Characterization of Critical Amino Acids in the Transport and Selectivity of the Plant Na+/H+ Exchanger Plasma Membrane SOS1
Abstract
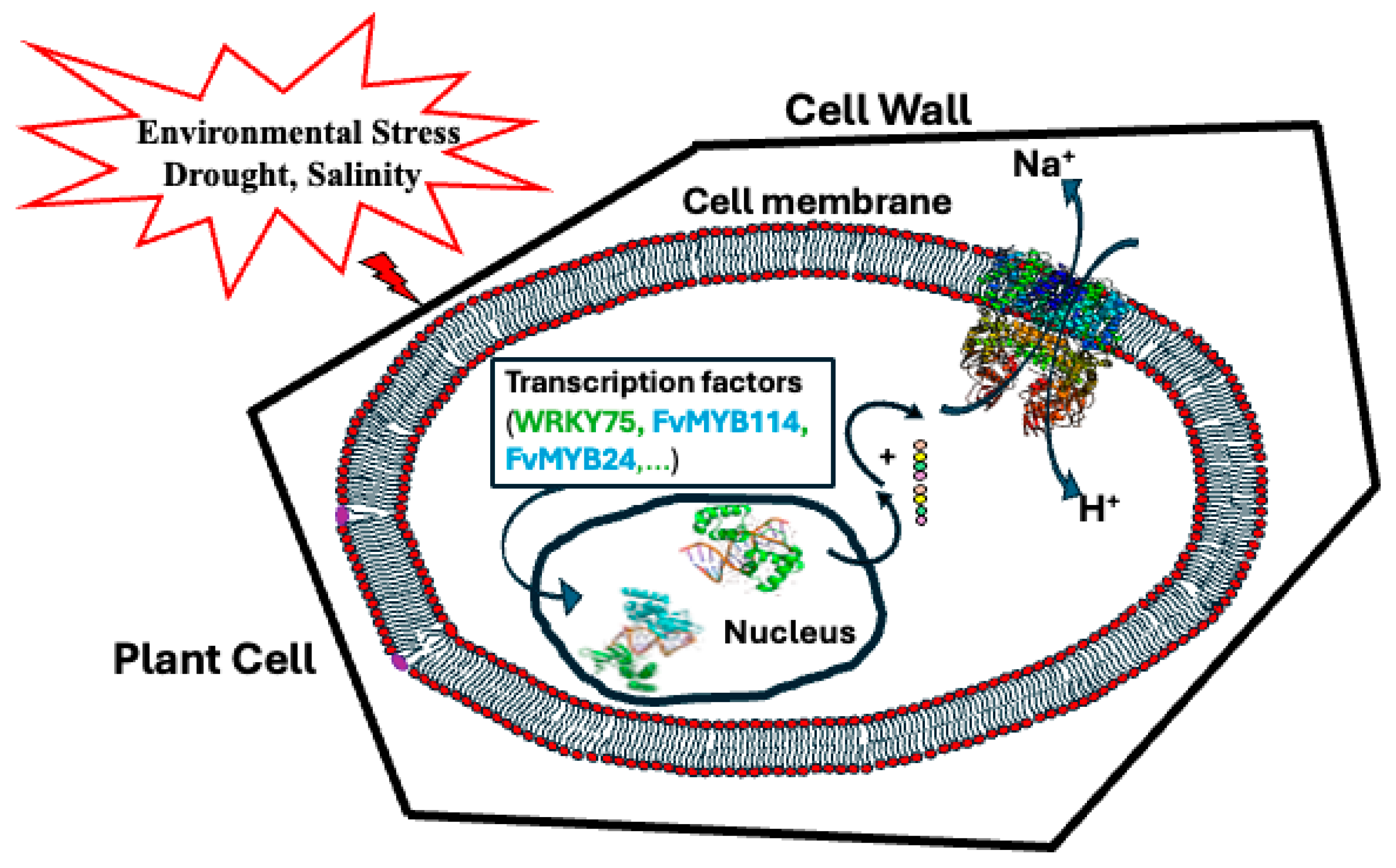
1. Introduction
2. Results
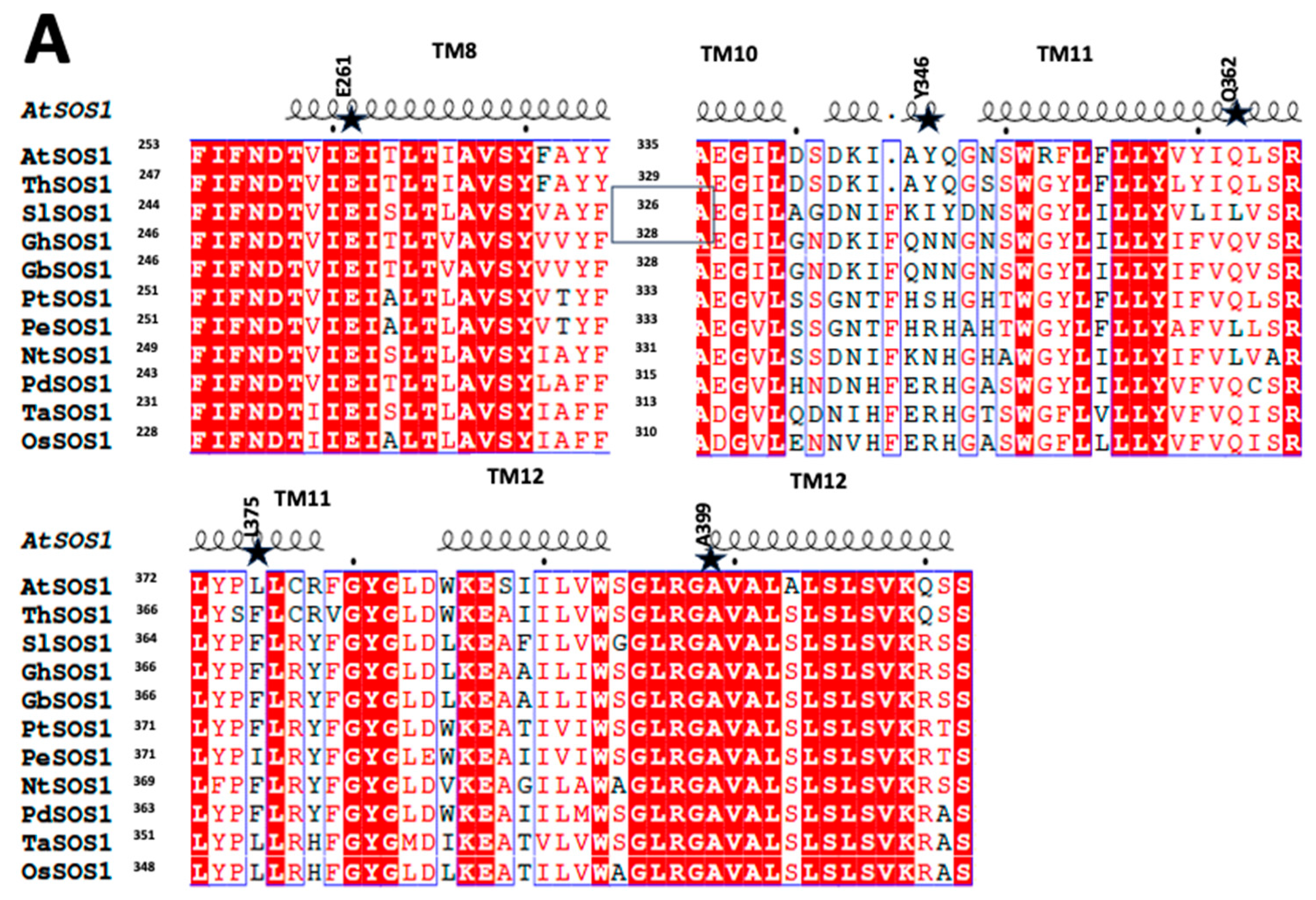
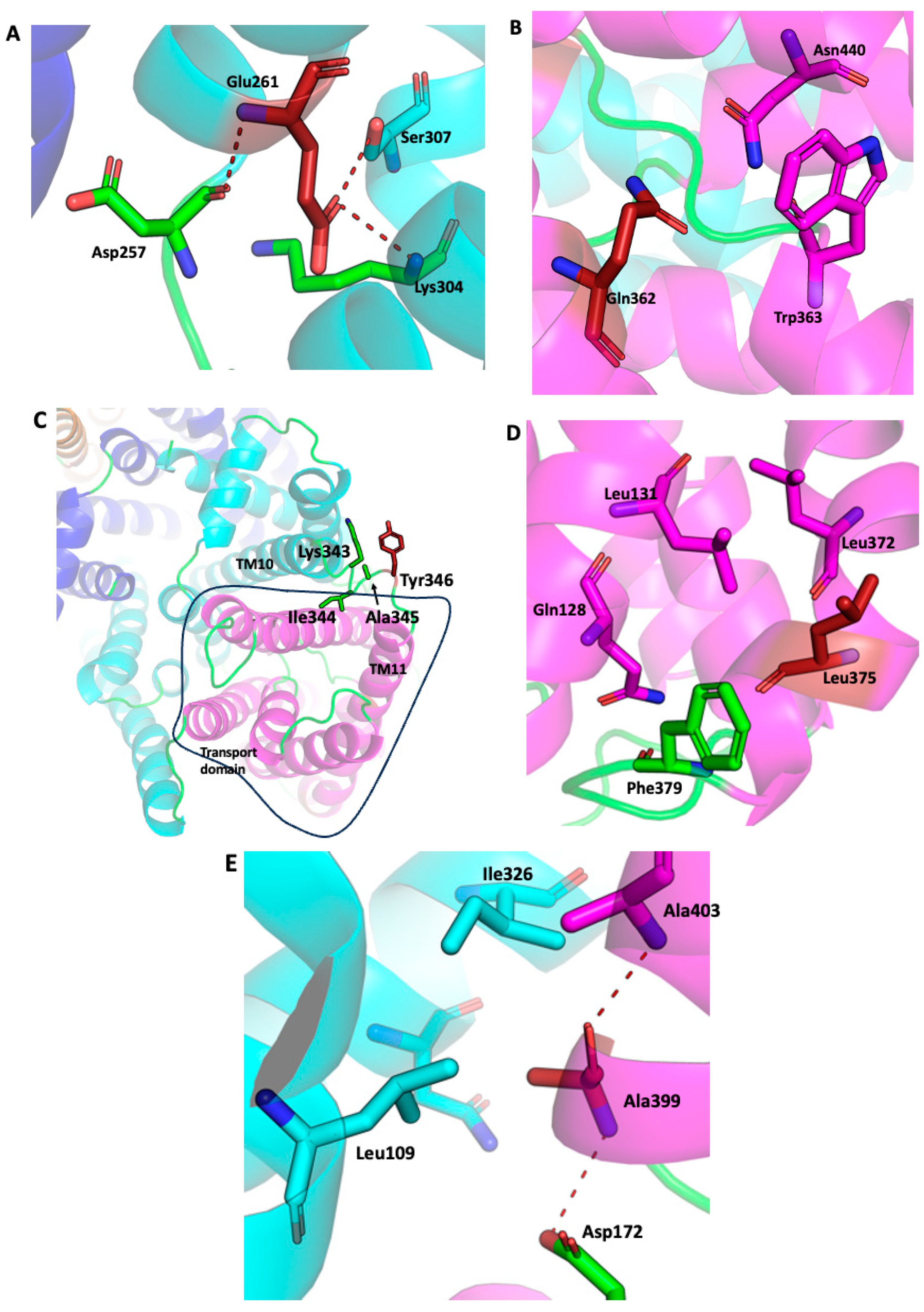
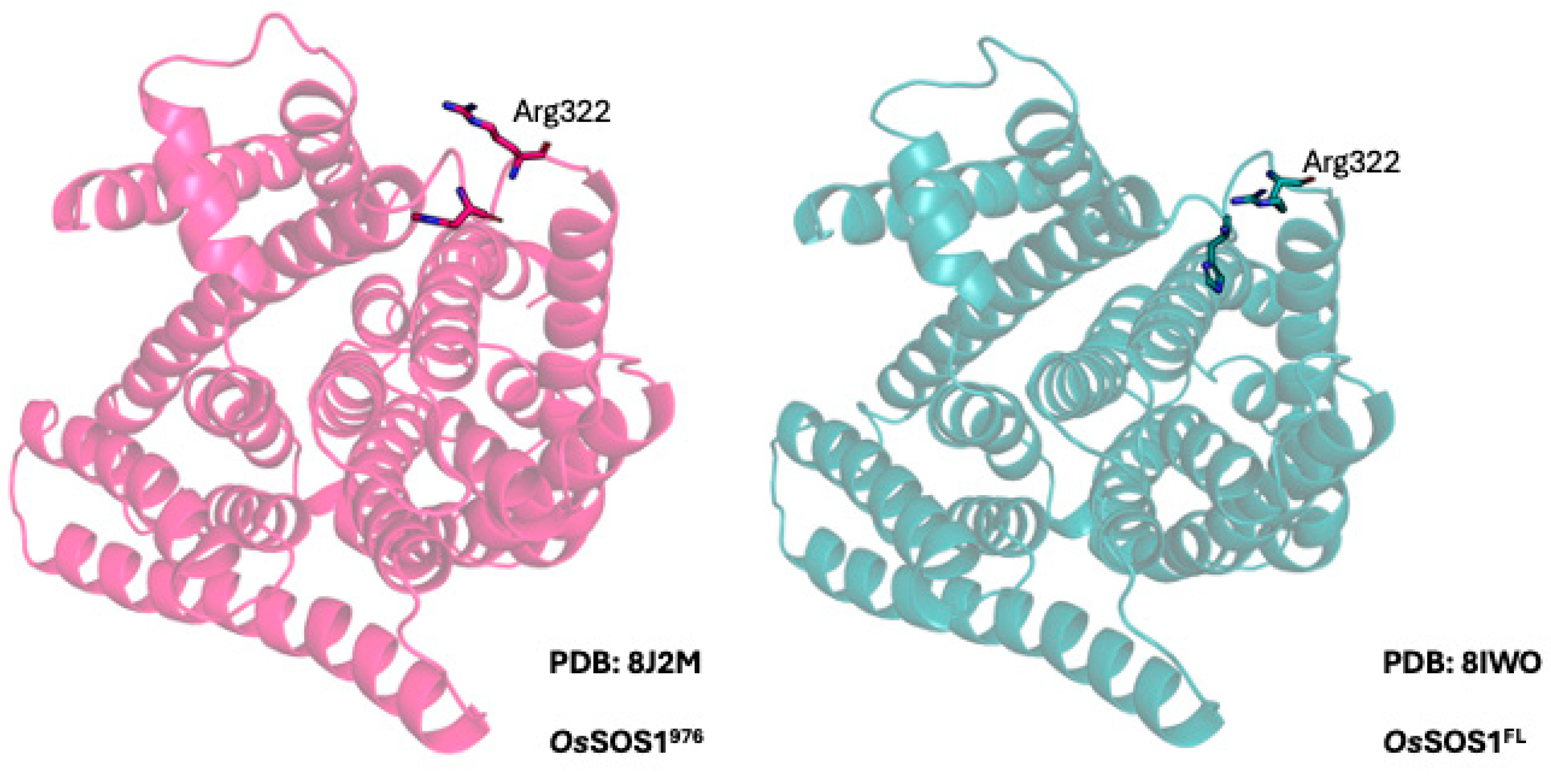
2.1. SOS1 Alignment and Modeling
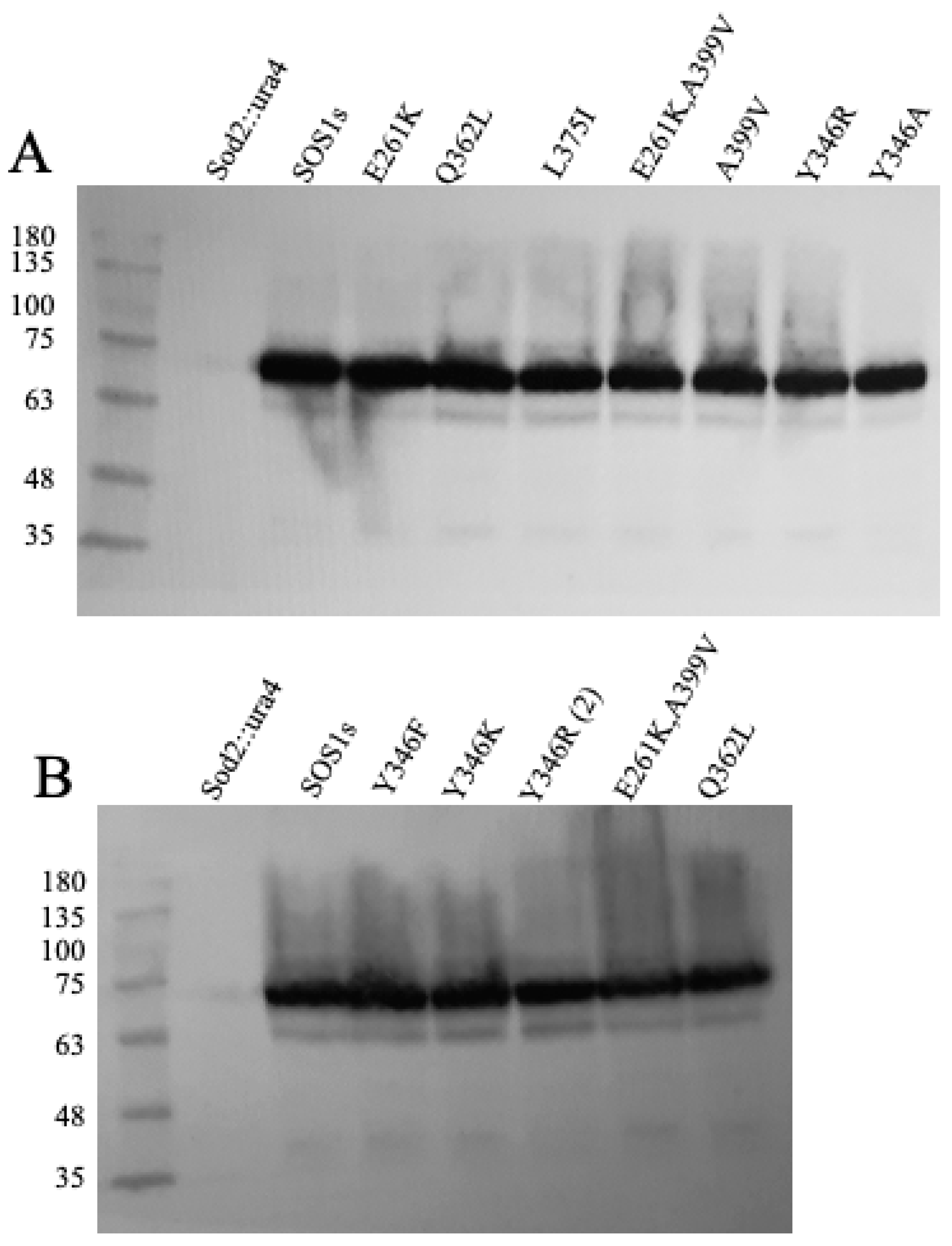
2.2. Expression of Wild Type and Mutant SOS1 in S. pombe
2.3. Salt Tolerance of Wild Type and Mutant SOS1 in S. pombe
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Materials
5.2. Plasmids and Site-Directed Mutagenesis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SOS | Salt Overlay Sensitive |
| SOS1s | Salt Overlay Sensitive 1 protein shortened at the C-terminus |
| At | Arabidopsis thaliana |
| TM | Transmembrane |
| GFP | Green fluorescent protein |
References
- Zhao, S.S.; Zhang, Q.K.; Liu, M.Y.; Zhou, H.P.; Ma, C.L.; Wang, P.P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Fu, H.Q.; Yang, Y.Q. How Plants Tolerate Salt Stress. Curr. Issues Mol. Biol. 2023, 45, 5914–5934. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Kumari, V.V.; Banerjee, P.; Verma, V.C.; Sukumaran, S.; Chandran, M.A.S.; Gopinath, K.A.; Venkatesh, G.; Yadav, S.K.; Singh, V.K.; Awasthi, N.K. Plant Nutrition: An Effective Way to Alleviate Abiotic Stress in Agricultural Crops. Int. J. Mol. Sci. 2022, 23, 8519. [Google Scholar] [CrossRef]
- Tarafdar, M.; Bahadur, V.; Rana, S.; Singh, R.K. A Review: Abiotic stress on transpiration, stomatal diffusive resistance and photosynthetic rate. Pharma Innov. J. 2022, 11, 1632–1635. [Google Scholar]
- Gao, Y.F.; Liu, J.K.; Yang, F.M.; Zhang, G.Y.; Wang, D.; Zhang, L.; Ou, Y.B.; Yao, Y.A. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant 2020, 168, 98–117. [Google Scholar] [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, J.; Li, Y.; Song, L.; Chen, D.; Liu, L.; Jiang, C.Z. A WRKY Protein, MfWRKY40, of Resurrection Plant Myrothamnus flabellifolia Plays a Positive Role in Regulating Tolerance to Drought and Salinity Stresses of Arabidopsis. Int. J. Mol. Sci. 2022, 23, 8145. [Google Scholar] [CrossRef]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef]
