. Biol. Cell 2013, 24, 1493–1503. [Google Scholar] [CrossRef]
- Gancedo, J.M.; Flores, C.-L.; Gancedo, C. The repressor Rgt1 and the cAMP-dependent protein kinases control the expression of the SUC2 gene in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2015, 1850, 1362–1367. [Google Scholar] [CrossRef]
- Roy, A.; Jouandot, D.; Cho, K.H.; Kim, J.-H. Understanding the mechanism of glucose-induced relief of Rgt1-mediated repression in yeast. FEBS Open Bio 2014, 4, 105–111. [Google Scholar] [CrossRef]
- Nicastro, R.; Tripodi, F.; Gaggini, M.; Castoldi, A.; Reghellin, V.; Nonnis, S.; Tedeschi, G.; Coccetti, P. Snf1 Phosphorylates Adenylate Cyclase and Negatively Regulates Protein Kinase A-dependent Transcription in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 24715–24726. [Google Scholar] [CrossRef] [PubMed]
- Al-Zain, A.; Schroeder, L.; Sheglov, A.; Ikui, A.E. Cdc6 degradation requires phosphodegron created by GSK-3 and Cdk1 for SCFCdc4 recognition in Saccharomyces cerevisiae. Mol. Biol. Cell 2015, 26, 2609–2619. [Google Scholar] [CrossRef]
- Reinke, A.; Chen, J.C.-Y.; Aronova, S.; Powers, T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J. Biol. Chem. 2006, 281, 31616–31626. [Google Scholar] [CrossRef] [PubMed]
- Kuranda, K.; Leberre, V.; Sokol, S.; Palamarczyk, G.; François, J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 2006, 61, 1147–1166. [Google Scholar] [CrossRef] [PubMed]
- Rathi, S.; Polat, I.; Pereira, G. The budding yeast GSK-3 homologue Mck1 is an essential component of the spindle position checkpoint. Open Biol. 2022, 12, 220203. [Google Scholar] [CrossRef]
- McQueen, J.; van Dyk, D.; Young, B.; Loewen, C.; Measday, V. The Mck1 GSK-3 kinase inhibits the activity of Clb2-Cdk1 post-nuclear division. Cell Cycle 2012, 11, 3421–3432. [Google Scholar] [CrossRef]
- Holt, L.J.; Tuch, B.B.; Villén, J.; Johnson, A.D.; Gygi, S.P.; Morgan, D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 2009, 325, 1682–1686. [Google Scholar] [CrossRef]
- Lyons, N.A.; Fonslow, B.R.; Diedrich, J.K.; Yates, J.R.; Morgan, D.O. Sequential primed kinases create a damage-responsive phosphodegron on Eco1. Nat. Struct. Mol. Biol. 2013, 20, 194–201. [Google Scholar] [CrossRef]
