Transcriptomic Analysis Reveals Key Pathways Influenced by HIV-2 Vpx
Abstract
:1. Introduction
2. Results
2.1. Differential Gene Expression Induced by Wild-Type Vpx
2.2. Effect of Vpx on Expression of HIV-1 Tat
2.3. RT-qPCR Results of SKOR2, U2AF1 and CASP3 Genes
2.4. CASP3 Activity Measurements
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Transfection of THP-1 Cells for Transcriptomic Analysis
4.3. Isolation of RNA for Transcriptomic Analysis
4.4. RNA-Seq Library Preparation and Sequencing
4.5. RNA-Seq Data Processing and Analysis
4.6. Gene Ontology Enrichment Analysis and Functional Annotation Analysis
4.7. Activation and Transfection of THP-1 Cells for Cytokine Measurements
4.8. Detection of Vpx Effect on the Expression of HIV-1 Tat by Western Blot
4.9. Infection of THP-1 Cells
4.10. Real-Time qPCR for SKOR2, U2AF1 and CASP3 Genes
4.11. Transfection of THP-1 and RNA Isolation for qPCR
4.12. Caspase 3 (CASP3) Activity Measurement
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CASP3 | Caspase 3 |
| CBP | CREB-binding protein |
| CCR5 | Chemokine coreceptor 5 |
| CD4 | Cluster of differentiation 4 |
| CORO7-PAM16 | Coronin 7–presequence translocase-associated motor 16 |
| Cul4 | Cullin 4 |
| DCAF1 | DDB1- and CUL4-associated factor 1 |
| DDB1 | Damage-specific DNA binding protein 1 |
| DEGs | Differentially expressed genes |
| DEPDC1 | DEP domain containing 1 |
| dGTPs | Deoxyguanosine triphosphate |
| dNTPs | Deoxyribonucleotide triphosphates |
| EDTA | Ethylenediaminetetraacetic acid |
| FAK | Focal adhesion kinase |
| FBS | Fetal bovine serum |
| FDR | False discovery rate |
| G1, G2, S | Gap-1, Gap-2, synthesis phase |
| GO | Gene ontology |
| HIV | Human immunodeficiency virus |
| HSP40 | Heat shock protein 40 |
| HUSH | Human silencing hub |
| IFNAs | Type-I alpha interferons |
| IL-1 | Interleukin 1 |
| IL-6 | Interleukin 6 |
| IRF5 | Interferon regulatory factor 5 |
| MDMs | Monocyte-derived macrophages |
| mRNA | Messenger ribonucleic acid |
| mTOR | Mechanistic target of rapamycin |
| MYD88 | Myeloid differentiation 88 |
| Nef | Negative effector |
| NF-κB | Nuclear factor kappa B |
| NOP58 | NOP58 ribonucleoprotein |
| Nup153 | Nucleoporin 153 |
| PCDHGC4 | Protocadherin gamma subfamily C |
| PCDHs | Protocadherins |
| PEI | Polyethylenimine |
| PI3K/AKT | Phosphoinositide 3-kinase/protein kinase B |
| PIC | Pre-integration complex |
| PLWHIV | People living with HIV |
| PMA | Phorbol 12-myristate 13-acetate |