- Shi, G.; Liu, G.; Liu, H.; Xu, N.; Yang, Q.; Song, Z.; Ye, W.; Wang, L. WRKY Transcriptional Factor IlWRKY70 from Iris laevigata Enhances Drought and Salinity Tolerances in Nicotiana tabacum. Int. J. Mol. Sci. 2023, 24, 16174. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, Y.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. A Novel Sweetpotato WRKY Transcription Factor, IbWRKY2, Positively Regulates Drought and Salt Tolerance in Transgenic Arabidopsis. Biomolecules 2020, 10, 506. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Xu, T.; Han, J.; Liu, W.; Wang, Y.; Li, X.; Sun, X.; Wang, X.; Li, T.; Yang, G. Overexpression of MxWRKY53 increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitro Cell. Dev. Biol. Plant 2022, 58, 266–278. [Google Scholar] [CrossRef]
- Banik, S.; Dutta, D. Membrane Proteins in Plant Salinity Stress Perception, Sensing, and Response. J. Membr. Biol. 2023, 256, 109–124. [Google Scholar] [CrossRef]
- Yang, W.; Liu, X.D.; Chi, X.J.; Wu, C.A.; Li, Y.Z.; Song, L.L.; Liu, X.M.; Wang, Y.F.; Wang, F.W.; Zhang, C.; et al. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 2011, 233, 219–229. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015, 82, 962–977. [Google Scholar] [CrossRef]
- Li, X.; Zhong, M.; Qu, L.; Yang, J.; Liu, X.; Zhao, Q.; Liu, X.; Zhao, X. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021, 310, 110983. [Google Scholar] [CrossRef]
- Wu, Y.; Li, X.; Zhang, J.; Zhao, H.; Tan, S.; Xu, W.; Pan, J.; Yang, F.; Pi, E. ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front. Plant Sci. 2022, 13, 1042084. [Google Scholar] [CrossRef]
- Lei, P.; Jiang, Y.; Zhao, Y.; Jiang, M.; Ji, X.; Ma, L.; Jin, G.; Li, J.; Zhang, S.; Kong, D.; et al. Functions of Basic Helix-Loop-Helix (bHLH) Proteins in the Regulation of Plant Responses to Cold, Drought, Salt, and Iron Deficiency: A Comprehensive Review. J. Agric. Food Chem. 2024, 72, 10692–10709. [Google Scholar] [CrossRef]
- Qiu, J.R.; Huang, Z.; Xiang, X.Y.; Xu, W.X.; Wang, J.T.; Chen, J.; Song, L.; Xiao, Y.; Li, X.; Ma, J.; et al. MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 2020, 20, 542. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, C.; Lan, H. A bHLH transcription factor from Chenopodium glaucum confers drought tolerance to transgenic maize by positive regulation of morphological and physiological performances and stress-responsive genes’ expressions. Mol. Breed. 2021, 41, 74. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhang, Z.; Ni, B.; Ding, H.; Liu, W.; Li, H.; Chai, L.; Yang, G. Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis. J. Plant Interact. 2018, 13, 433–441. [Google Scholar] [CrossRef]