- Seoane, A.I.; Morgan, D.O. Firing of Replication Origins Frees Dbf4-Cdc7 to Target Eco1 for Destruction. Curr. Biol. 2017, 27, 2849–2855.e2. [Google Scholar] [CrossRef]
- Drechsler, H.; Tan, A.N.; Liakopoulos, D. Yeast GSK-3 kinase regulates astral microtubule function through phosphorylation of the microtubule-stabilizing kinesin Kip2. J. Cell Sci. 2015, 128, 3910–3921. [Google Scholar] [CrossRef]
- Linding, R.; Jensen, L.J.; Ostheimer, G.J.; van Vugt, M.A.T.M.; Jørgensen, C.; Miron, I.M.; Diella, F.; Colwill, K.; Taylor, L.; Elder, K.; et al. Systematic discovery of in vivo phosphorylation networks. Cell 2007, 129, 1415–1426. [Google Scholar] [CrossRef]
- Sutherland, C. What Are the bona fide GSK3 Substrates? Int. J. Alzheimers Dis. 2011, 2011, 505607. [Google Scholar] [CrossRef]
- Kockeritz, L.; Doble, B.; Patel, S.; Woodgett, J.R. Glycogen synthase kinase-3—An overview of an over-achieving protein kinase. Curr. Drug Targets 2006, 7, 1377–1388. [Google Scholar] [CrossRef]
- Hajka, D.; Budziak, B.; Pietras, Ł.; Duda, P.; McCubrey, J.A.; Gizak, A. GSK3 as a Regulator of Cytoskeleton Architecture: Consequences for Health and Disease. Cells 2021, 10, 2092. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, T.; Waterman-Storer, C.M. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J. Cell Biol. 2005, 169, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.L.; McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 1999, 15, 1541–1553. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A.; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Wright, R. Transmission Electron Microscopy of Yeast. Microsc. Res. Tech. 2000, 51, 496–510. [Google Scholar] [CrossRef]
- Oka, T.; Jigami, Y. Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. FEBS J. 2006, 273, 2645–2657. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
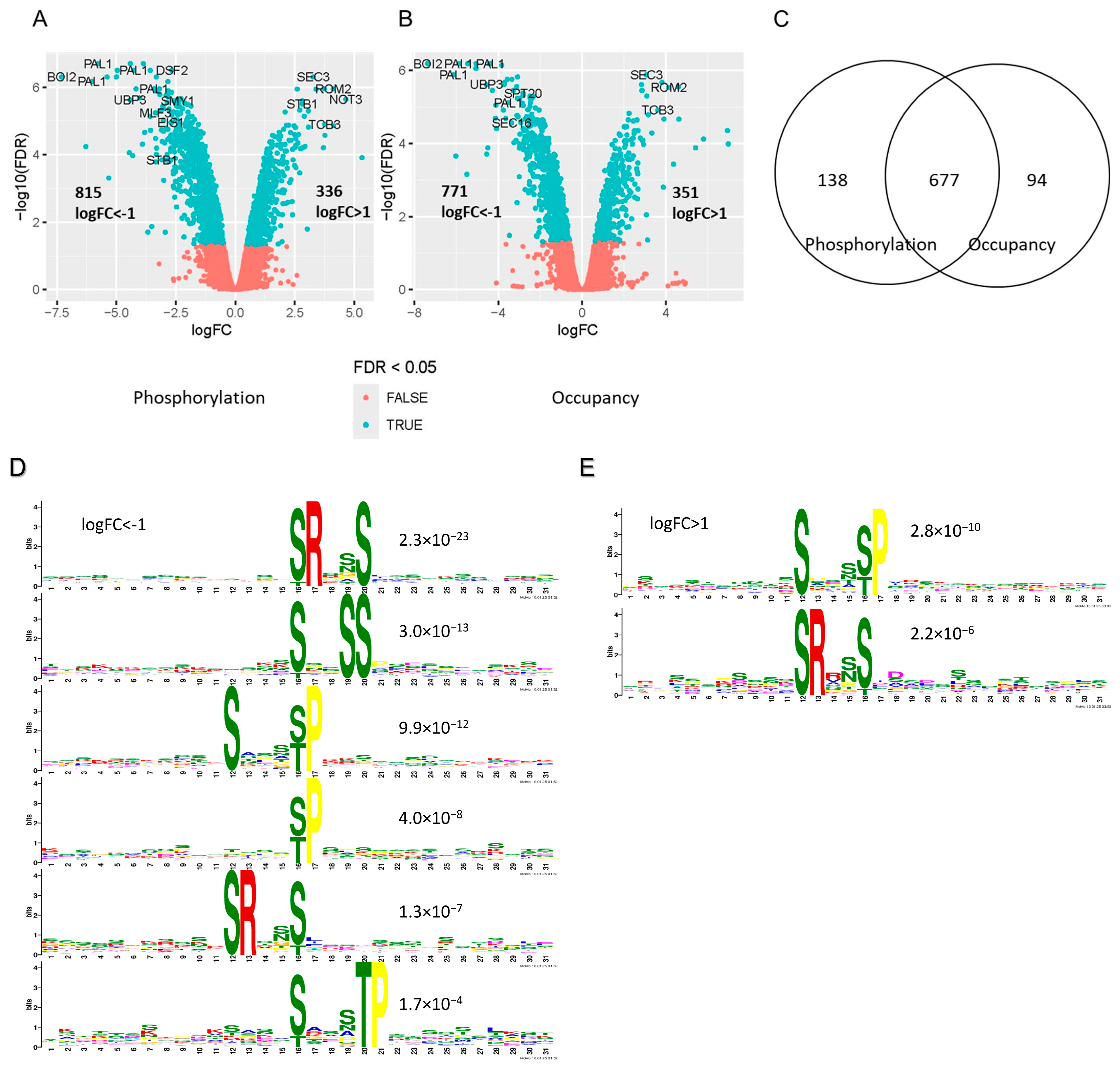
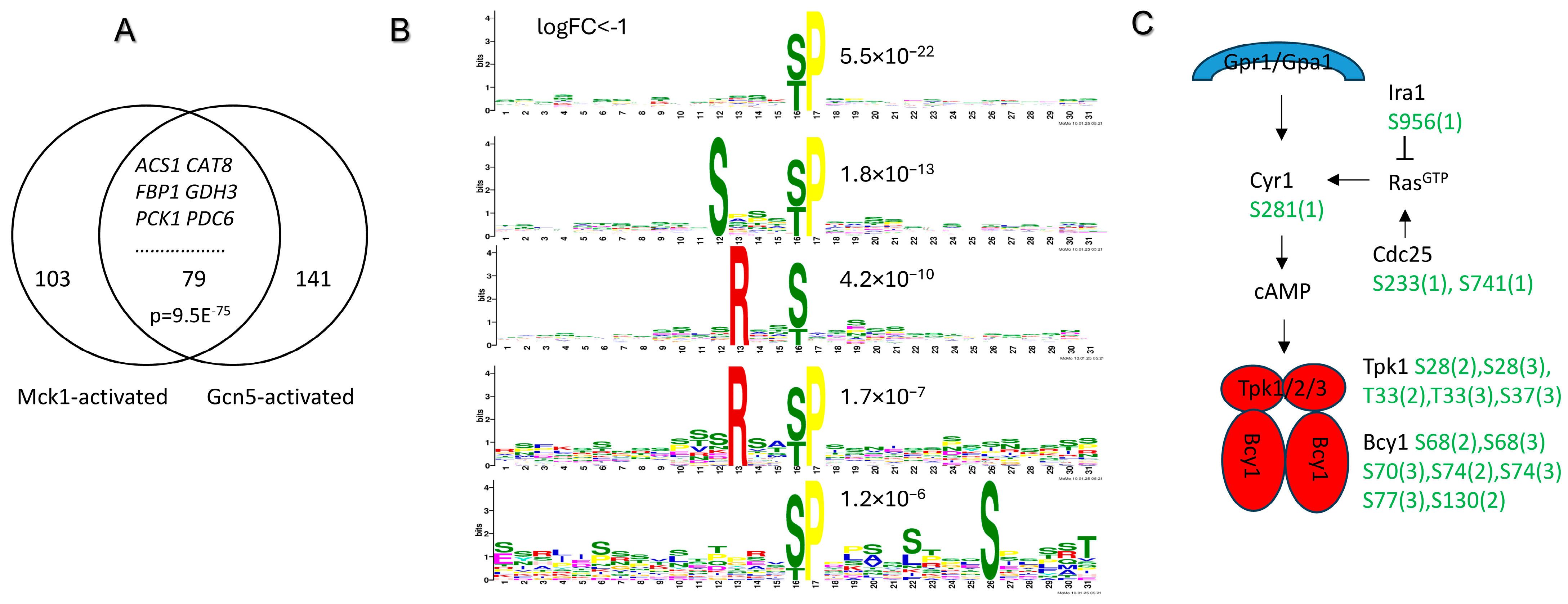
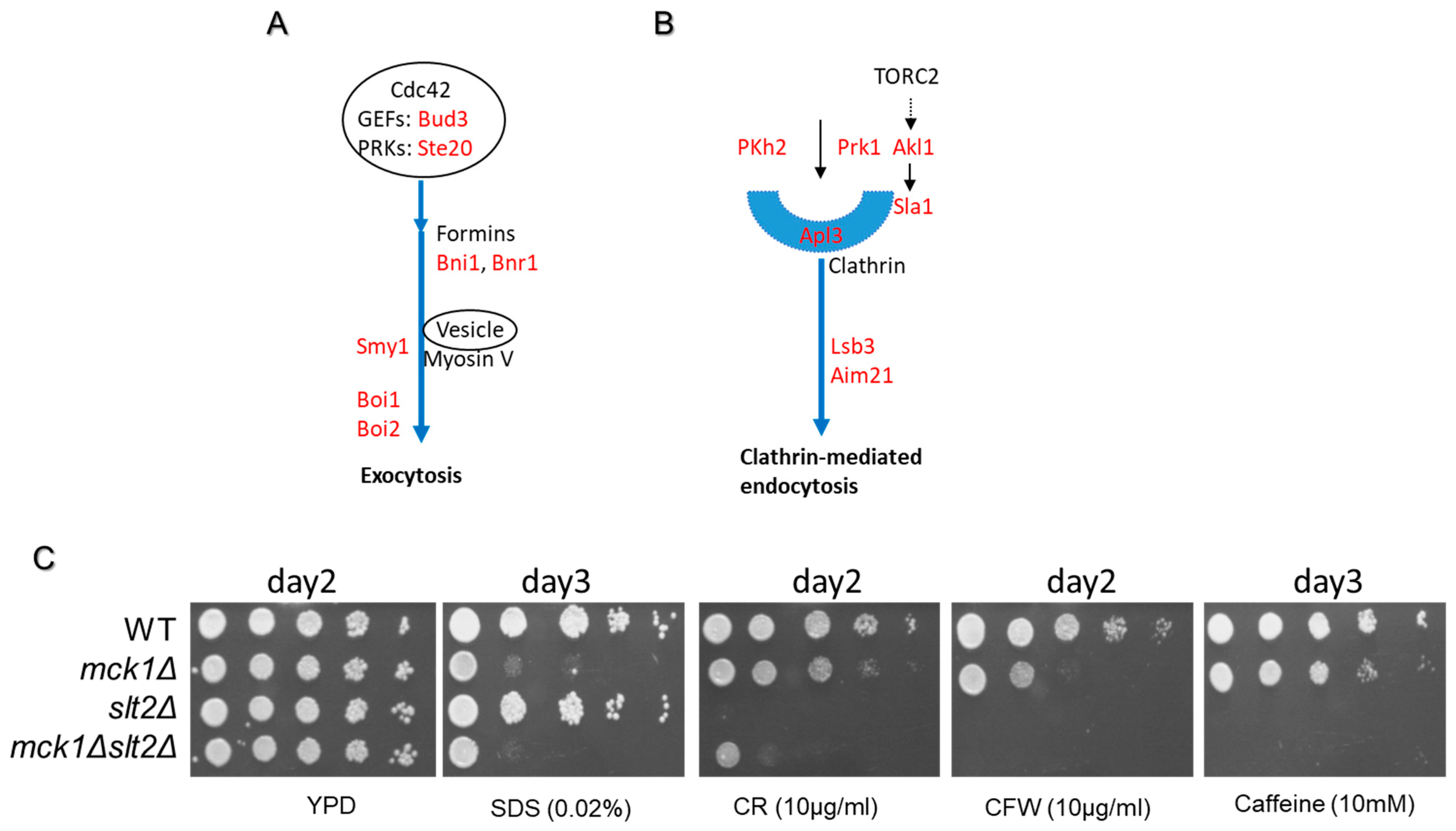
| Functional Category | Gene Name | log2FC RNA | log2FC Protein | Protein Name |
|---|---|---|---|---|
| Stress response | CTA1 * | −0.67 | −1.59 | Catalase A |
| CTT1 | −1.24 | −0.69 | Catalase T | |
| GPX1 | −0.73 | −0.71 | Glutathione peroxidase | |