| PPDE | Posterior probabilities of genes being differentially expressed |
| PYK2 | Proline-rich tyrosine kinase 2 |
| Rbx1 | Ring box protein1 containing E3 ubiquitin ligase complex |
| RIN | RNA integrity number |
| RPMI | Roswell Park Memorial Institute |
| RT | Reverse transcriptase |
| SAMHD1 | SAM and HD domain containing deoxynucleoside triphosphate Triphosphohydrolase 1 |
| SIV/sm | Simian immunodeficiency virus of sooty mangabeys |
| SIV | Simian immunodeficiency virus |
| SKOR2 | SKI family transcriptional co-repressor 2 |
| SMAD | Suppressor of mothers against decapentaplegic |
| snRNP | Small nuclear ribonucleic proteins |
| SNX14 | Sorting nexin 14 |
| Tat | Trans-activator |
| TGFBR1 | Transforming growth factor-β receptor 1 |
| TGF-β | Transforming growth factor β |
| TNF | Tumor necrosis factor |
| U2AF | U2 small nuclear RNA auxiliary factor 1 |
| U2AF1L5 | U2 small nuclear RNA auxiliary factor 1 like 5 |
| Vif | Viral infectivity factor |
| Vpr | Viral protein R |
| Vpu | Viral protein U |
| Vpx | Viral protein X |
| Wnt/β | Wingless-related integration site and beta pathway |
References
- Zagury, J.F.; Franchini, G.; Reitz, M.; Collalti, E.; Starcich, B.; Hall, L.; Fargnoli, K.; Jagodzinski, L.; Guo, H.G.; Laure, F.; et al. Genetic variability between isolates of human immunodeficiency virus (HIV) type 2 is comparable to the variability among HIV type 1. Proc. Natl. Acad. Sci. USA 1988, 85, 5941–5945. [Google Scholar] [CrossRef]
- Vidya Vijayan, K.K.; Karthigeyan, K.P.; Tripathi, S.P.; Hanna, L.E. Pathophysiology of CD4+ T-cell depletion in HIV-1 and HIV-2 infections. Front. Immunol. 2017, 8, 580. [Google Scholar] [CrossRef] [PubMed]
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 15035. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Emerman, M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe 2008, 3, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Segeral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. Samhd1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef]
- Strebel, K. HIV accessory proteins versus host restriction factors. Curr. Opin. Virol. 2013, 3, 692–699. [Google Scholar] [CrossRef]
- Tristem, M.; Marshall, C.; Karpas, A.; Petrik, J.; Hill, F. Origin of vpx in lentiviruses. Nature 1990, 347, 341–342. [Google Scholar] [CrossRef]
- Fujita, M.; Otsuka, M.; Nomaguchi, M.; Adachi, A. Multifaceted activity of HIV vpr/vpx proteins: The current view of their virological functions. Rev. Med. Virol. 2010, 20, 68–76. [Google Scholar] [CrossRef]