- Liu, W.; Wang, T.; Wang, Y.; Liang, X.; Han, J.; Han, D. MbMYBC1, a M. baccata MYB transcription factor, contribute to cold and drought stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 2023, 14, 1141446. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Han, J.; Guo, B.; Zhao, W.; Zhou, S.; Zhou, C.; Zhang, L.; Li, X.; Han, D. MbICE1 Confers Drought and Cold Tolerance through Up-Regulating Antioxidant Capacity and Stress-Resistant Genes in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 16072. [Google Scholar] [CrossRef]
- Li, W.; Li, P.; Chen, H.; Zhong, J.; Liang, X.; Wei, Y.; Zhang, L.; Wang, H.; Han, D. Overexpression of a Fragaria vesca 1R-MYB Transcription Factor Gene (FvMYB114) Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 5261. [Google Scholar] [CrossRef]
- Li, W.; Wei, Y.; Zhang, L.; Wang, Y.; Song, P.; Li, X.; Han, D. FvMYB44, a Strawberry R2R3-MYB Transcription Factor, Improved Salt and Cold Stress Tolerance in Transgenic Arabidopsis. Agronomy 2023, 13, 1051. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long- distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef]
- Salazar, O.R.; Chen, K.; Melino, V.J.; Reddy, M.P.; Hribova, E.; Cizkova, J.; Berankova, D.; Arciniegas Vega, J.P.; Caceres Leal, L.M.; Aranda, M.; et al. SOS1 tonoplast neo-localization and the RGG protein SALTY are important in the extreme salinity tolerance of Salicornia bigelovii. Nat. Commun. 2024, 15, 4279. [Google Scholar] [CrossRef]
- Ramakrishna, P.; Gamez-Arjona, F.M.; Bellani, E.; Martin-Olmos, C.; Escrig, S.; De Bellis, D.; De Luca, A.; Pardo, J.M.; Quintero, F.J.; Genoud, C.; et al. Elemental cryo-imaging reveals SOS1-dependent vacuolar sodium accumulation. Nature 2025, 637, 1228–1233. [Google Scholar] [CrossRef]
- Lu, K.K.; Song, R.F.; Guo, J.X.; Zhang, Y.; Zuo, J.X.; Chen, H.H.; Liao, C.Y.; Hu, X.Y.; Ren, F.; Lu, Y.T.; et al. CycC1;1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. Plant Cell 2023, 35, 2570–2591. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shi, M.; Zhang, Y.; Xie, X.; Sun, P.; Fang, C.; Zhao, J. FvMYB24, a strawberry R2R3-MYB transcription factor, improved salt stress tolerance in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 2021, 569, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guo, J.; Chen, X.; Zhou, Y.; Pei, Y.; Chen, L.; Ul Haq, S.; Lu, M.; Gong, H.; Chen, R. Pepper bHLH transcription factor CabHLH035 contributes to salt tolerance by modulating ion homeostasis and proline biosynthesis. Hortic. Res. 2022, 9, uhac203. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, J.; Meng, X.; Fang, X.; Xia, M.; Zhang, J.; Cao, S.; Fan, T. Linker histone variant HIS1-3 and WRKY1 oppositely regulate salt stress tolerance in Arabidopsis. Plant Physiol. 2022, 189, 1833–1847. [Google Scholar] [CrossRef]