| HSP12 | −0.89 | −0.88 | Heat shock protein | |
| SIP18 | −5.50 | −5.50 | Phospholipid-binding hydrophilin | |
| SPG4 | −1.19 | −1.72 | Stationary phase protein | |
| SSA3 | −3.64 | −1.70 | Heat shock protein | |
| SSA4 | −0.66 | −0.82 | Heat shock protein | |
| Mitochondrial function | ACO1 | −1.14 | −0.85 | Aconitate hydratase |
| CYB2 * | −1.71 | −1.74 | Cytochrome b | |
| DLD1 * | −1.05 | −1.04 | D-lactate dehydrogenase | |
| GUT2 * | −1.20 | −0.84 | Glycerol−3-phosphate dehydrogenase | |
| ICL2 * | −1.25 | −2.47 | 2-methylisocitrate lyase | |
| IDH1 | −0.59 | −0.70 | Isocitrate dehydrogenase subunit | |
| IDP2 * | −1.20 | −1.32 | Isocitrate dehydrogenase | |
| MBR1 | −0.59 | −1.28 | Mitochondrial biogenesis regulation protein | |
| NDE2 | −1.18 | −1.28 | External NADH-ubiquinone oxidoreductase | |
| Gluconeogenesis | FBP1 * | −1.71 | −3.58 | Fructose−1,6-bisphosphatase |
| PCK1 * | −3.04 | −3.35 | Phosphoenolpyruvate carboxykinase | |
| CAT8 | −0.52 | −0.83 | Transcription activator | |
| Pentose phosphate pathway | GND2 | −2.89 | −1.57 | 6-phosphogluconate dehydrogenase |
| TKL2 | −2.97 | −2.62 | Transketolase 2 | |
| Peroxisomal function | MLS1 * | −0.85 | −1.60 | Malate synthase |
| YPL113C | −0.80 | −0.66 | Glyoxylate reductase | |
| FOX2 * | −0.98 | −1.11 | 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase | |
| Other metabolic processes | ACS1 * | −0.89 | −1.69 | Acetyl-coenzyme A synthetase |
| ADH2 * | −1.37 | −1.11 | Alcohol dehydrogenase | |
| AGX1 | −1.08 | −1.98 | Alanine-glyoxylate aminotransferase | |
| GUT1 * | −0.80 | −0.86 | Glycerol kinase | |
| HXT5 | −4.42 | −2.63 | Glucose transporter | |
| INO1 | −1.16 | −1.34 | Inositol-3-phosphate synthase | |
| NQM1 | −1.82 | −0.88 | Transaldolase | |
| RGI2 * | −1.85 | −1.38 | Respiratory growth induced protein | |
| YKL107W | −2.46 | −4.24 | NADH-dependent aldehyde reductase | |
| Other cellular processes | AMS1 | −0.78 | −0.70 | Alpha-mannosidase |
| ATG34 | −0.70 | −0.62 | Autophagy-related protein | |
| GAC1 | −0.63 | −0.77 | Regulatory subunit for Glc7p | |
| HBT1 | −2.28 | −2.48 | Shmoo tip protein | |
| MUB1 | −0.60 | −0.79 | MYND-type zinc finger protein | |