- Planelles, V.; Barker, E. Roles of vpr and vpx in modulating the virus-host cell relationship. Mol. Asp. Med. 2010, 31, 398–406. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Taya, K.; Nakayama, E.E.; Shioda, T. Moderate restriction of macrophage-tropic human immunodeficiency virus type 1 by samhd1 in monocyte-derived macrophages. PLoS ONE 2014, 9, e90969. [Google Scholar] [CrossRef] [PubMed]
- Baldauf, H.M.; Stegmann, L.; Schwarz, S.M.; Ambiel, I.; Trotard, M.; Martin, M.; Burggraf, M.; Lenzi, G.M.; Lejk, H.; Pan, X.; et al. Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc. Natl. Acad. Sci. USA 2017, 114, 2729–2734. [Google Scholar] [CrossRef] [PubMed]
- Hrecka, K.; Hao, C.; Gierszewska, M.; Swanson, S.K.; Kesik-Brodacka, M.; Srivastava, S.; Florens, L.; Washburn, M.P.; Skowronski, J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the samhd1 protein. Nature 2011, 474, 658–661. [Google Scholar] [PubMed]
- McAllery, S.A.; Ahlenstiel, C.L.; Suzuki, K.; Symonds, G.P.; Kelleher, A.D.; Turville, S.G. The feasibility of incorporating vpx into lentiviral gene therapy vectors. Mol. Ther. Methods Clin. Dev. 2016, 5, 16066. [Google Scholar] [PubMed]
- Gao, Y.; Ju, Y.; Ren, X.; Zhang, L.; Yin, X. Enhanced infection efficiency and cytotoxicity mediated by vpx-containing lentivirus in chimeric antigen receptor macrophage (car-m). Heliyon 2023, 9, e21886. [Google Scholar]
- Zhang, K.; Lv, D.W.; Li, R. Conserved herpesvirus protein kinases target samhd1 to facilitate virus replication. Cell Rep. 2019, 28, 449–459.e5. [Google Scholar]
- Chen, Z.; Zhu, M.; Pan, X.; Zhu, Y.; Yan, H.; Jiang, T.; Shen, Y.; Dong, X.; Zheng, N.; Lu, J.; et al. Inhibition of hepatitis b virus replication by samhd1. Biochem. Biophys. Res. Commun. 2014, 450, 1462–1468. [Google Scholar] [CrossRef]
- Gramberg, T.; Kahle, T.; Bloch, N.; Wittmann, S.; Mullers, E.; Daddacha, W.; Hofmann, H.; Kim, B.; Lindemann, D.; Landau, N.R. Restriction of diverse retroviruses by samhd1. Retrovirology 2013, 10, 26. [Google Scholar] [CrossRef]
- Bonifati, S.; Daly, M.B.; St Gelais, C.; Kim, S.H.; Hollenbaugh, J.A.; Shepard, C.; Kennedy, E.M.; Kim, D.H.; Schinazi, R.F.; Kim, B.; et al. Samhd1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic thp-1 cells. Virology 2016, 495, 92–100. [Google Scholar]
- Daddacha, W.; Koyen, A.E.; Bastien, A.J.; Head, P.E.; Dhere, V.R.; Nabeta, G.N.; Connolly, E.C.; Werner, E.; Madden, M.Z.; Daly, M.B.; et al. Samhd1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep. 2017, 20, 1921–1935. [Google Scholar] [CrossRef]