- Du, N.; Xue, L.; Xue, D.; Dong, X.; Yang, Q.; Shah Jahan, M.; Guo, H.; Fu, R.; Wang, Y.; Piao, F. The transcription factor SlNAP1 increases salt tolerance by modulating ion homeostasis and ROS metabolism in Solanum lycopersicum. Gene 2023, 849, 146906. [Google Scholar] [CrossRef]
- Dai, W.; Wang, M.; Gong, X.; Liu, J.H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef]
- Sze, H.; Chanroj, S. Plant Endomembrane Dynamics: Studies of K+/H+ Antiporters Provide Insights on the Effects of pH and Ion Homeostasis. Plant Physiol. 2018, 177, 875–895. [Google Scholar] [CrossRef]
- Rodriguez-Rosales, M.P.; Galvez, F.J.; Huertas, R.; Aranda, M.N.; Baghour, M.; Cagnac, O.; Venema, K. Plant NHX cation/proton antiporters. Plant Signal. Behav. 2009, 4, 265–276. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New Insights on Plant Salt Tolerance Mechanisms and Their Potential Use for Breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Oh, D.H.; Zahir, A.; Yun, D.J.; Bressan, R.A.; Bohnert, H.J. SOS1 and halophytism. Plant Signal. Behav. 2009, 4, 1081–1083. [Google Scholar] [CrossRef] [PubMed]
- Olias, R.; Eljakaoui, Z.; Li, J.; De Morales, P.A.; Marin-Manzano, M.C.; Pardo, J.M.; Belver, A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant. Cell Environ. 2009, 32, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Olias, R.; Eljakaoui, Z.; Pardo, J.M.; Belver, A. The Na+/H+ exchanger SOS1 controls extrusion and distribution of Na+ in tomato plants under salinity conditions. Plant Signal. Behav. 2009, 4, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.M.; Hirschi, K.D.; Sze, H. Plants pass the salt. Trends Plant Sci. 2003, 8, 200–201. [Google Scholar] [CrossRef]
- Sathee, L.; Sairam, R.K.; Chinnusamy, V.; Jha, S.K. Differential transcript abundance of salt overly sensitive (SOS) pathway genes is a determinant of salinity stress tolerance of wheat. Acta Physiol. Plant. 2015, 37, 169. [Google Scholar] [CrossRef]
- Yue, Y.; Zhang, M.; Zhang, J.; Duan, L.; Li, Z. SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 2012, 169, 255–261. [Google Scholar] [CrossRef]