| FMP45 | −3.84 | −2.06 | SUR7 family protein | |
| NAT4 | −0.65 | −0.82 | Histone-specific N-acetyltransferase | |
| PNS1 | −0.76 | −0.83 | Putative choline transporter | |
| PRY1 | −0.63 | −1.65 | Lipid binding protein | |
| UIP4 | −0.64 | −0.60 | Protein required for nuclear envelope integrity | |
| XBP1 | −1.46 | −2.17 | Transcriptional repressor induced by stress or starvation | |
| Unknown | YBR241C | −0.61 | −1.51 | Putative Transporter |
| YDL199C | −0.61 | −1.07 | Putative transporter | |
| YGR067C * | −0.62 | −1.33 | Zinc finger protein | |
| YGR201C | −1.01 | −0.60 | Putative elongation factor | |
| YHR033W | −2.94 | −2.67 | Uncharacterized | |
| YKL065W-A | −3.16 | −3.27 | Uncharacterized | |
| YNL195C | −0.69 | −1.00 | Uncharacterized |
| GO Terms | Gene Name | Function | log2FC < −1 (Multiplicity) | log2FC > 1 (Multiplicity) |
|---|---|---|---|---|
| Signaling * | LCB5 | Minor sphingoid long-chain base kinase | S160(2); S164(2) | |
| MDS3 | Negative regulator of early meiotic gene expression | S602(2); S603(2); S698(1); S698(2); S702(1) | S606(1) | |
| PKH2 | Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis | S988(3); S990(3); S992(3); T997(2); S1001(2); S1003(1); S1003(2); S1005(2) | ||
| RCN1 | Noncompetitive calcineurin inhibitor involved in calcium-mediated signaling | S113(2); S117(2); S117(1) | ||
| RTC1 | Subunit of SEACAT inhibiting the TORC1 inhibitory role of the Iml1p/SEACIT subcomplex | S1129(2); S1133(2) | S1133(1) | |
| SIP1 | One of three alternate beta-subunits of the Snf1p kinase complex | S377(2); S381(2); S385(1) | ||
| SIP2 | One of three alternate beta subunits of the Snf1 kinase complex | S66(2); S70(2); S133(2); S133(3); S136(2); S136(3); S137(3) | S137(1) | |
| SLN1 | Histidine phosphotransfer kinase | S386(2); S390(2) | ||
| Pol II transcritption & | ASH1 | Component of the Rpd3L histone deacetylase complex | T87(2); S91(2); S91(3); T104(3); S108(3) | |
| DCP2 | Catalytic subunit of Dcp1p-Dcp2p decapping enzyme complex | S724(2); S725(1); S725(2); S728(2); S729(2) | S729(1) | |
| EAF7 | Subunit of nuclear NuA4 histone acetyltransferase complex | S393(1); S393(2);T396(2); S397(2) | T396(1); S397(1) | |