- Kapoor-Vazirani, P.; Rath, S.K.; Liu, X.; Shu, Z.; Bowen, N.E.; Chen, Y.; Haji-Seyed-Javadi, R.; Daddacha, W.; Minten, E.V.; Danelia, D.; et al. Samhd1 deacetylation by sirt1 promotes DNA end resection by facilitating DNA binding at double-strand breaks. Nat. Commun. 2022, 13, 6707. [Google Scholar] [CrossRef] [PubMed]
- Fink, D.L.; Cai, J.; Whelan, M.V.X.; Monit, C.; Maluquer de Motes, C.; Towers, G.J.; Sumner, R.P. HIV-2/siv vpx antagonises nf-kappab activation by targeting p65. Retrovirology 2022, 19, 2. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Guney, M.H.; Kim, K.; Goh, S.L.; McCauley, S.; Dauphin, A.; Diehl, W.E.; Luban, J. Primate immunodeficiency virus proteins vpx and vpr counteract transcriptional repression of proviruses by the hush complex. Nat. Microbiol. 2018, 3, 1354–1361. [Google Scholar] [CrossRef]
- Cheng, X.; Belshan, M.; Ratner, L. Hsp40 facilitates nuclear import of the human immunodeficiency virus type 2 vpx-mediated preintegration complex. J. Virol. 2008, 82, 1229–1237. [Google Scholar] [CrossRef]
- Mahalingam, S.; Van Tine, B.; Santiago, M.L.; Gao, F.; Shaw, G.M.; Hahn, B.H. Functional analysis of the simian immunodeficiency virus vpx protein: Identification of packaging determinants and a novel nuclear targeting domain. J. Virol. 2001, 75, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Raja, S.; Mahalingam, S. Viral protein x unlocks the nuclear pore complex through a human nup153-dependent pathway to promote nuclear translocation of the lentiviral genome. Mol. Biol. Cell 2020, 31, 304–317. [Google Scholar] [CrossRef]
- Singhal, P.K.; Rajendra Kumar, P.; Subba Rao, M.R.; Mahalingam, S. Nuclear export of simian immunodeficiency virus vpx protein. J. Virol. 2006, 80, 12271–12282. [Google Scholar] [CrossRef]
- Belshan, M.; Mahnke, L.A.; Ratner, L. Conserved amino acids of the human immunodeficiency virus type 2 vpx nuclear localization signal are critical for nuclear targeting of the viral preintegration complex in non-dividing cells. Virology 2006, 346, 118–126. [Google Scholar] [CrossRef] [PubMed]
- McCulley, A.; Ratner, L. HIV-2 viral protein x (vpx) ubiquitination is dispensable for ubiquitin ligase interaction and effects on macrophage infection. Virology 2012, 427, 67–75. [Google Scholar] [CrossRef]
- Cheng, X.; Ratner, L. HIV-2 vpx protein interacts with interferon regulatory factor 5 (irf5) and inhibits its function. J. Biol. Chem. 2014, 289, 9146–9157. [Google Scholar] [CrossRef]