- Shi, H.; Lee, B.H.; Wu, S.J.; Zhu, J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, Z.Z.; Zhou, X.F.; Yin, H.B.; Li, X.; Xin, X.F.; Hong, X.H.; Zhu, J.K.; Gong, Z. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef]
- Duscha, K.; Martins Rodrigues, C.; Muller, M.; Wartenberg, R.; Fliegel, L.; Deitmer, J.W.; Jung, M.; Zimmermann, R.; Neuhaus, H.E. 14-3-3 Proteins and Other Candidates form Protein-Protein Interactions with the Cytosolic C-terminal End of SOS1 Affecting Its Transport Activity. Int. J. Mol. Sci. 2020, 21, 3334. [Google Scholar] [CrossRef]
- El Mahi, H.; Perez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gamez-Arjona, F.; Fernandez, J.L.; Bundo, M.; Mendoza, I.; Mieulet, D.; et al. A Critical Role of Sodium Flux via the Plasma Membrane Na+/H+ Exchanger SOS1 in the Salt Tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]
- Oh, D.H.; Leidi, E.; Zhang, Q.; Hwang, S.M.; Li, Y.; Quintero, F.J.; Jiang, X.; D’Urzo, M.P.; Lee, S.Y.; Zhao, Y.; et al. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009, 151, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, C.; Wang, P.; Ma, Q.; Bao, A.K.; Zhang, J.L.; Wang, S.M. The Effect of AtHKT1;1 or AtSOS1 Mutation on the Expressions of Na+ or K+ Transporter Genes and Ion Homeostasis in Arabidopsis thaliana under Salt Stress. Int. J. Mol. Sci. 2019, 20, 1085. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.; Ding, L.; Zhu, J.K. SOS1, a Genetic Locus Essential for Salt Tolerance and Potassium Acquisition. Plant Cell 1996, 8, 617–627. [Google Scholar] [CrossRef]
- Jia, Z.-P.; McCullough, N.; Martel, R.; Hemmingsen, S.; Young, P.G. Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J. 1992, 11, 1631–1640. [Google Scholar] [CrossRef]
- Ullah, A.; Kemp, G.; Lee, B.; Alves, C.; Young, H.; Sykes, B.D.; Fliegel, L. Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2. J. Biol. Chem. 2013, 288, 24609–24624. [Google Scholar] [CrossRef]
- Ullah, A.; Dutta, D.; Fliegel, L. Expression and characterization of the SOS1 Arabidopsis salt tolerance protein. Mol. Cell Biochem. 2016, 415, 133–143. [Google Scholar] [CrossRef]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.J.; Zhu, J.K.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef]