| HAA1 | Transcriptional activator involved in adaptation to weak acid stress | S413(2); S417(2) | ||
| NOT3 | Component of the CCR4-NOT core complex | S303(2); S304(2); T305(2); S442(2); S442(3); S442(1); S446(2); S446(3); S450(2); S450(3); T454(3) | S307(1) | |
| NOT5 | Component of the CCR4-NOT core complex | S271(2); S273(3); S275(2); S275(3); S302(1); S302(2); T306(2) | ||
| ROX3 | Subunit of the RNA polymerase II mediator complex | S200(1); S200(3); T204(3) | T204(1) | |
| SGF29 | Component of the HAT/Core module of the SAGA, SLIK, and ADA complexes | S83(2); S83(1) | T87(1) | |
| SMY2 | ER membrane protein involved in ER-to-Golgi vesicle-mediated transport | S80(2); S80(3); T82(3); S83(2); S83(3); S84(2); S84(3) | ||
| SPT20 | Subunit of the SAGA transcriptional regulatory complex | S593(1); S593(2); S595(1); S595(2); T597(2) | ||
| STB3 | Transcription activator involved in positive regulation of transcription by glucose | S337(1); S337(2); S341(2) | ||
| TAF5 | subunit of SAGA and transcription factor TFIID complex | S411(2); S411(3); S411(1); S414(2); S414(3); S415(2); S415(3) | S414(1); S415(1) | |
| WAR1 | Transcription factor; binds to a weak acid response element to induce transcription of PDR12 and FUN34 | S124(2); T128(2) | ||
| Polarized growth | AIM21 | Subunit of a complex associating with actin filaments | S145(2); S149(2) | |
| AKL1 | Ser/Thr protein kinase negatively regulating endocytosis | S403(2); S407(2) | ||
| APL3 | Alpha-adaptin | S723(2) | T727(1) | |
| BCK1 * | MAPKKK in the PKC1 signaling pathway | S505(2); S509(2) | ||
| BNI5 | Linker protein for recruitment of myosin to the bud neck | S270(1); S270(2); S273(2); T274(2) | T274(1) | |
| BOI1 | Protein involved in polar growth | S574(2); S574(1); S578(2) | ||
| BOI2 | Protein involved in polar growth | T612(2); S615(2); S615(3); S616(3); S617(2); S617(3); S619(3); S620(3); S637(3); S639(2); S639(3); T641(3); S642(2); S642(3); S643(3) | ||
| DSF2 | Deletion suppressor of mpt5 mutation; relocalizes from bud neck to cytoplasm upon DNA replication stress | S391(2); S395(2) | ||
| KRE6 | Glucosyl hydrolase required for beta-1,6-glucan biosynthesis | S108(2); S108(1); S112(2) | ||
| LSB3 | Protein involved in actin cortical patch localization | S377(2); T393(2); S397(2); S397(3) | S381(1) | |