- Mahdi, M.; Szojka, Z.; Motyan, J.A.; Tozser, J. Inhibitory effects of HIV-2 vpx on replication of HIV-1. J. Virol. 2018, 92, e00554-18. [Google Scholar] [PubMed]
- Veenhuis, R.T.; Abreu, C.M.; Costa, P.A.G.; Ferreira, E.A.; Ratliff, J.; Pohlenz, L.; Shirk, E.N.; Rubin, L.H.; Blankson, J.N.; Gama, L.; et al. Monocyte-derived macrophages contain persistent latent HIV reservoirs. Nat. Microbiol. 2023, 8, 833–844. [Google Scholar] [PubMed]
- Wallis, Z.K.; Williams, K.C. Monocytes in HIV and siv infection and aging: Implications for inflamm-aging and accelerated aging. Viruses 2022, 14, 409. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, A.D.; Bouazzi, R.; Afzal, S.; Gelpi, M.; Benfield, T.; Hogh, J.; Thomsen, M.T.; Troseid, M.; Nordestgaard, B.G.; Nielsen, S.D. Monocyte count and soluble markers of monocyte activation in people living with HIV and uninfected controls. BMC Infect. Dis. 2022, 22, 451. [Google Scholar] [CrossRef]
- Shan, M.; Su, Y.; Kang, W.; Gao, R.; Li, X.; Zhang, G. Aberrant expression and functions of protocadherins in human malignant tumors. Tumour Biol. 2016, 37, 12969–12981. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Meng, S.; Han, M.H.; Lin, C.; Wang, X. Alpha- and gamma-protocadherins negatively regulate pyk2. J. Biol. Chem. 2009, 284, 2880–2890. [Google Scholar] [CrossRef]
- Keeler, A.B.; Schreiner, D.; Weiner, J.A. Protein kinase c phosphorylation of a gamma-protocadherin c-terminal lipid binding domain regulates focal adhesion kinase inhibition and dendrite arborization. J. Biol. Chem. 2015, 290, 20674–20686. [Google Scholar] [PubMed]
- Dallosso, A.R.; Oster, B.; Greenhough, A.; Thorsen, K.; Curry, T.J.; Owen, C.; Hancock, A.L.; Szemes, M.; Paraskeva, C.; Frank, M.; et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene 2012, 31, 4409–4419. [Google Scholar]
- Pancho, A.; Aerts, T.; Mitsogiannis, M.D.; Seuntjens, E. Protocadherins at the crossroad of signaling pathways. Front. Mol. Neurosci. 2020, 13, 117. [Google Scholar]
- Bao, J.; Ye, J.; Xu, J.; Liu, S.; Wang, L.; Li, Z.; Li, Q.; Liu, F.; He, X.; Zou, H.; et al. Comprehensive rna-seq reveals molecular changes in kidney malignancy among people living with HIV. Mol. Ther. Nucleic Acids 2022, 29, 91–101. [Google Scholar]
- Arokium, H.; Kamata, M.; Chen, I. Virion-associated vpr of human immunodeficiency virus type 1 triggers activation of apoptotic events and enhances fas-induced apoptosis in human t cells. J. Virol. 2009, 83, 11283–11297. [Google Scholar]