- Meng, K.; Wu, Y. Footprints of divergent evolution in two Na+/H+ type antiporter gene families (NHX and SOS1) in the genus Populus. Tree Physiol. 2018, 38, 813–824. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Tang, L.H.; Nie, J.W.; Zhang, C.R.; Han, X.; Li, Q.Y.; Qin, L.; Wang, M.H.; Huang, X.; Yu, F.; et al. Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na+/H+ antiporter. Nat. Plants 2023, 9, 1924–1936. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, C.; Chen, Q.; Xie, Q.; Gao, Y.; He, L.; Li, Y.; Dong, Y.; Jiang, X.; Zhao, Y. Architecture and autoinhibitory mechanism of the plasma membrane Na+/H+ antiporter SOS1 in Arabidopsis. Nat. Commun. 2023, 14, 4487. [Google Scholar] [CrossRef]
- Nunez-Ramirez, R.; Sanchez-Barrena, M.J.; Villalta, I.; Vega, J.F.; Pardo, J.M.; Quintero, F.J.; Martinez-Salazar, J.; Albert, A. Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na+/H+ antiporter. J. Mol. Biol. 2012, 424, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Ullah, A.; Bibi, S.; Fliegel, L. Functional Analysis of Conserved Transmembrane Charged Residues and a Yeast Specific Extracellular Loop of the Plasma Membrane Na+/H+ Antiporter of Schizosaccharomyces pombe. Sci. Rep. 2019, 9, 6191. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Shin, K.; Rainey, J.K.; Fliegel, L. Transmembrane Segment XI of the Na+/H+ Antiporter of S. pombe is a Critical Part of the Ion Translocation Pore. Sci. Rep. 2017, 7, 12793. [Google Scholar] [CrossRef]
- Dibrov, P.; Young, P.G.; Fliegel, L. Functional analysis of amino acid residues essential for activity in the Na+/H+ exchanger of fission yeast. Biochemistry 1998, 36, 8282–8288. [Google Scholar] [CrossRef]
- Dutta, D.; Fliegel, L. Structure and function of yeast and fungal Na+/H+ antiporters. IUBMB Life 2018, 70, 23–31. [Google Scholar] [CrossRef]
- Sugiura, Y.; Saito, Y.; Endo, T.; Makita, Y. Effect of the Ionic Radius of Alkali Metal Ions on Octacalcium Phosphate Formation via Different Substitution Modes. Cryst. Growth Des. 2019, 19, 4162–4171. [Google Scholar] [CrossRef]
- Cao, S.; Yuan, L.; Zhai, H.; Liu, C.; He, S.; Liu, Q. Overexpression of SOS genes enhanced salt tolerance in sweetpotato. J. Integr. Agric. 2012, 11, 378–386. [Google Scholar]
- Dutta, D.; Esmaili, M.; Overduin, M.; Fliegel, L. Expression and detergent free purification and reconstitution of the plant plasma membrane Na+/H+ antiporter SOS1 overexpressed in Pichia pastoris. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183111. [Google Scholar] [CrossRef]
- Hahnenberger, K.M.; Jia, Z.; Young, P.G. Functional expression of the Schizosaccharomyces pombe Na+/H+ antiporter gene, sod2, in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5031–5036. [Google Scholar] [CrossRef]