| PAL1 | Protein of unknown function thought to be involved in endocytosis | S186(1); S186(2); S186(3); S189(2); S189(3); S190(2); S190(3) | ||
| PRK1 | Ser/Thr protein kinase regulating the organization and function of the actin cytoskeleton | S533(1); S533(3); T537(3); S540(3) | T537(2); S540(2) | |
| SEC31 | Component of the Sec13p-Sec31p complex of the COPII vesicle coat | S988(2); S988(3); S988(1); S992(3) | S992(1) | |
| SKG1 | Transmembrane protein with a role in cell wall polymer composition | T212(2); T212(3); S215(2); S215(3); S216(2); S216(3) | ||
| SLA1 | Cytoskeletal protein binding protein | S473(3); S476(3); S477(3) | ||
| SMY1 | Kinesin-like myosin passenger-protein | S566(2); S566(1); S570(2) | S570(1) | |
| TAO3 | Component of the RAM signaling network involved in regulation of Ace2p activity and cellular morphogenesis | S318(2) | S322(1) | |
| Polarized growth and cell cycle | BIK1 | Microtubule-associated protein | T85(1); T85(2); T85(3); S86(1); S86(2); S86(3); T89(2); T89(3); T90(3) | |
| BNI1 | Formin; polarisome component | S75(1); S75(2); S79(2); S257(2); S1334(1); S1334(3); T1337(3); S1338(1); S1338(2); S1338(3) | ||
| BNR1 | Formin nucleating the formation of linear actin filaments | S604(2); S604(3); S608(3) | ||
| BUD3 | Guanine nucleotide exchange factor (GEF) for Cdc42p | T1026(3); S1029(3); S1030(3) | ||
| CDC55 * | Regulatory subunit B of protein phosphatase 2A | S145(1); S145(2); S149(2) | ||
| CYK3 | SH3-domain protein located in the bud neck and cytokinetic actin ring | S118(2); S122(2) | S122(1) | |
| GIN4 * | Protein kinase involved in bud growth and assembly of the septin ring | S947(3); S949(3) | S950(1); S951(1) | |
| HSL1 * | Ser/thr protein kinase involved in the G2/M transition | S1325(2); S1328(2); S1329(2) | ||
| KAR9 * | Spindle pole protein | T586(2); T590(2) | ||
| KIN4 * | Serine/threonine protein kinase inhibiting the mitotic exit network (MEN) when the spindle position checkpoint (SPOC) is activated | S384(2); S384(3); S388(2); S388(3) | ||
| STE20 * | MAP kinase kinase kinase kinase involved in pheromone signaling, bud site selection, regulation of mitotic exit and others | S524(1); S524(2); T528(2) | ||