- Garden, G.A.; Budd, S.L.; Tsai, E.; Hanson, L.; Kaul, M.; D’Emilia, D.M.; Friedlander, R.M.; Yuan, J.; Masliah, E.; Lipton, S.A. Caspase cascades in human immunodeficiency virus-associated neurodegeneration. J. Neurosci. 2002, 22, 4015–4024. [Google Scholar] [CrossRef] [PubMed]
- Kruman, I.I.; Nath, A.; Mattson, M.P. HIV-1 protein tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp. Neurol. 1998, 154, 276–288. [Google Scholar] [PubMed]
- Akari, H.; Bour, S.; Kao, S.; Adachi, A.; Strebel, K. The human immunodeficiency virus type 1 accessory protein vpu induces apoptosis by suppressing the nuclear factor kappab-dependent expression of antiapoptotic factors. J. Exp. Med. 2001, 194, 1299–1311. [Google Scholar] [PubMed]
- Sorgel, S.; Fraedrich, K.; Votteler, J.; Thomas, M.; Stamminger, T.; Schubert, U. Perinuclear localization of the HIV-1 regulatory protein vpr is important for induction of g2-arrest. Virology 2012, 432, 444–451. [Google Scholar]
- Eskandari, E.; Eaves, C.J. Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J. Cell Biol. 2022, 221, e202201159. [Google Scholar] [CrossRef]
- Tecalco-Cruz, A.C.; Rios-Lopez, D.G.; Vazquez-Victorio, G.; Rosales-Alvarez, R.E.; Macias-Silva, M. Transcriptional cofactors ski and snon are major regulators of the tgf-beta/smad signaling pathway in health and disease. Signal Transduct. Target. Ther. 2018, 3, 15. [Google Scholar]
- Chinnapaiyan, S.; Dutta, R.K.; Nair, M.; Chand, H.S.; Rahman, I.; Unwalla, H.J. Tgf-beta1 increases viral burden and promotes HIV-1 latency in primary differentiated human bronchial epithelial cells. Sci. Rep. 2019, 9, 12552. [Google Scholar]
- Stettner, M.R.; Nance, J.A.; Wright, C.A.; Kinoshita, Y.; Kim, W.K.; Morgello, S.; Rappaport, J.; Khalili, K.; Gordon, J.; Johnson, E.M. Smad proteins of oligodendroglial cells regulate transcription of jc virus early and late genes coordinately with the tat protein of human immunodeficiency virus type 1. J. Gen. Virol. 2009, 90, 2005–2014. [Google Scholar]
- Yim, L.Y.; Lam, K.S.; Luk, T.Y.; Mo, Y.; Lu, X.; Wang, J.; Cheung, K.W.; Lui, G.C.Y.; Chan, D.P.C.; Wong, B.C.K.; et al. Transforming growth factor beta signaling promotes HIV-1 infection in activated and resting memory CD4+ T cells. J. Virol. 2023, 97, e0027023. [Google Scholar]
- Sawaya, B.E.; Thatikunta, P.; Denisova, L.; Brady, J.; Khalili, K.; Amini, S. Regulation of tnfalpha and tgfbeta-1 gene transcription by HIV-1 tat in cns cells. J. Neuroimmunol. 1998, 87, 33–42. [Google Scholar] [PubMed]