- Rimon, A.; Amartely, H.; Padan, E. The crossing of two unwound transmembrane regions that is the hallmark of the NhaA structural fold is critical for antiporter activity. Sci. Rep. 2024, 14, 5915. [Google Scholar] [CrossRef]
- Beauchamp, D.L.; Khajehpour, M. The effect of lithium ions on the hydrophobic effect: Does lithium affect hydrophobicity differently than other ions? Biophys. Chem. 2012, 163–164, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Francisco, J.S.; Zeng, X.C. Unraveling the mechanism of selective ion transport in hydrophobic subnanometer channels. Proc. Natl. Acad. Sci. USA 2015, 112, 10851–10856. [Google Scholar] [CrossRef] [PubMed]
- Fliegel, L. Functional analysis of conserved polar residues important for salt tolerance of the Na+/H+ exchanger of Schizosaccharomyces pombe. Mol. Cell. Biochem. 2004, 43, 16477–16486. [Google Scholar]
- Fliegel, L. The Na+/H+ exchanger isoform 1. Int. J. Biochem. Cell Biol. 2005, 37, 33–37. [Google Scholar] [CrossRef]
- Ullah, A.; El-Magd, R.A.; Fliegel, L. Functional role and analysis of cysteine residues of the salt tolerance protein Sod2. Mol. Cell. Biochem. 2014, 386, 85–98. [Google Scholar] [CrossRef]
- Ndayizeye, M.; Touret, N.; Fliegel, L. Proline 146 is critical to the structure, function and targeting of sod2, the Na+/H+ exchanger of Schizosaccharomyces pombe. Biochim. Biophys. Acta 2009, 1788, 983–992. [Google Scholar] [CrossRef]
- Fliegel, L.; Wiebe, C.; Chua, G.; Young, P.G. Functional expression and cellular localization of the Na+/H+ exchanger Sod2 of the fission yeast Schizosaccharomyces pombe. Can. J. Physiol. Pharmacol. 2005, 83, 565–572. [Google Scholar] [CrossRef]
- Slepkov, E.R.; Chow, S.; Lemieux, M.J.; Fliegel, L. Proline residues in transmembrane segment IV are critical for activity, expression and targeting of the Na+/H+ exchanger isoform 1. Biochem. J. 2004, 379, 31–38. [Google Scholar] [CrossRef]
- Silva, N.L.; Wang, H.; Harris, C.V.; Singh, D.; Fliegel, L. Characterization of the Na+/H+ exchanger in human choriocarcinoma (BeWo) cells. Pflugers Archiv Eur. J. Physiol. 1997, 433, 792–802. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 82, 82–92. [Google Scholar]

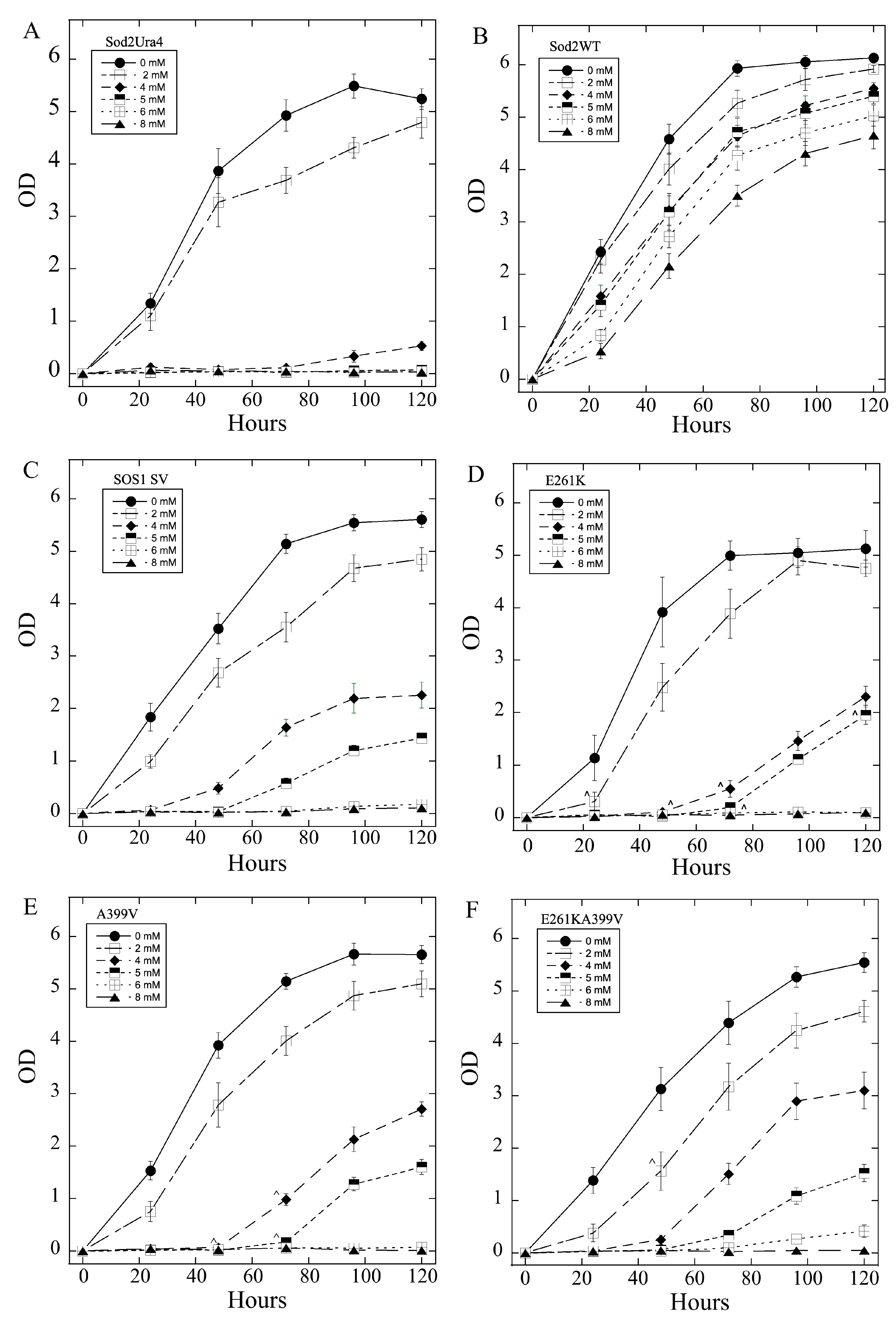
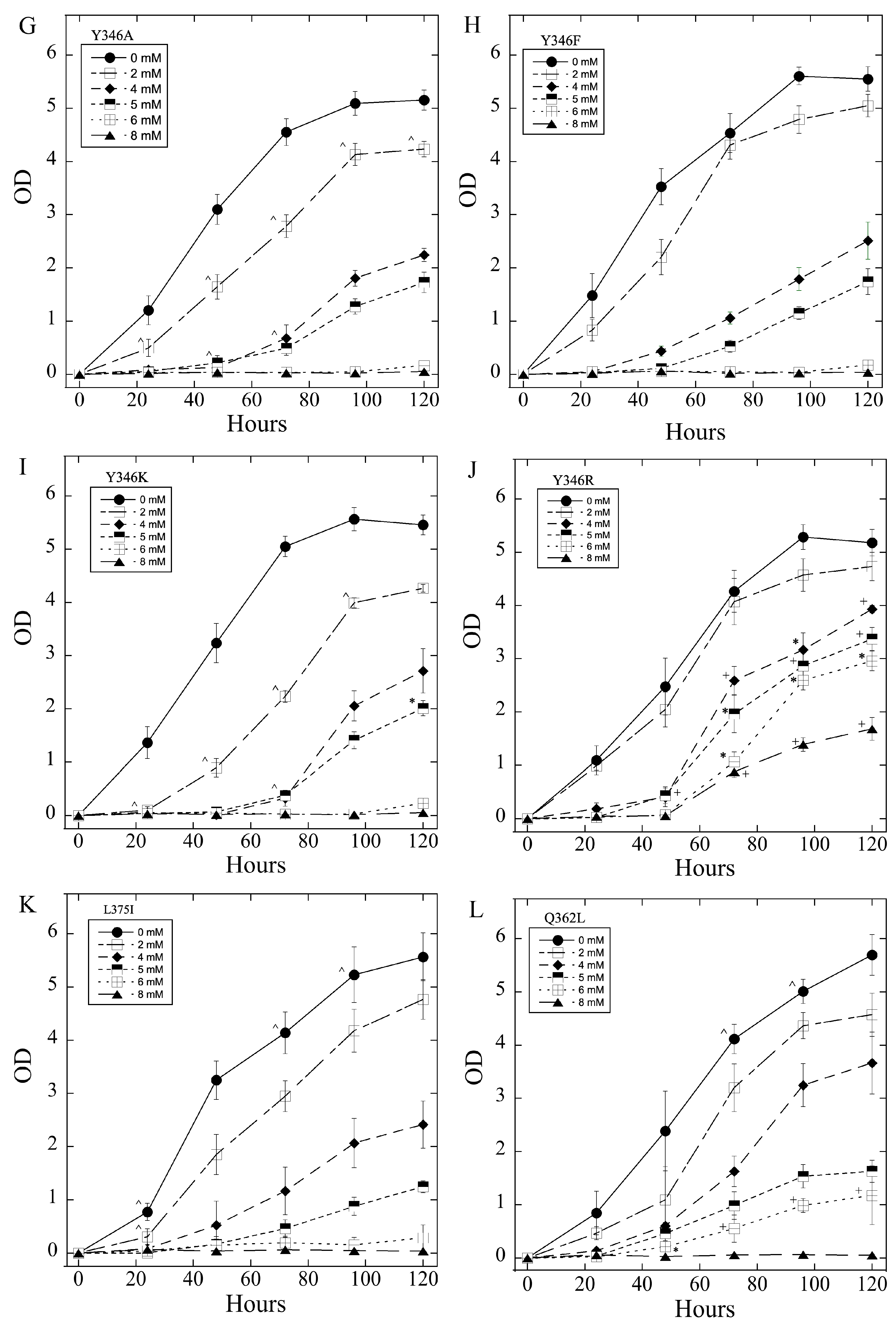
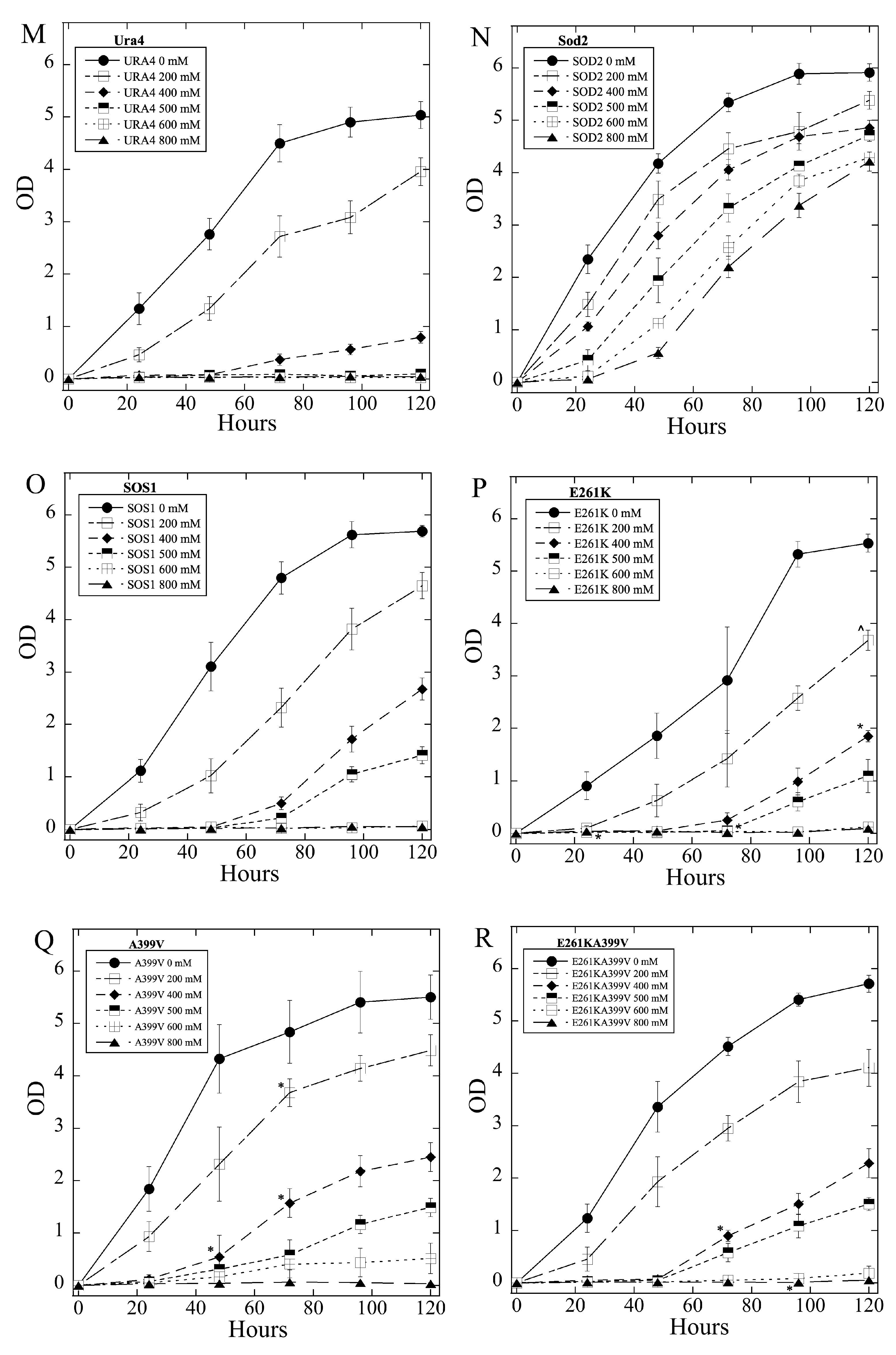
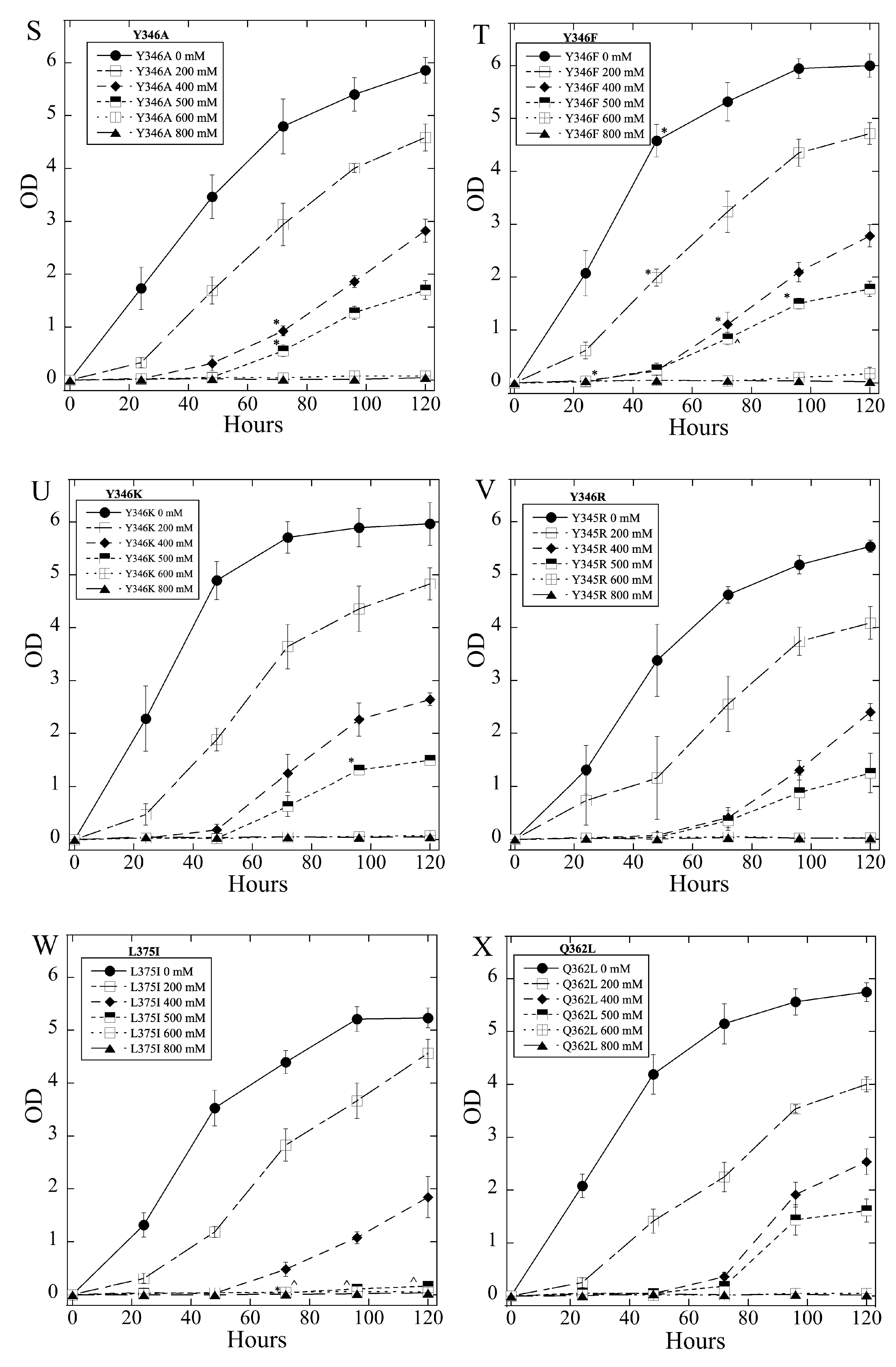
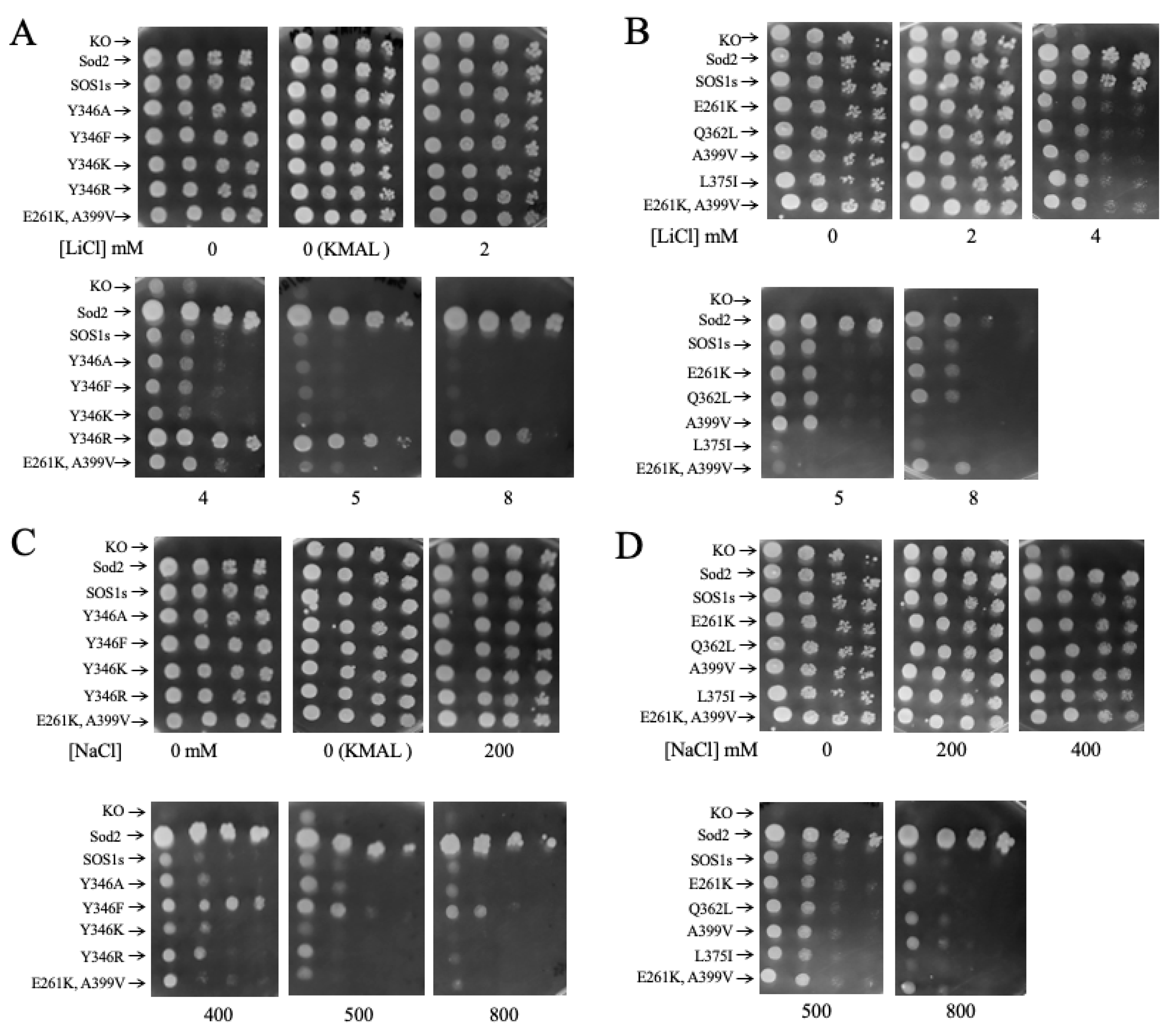
| A. Growth Effects in Liquid Media Relative to sSOS1. | ||||