| Cell cycle | ACE2 & | Transcription factor required for septum destruction after cytokinesis | S249(3); S253(3) | S253(1) |
| BCK2 | Serine/threonine-rich protein involved in PKC1 signaling pathway | S575(2); T579(2) | ||
| CDC4 | F-box protein required for both the G1/S and G2/M phase transitions | S71(2); S71(3); S74(2); S74(3); T75(2); T75(3) | S74(1); T75(1) | |
| IGO1 | Protein required for initiation of the G0 program | S7(1); S11(2) | ||
| LTE1 | Protein similar to GDP/GTP exchange factors | S850(2); S854(2) | S854(1) | |
| MCM3 | Protein involved in DNA replication | S777(2); S777(1); S779(2); S779(3) | S779(1); S781(1) | |
| RFM1 & | Component of the Sum1p-Rfm1p-Hst1p complex | S211(2); S215(2) | ||
| SLD2 | DNA-binding subunit of the DNA replication preinitiation complex | S124(2); S128(2) | S128(1) | |
| VHS2 | Regulator of septin dynamics | S84(2) | S88(1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Tang, Y.; Zhou, H.; Li, K.; West, J.A.; Griffin, J.L.; Lilley, K.S.; Zhang, N. The Yeast Gsk-3 Kinase Mck1 Is Necessary for Cell Wall Remodeling in Glucose-Starved and Cell Wall-Stressed Cells. Int. J. Mol. Sci. 2025, 26, 3534. https://doi.org/10.3390/ijms26083534
Zhang F, Tang Y, Zhou H, Li K, West JA, Griffin JL, Lilley KS, Zhang N. The Yeast Gsk-3 Kinase Mck1 Is Necessary for Cell Wall Remodeling in Glucose-Starved and Cell Wall-Stressed Cells. International Journal of Molecular Sciences. 2025; 26(8):3534. https://doi.org/10.3390/ijms26083534
Chicago/Turabian StyleZhang, Fan, Yingzhi Tang, Houjiang Zhou, Kaiqiang Li, James A. West, Julian L. Griffin, Kathryn S. Lilley, and Nianshu Zhang. 2025. "The Yeast Gsk-3 Kinase Mck1 Is Necessary for Cell Wall Remodeling in Glucose-Starved and Cell Wall-Stressed Cells" International Journal of Molecular Sciences 26, no. 8: 3534. https://doi.org/10.3390/ijms26083534
APA StyleZhang, F., Tang, Y., Zhou, H., Li, K., West, J. A., Griffin, J. L., Lilley, K. S., & Zhang, N. (2025). The Yeast Gsk-3 Kinase Mck1 Is Necessary for Cell Wall Remodeling in Glucose-Starved and Cell Wall-Stressed Cells. International Journal of Molecular Sciences, 26(8), 3534. https://doi.org/10.3390/ijms26083534