- Boby, N.; Ransom, A.; Pace, B.T.; Williams, K.M.; Mabee, C.; Das, A.; Srivastav, S.K.; Porter, E.; Pahar, B. Enhanced intestinal tgf-beta/smad-dependent signaling in simian immunodeficiency virus infected rhesus macaques. Cells 2021, 10, 806. [Google Scholar] [PubMed]
- Mirzaei, H.; Faghihloo, E. Viruses as key modulators of the tgf-beta pathway; a double-edged sword involved in cancer. Rev. Med. Virol. 2018, 28, e1967. [Google Scholar]
- Wang, J.; Guan, E.; Roderiquez, G.; Norcross, M.A. Synergistic induction of apoptosis in primary CD4+ T cells by macrophage-tropic HIV-1 and tgf-beta1. J. Immunol. 2001, 167, 3360–3366. [Google Scholar]
- Desai, S.; Landay, A. Early immune senescence in HIV disease. Curr. HIV/AIDS Rep. 2010, 7, 4–10. [Google Scholar] [PubMed]
- Appay, V.; Sauce, D. Immune activation and inflammation in HIV-1 infection: Causes and consequences. J. Pathol. 2008, 214, 231–241. [Google Scholar]
- Bauer, M.E.; Fuente Mde, L. The role of oxidative and inflammatory stress and persistent viral infections in immunosenescence. Mech. Ageing Dev. 2016, 158, 27–37. [Google Scholar]
- Hoshino, S.; Konishi, M.; Mori, M.; Shimura, M.; Nishitani, C.; Kuroki, Y.; Koyanagi, Y.; Kano, S.; Itabe, H.; Ishizaka, Y. HIV-1 vpr induces tlr4/myd88-mediated il-6 production and reactivates viral production from latency. J. Leukoc. Biol. 2010, 87, 1133–1143. [Google Scholar]
- Mohamed, A.; Bakir, T.; Al-Hawel, H.; Al-Sharif, I.; Bakheet, R.; Kouser, L.; Murugaiah, V.; Al-Mozaini, M. HIV-2 vpx neutralizes host restriction factor samhd1 to promote viral pathogenesis. Sci. Rep. 2021, 11, 20984. [Google Scholar]
- Vermeire, J.; Roesch, F.; Sauter, D.; Rua, R.; Hotter, D.; Van Nuffel, A.; Vanderstraeten, H.; Naessens, E.; Iannucci, V.; Landi, A.; et al. HIV triggers a cgas-dependent, vpu- and vpr-regulated type i interferon response in CD4+ T cells. Cell Rep. 2016, 17, 413–424. [Google Scholar]
- Sertznig, H.; Hillebrand, F.; Erkelenz, S.; Schaal, H.; Widera, M. Behind the scenes of HIV-1 replication: Alternative splicing as the dependency factor on the quiet. Virology 2018, 516, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Dlamini, Z.; Hull, R. Can the HIV-1 splicing machinery be targeted for drug discovery? HIV AIDS 2017, 9, 63–75. [Google Scholar] [CrossRef]