| Mutant | Growth in 2–5 mM LiCl | Growth in 6, 8 LiCl mM | Growth in 200–500 mM NaCl | Growth in 600, 800 mM NaCl |
| E261K | − | + | − | = |
| A399V | − | = | + | = |
| E261KA399V | − | = | + | = |
| Y346A | −,− | = | + | = |
| Y346F | = | = | +, ^ | = |
| Y346K | −,− | − | = | + |
| Y346R | +++ | +++ | = | = |
| L375I | =, * | = | −,− | − |
| Q362L | =, * | +, * | = | = |
| B. Growth Effects on Solid Media. | ||||
| Mutant | Growth in 2, 4 mM LiCl | Growth in 5, 8 LiCl mM | Growth in 200, 400 mM NaCl | Growth in 500, 800 mM NaCl |
| E261K | − | = | = | = |
| A399V | − | − | = | = |
| E261KA399V | − | − | = | = |
| Y346A | = | = | = | = |
| Y346F | = | = | + | + |
| Y346K | = | = | = | = |
| Y346R | ++ | +++ | = | = |
| L375I | = | −,− | = | = |
| Q362L | = | = | = | = |
| Mutation | Oligonucleotide | Restriction Site |
|---|---|---|
| E261K | CAATGACACTGTtATAaAGGATTACTCTTACAATTGC | Psi1 |
| Y346A | AGTGATAAGATTGCCgcaCAAGGGAAcTCATGGCGATTTC | -EcoR1 |
| Y346F | AGTGATAAGATTGCCTtcCAAGGGAAcTCATGG | -EcoR1 |
| Y346K | AGTGATAAGATTGCCaagCAAGGGAAcTCATGGCGATTTC | -EcoR1 |
| Y346R | GATAAGATTGCCcgCCAAGGGAAcTCATGGCGATTTC | -EcoR1 |
| L375I | GGAGTTCTATATCCAaTTcTgTGcagaTTTGGCTATGGTTTG | BsgI |
| Q362L | CTATACGTTTACATCCtcCTcTCGCGTGTTGTTG | BseR1 |
| A399V | GGTTTGAGGGGCgTcGTGGCTCTTGCAC | -Bts1 |
| E261KA399V | CAATGACACTGTtATAaAGATTACTCTTACAATTGCGGTTTGAGGGGCgTcGTGGCTCTTGCAC | Psi and Bts1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, A.; Dutta, D.; Fliegel, L. Characterization of Critical Amino Acids in the Transport and Selectivity of the Plant Na+/H+ Exchanger Plasma Membrane SOS1. Int. J. Mol. Sci. 2025, 26, 3518. https://doi.org/10.3390/ijms26083518
Ullah A, Dutta D, Fliegel L. Characterization of Critical Amino Acids in the Transport and Selectivity of the Plant Na+/H+ Exchanger Plasma Membrane SOS1. International Journal of Molecular Sciences. 2025; 26(8):3518. https://doi.org/10.3390/ijms26083518
Chicago/Turabian StyleUllah, Asad, Debajyoti Dutta, and Larry Fliegel. 2025. "Characterization of Critical Amino Acids in the Transport and Selectivity of the Plant Na+/H+ Exchanger Plasma Membrane SOS1" International Journal of Molecular Sciences 26, no. 8: 3518. https://doi.org/10.3390/ijms26083518
APA StyleUllah, A., Dutta, D., & Fliegel, L. (2025). Characterization of Critical Amino Acids in the Transport and Selectivity of the Plant Na+/H+ Exchanger Plasma Membrane SOS1. International Journal of Molecular Sciences, 26(8), 3518. https://doi.org/10.3390/ijms26083518