- Erkelenz, S.; Hillebrand, F.; Widera, M.; Theiss, S.; Fayyaz, A.; Degrandi, D.; Pfeffer, K.; Schaal, H. Balanced splicing at the tat-specific HIV-1 3′ss A3 is critical for HIV-1 replication. Retrovirology 2015, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Yukl, S.A.; Kaiser, P.; Kim, P.; Telwatte, S.; Joshi, S.K.; Vu, M.; Lampiris, H.; Wong, J.K. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018, 10, eaap9927. [Google Scholar] [CrossRef]
- Lassen, K.; Han, Y.; Zhou, Y.; Siliciano, J.; Siliciano, R.F. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 2004, 10, 525–531. [Google Scholar] [CrossRef]
- Deguine, J.; Barton, G.M. Myd88: A central player in innate immune signaling. F1000Prime Rep. 2014, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Lee, H.L.; Pacchia, A.L.; Ron, Y.; Dougherty, J.P. A HIV-2-based self-inactivating vector for enhanced gene transduction. J. Biotechnol. 2007, 127, 745–757. [Google Scholar] [CrossRef]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of crispr-cas9 protein and sgrna to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1334. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. Multiqc: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; White, K.P. Rna-seq differential expression studies: More sequence or more replication? Bioinformatics 2014, 30, 301–304. [Google Scholar]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. Ebseq: An empirical bayes hierarchical model for inference in rna-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Albert, J.; Naucler, A.; Bottiger, B.; Broliden, P.A.; Albino, P.; Ouattara, S.A.; Bjorkegren, C.; Valentin, A.; Biberfeld, G.; Fenyo, E.M. Replicative capacity of HIV-2, like HIV-1, correlates with severity of immunodeficiency. Aids 1990, 4, 291–295. [Google Scholar]
- Ozkaya Sahin, G.; Mansson, F.; Palm, A.A.; Vincic, E.; da Silva, Z.; Medstrand, P.; Norrgren, H.; Fenyo, E.M.; Jansson, M. Frequent intratype neutralization by plasma immunoglobulin a identified in HIV type 2 infection. AIDS Res. Hum. Retroviruses 2013, 29, 470–478. [Google Scholar] [PubMed]
- Chopra, P.; Gupta, S.; Dastidar, S.G.; Ray, A. Development of cell death-based method for the selectivity screening of caspase-1 inhibitors. Cytotechnology 2009, 60, 77. [Google Scholar]
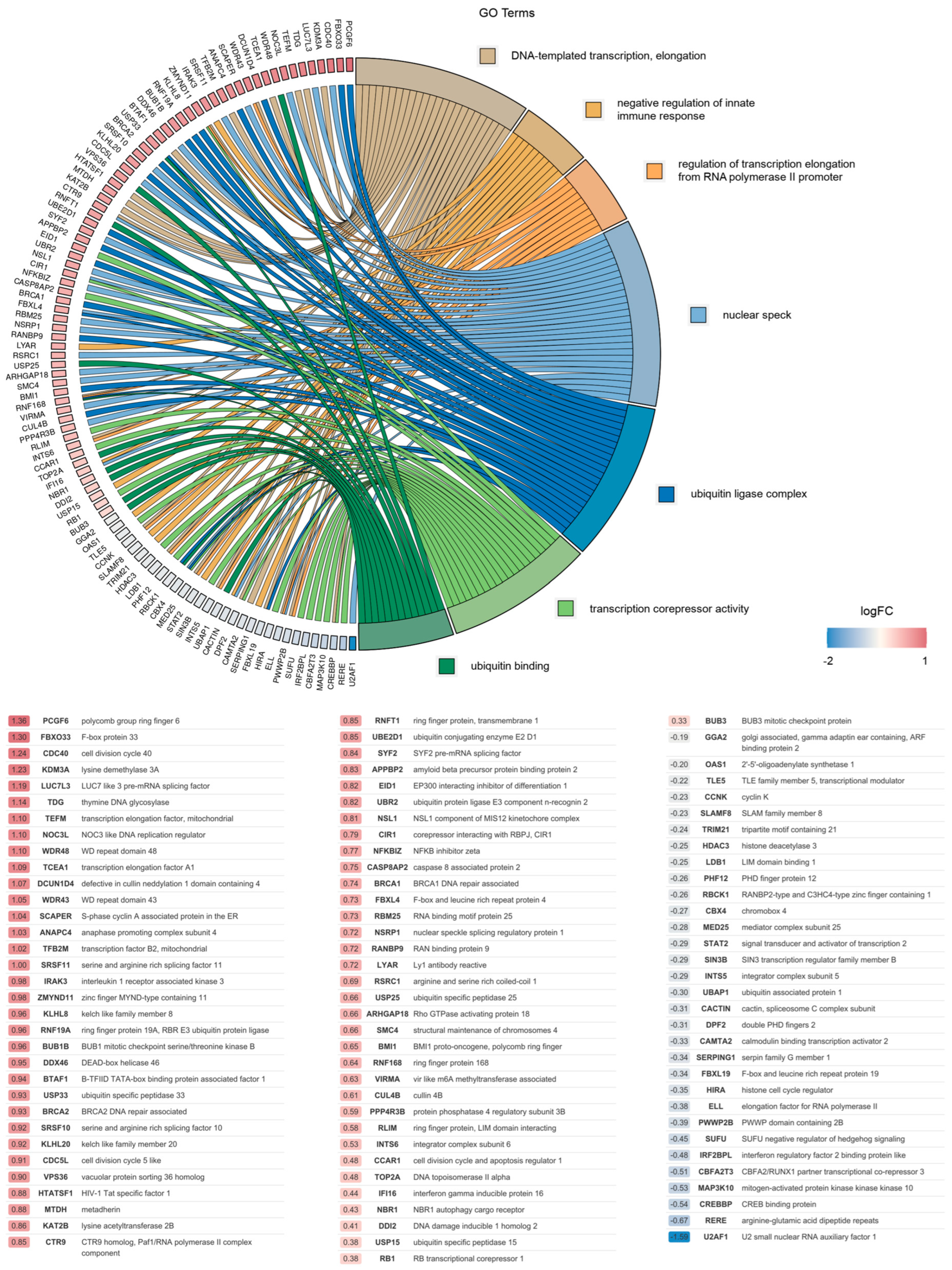


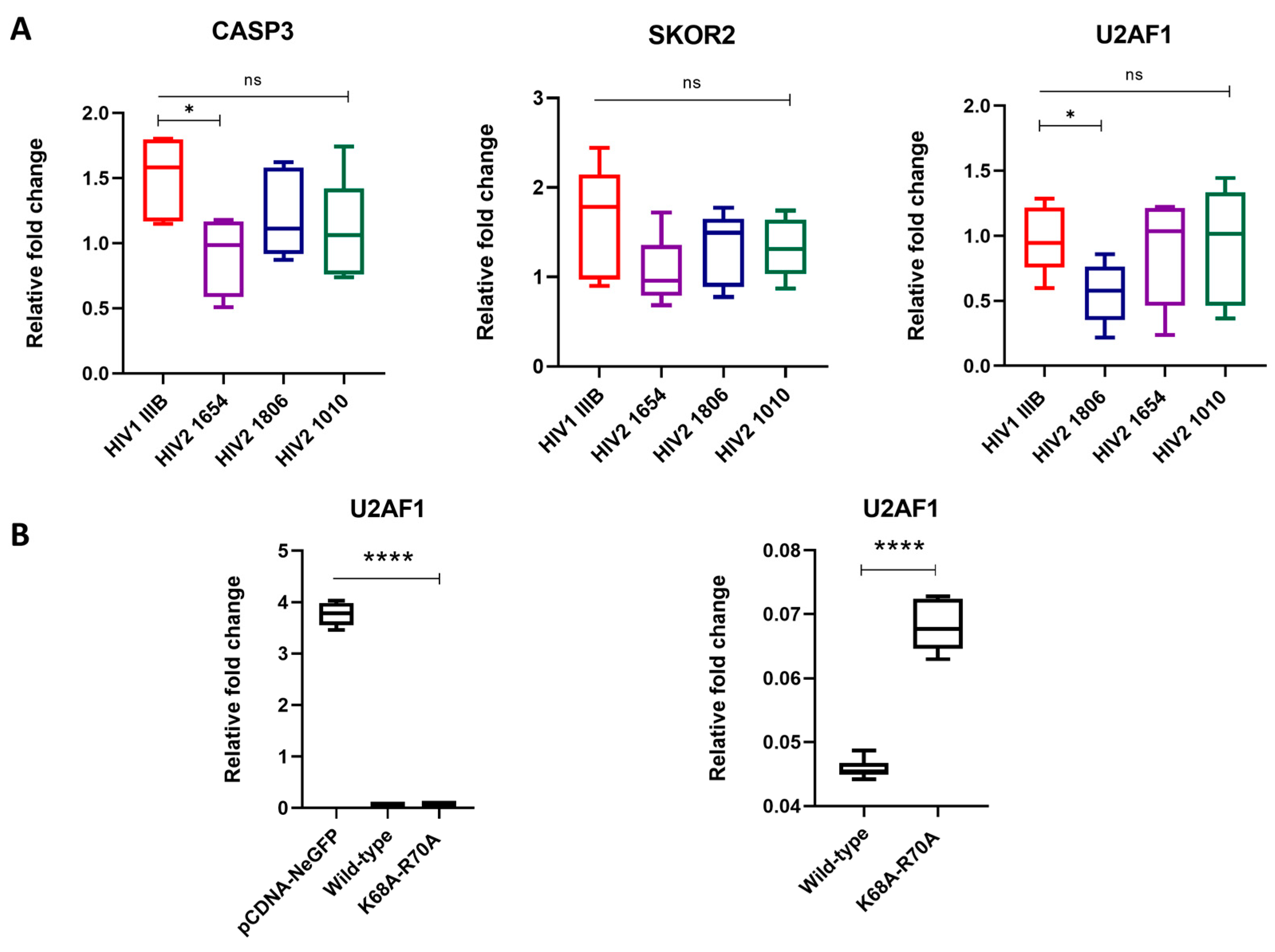
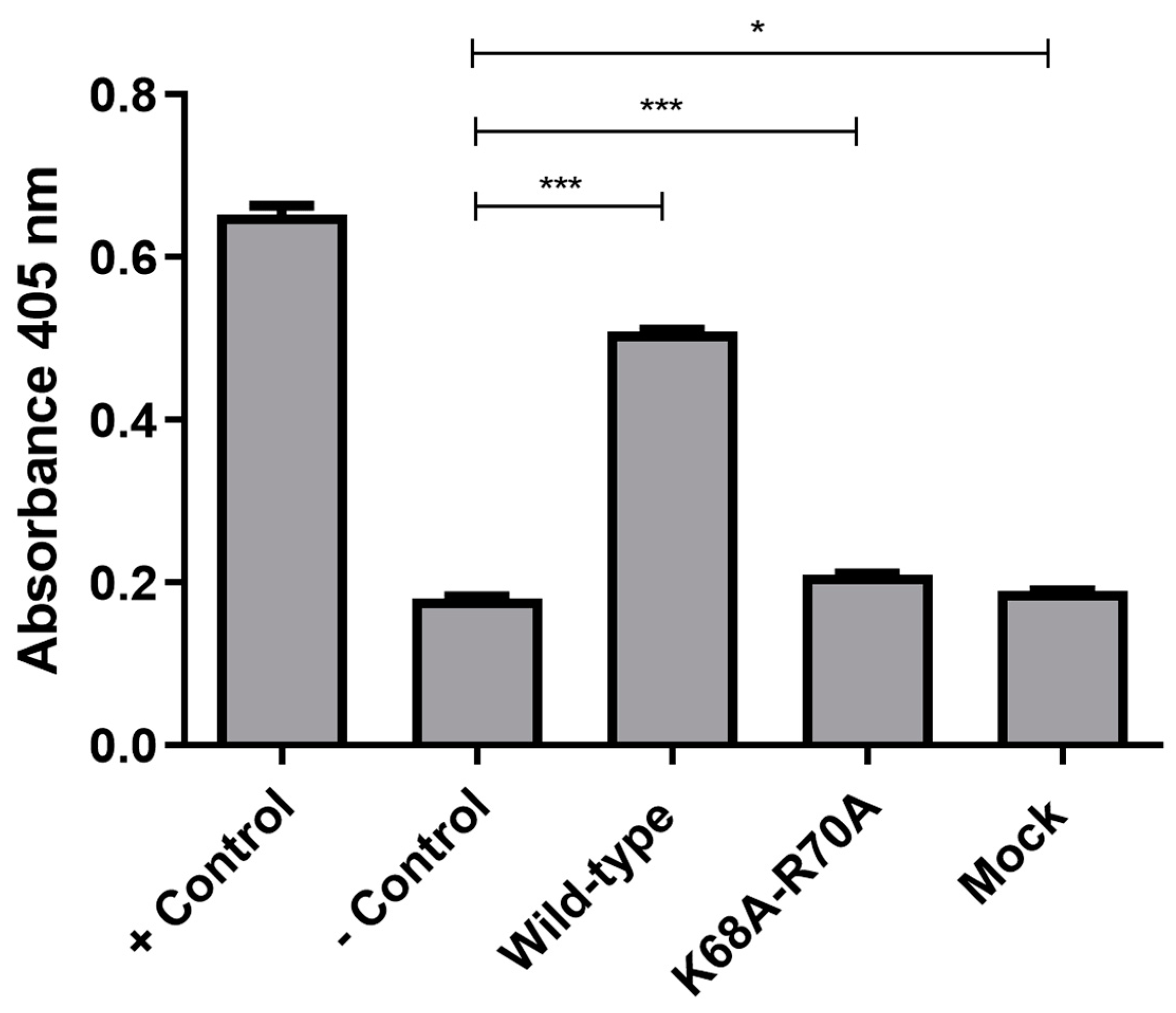
| Pattern | Relation of Expression Levels |
|---|---|
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szojka, Z.I.; Kunkli, B.; Kiarie, I.W.; Linkner, T.R.; Al-Muffti, A.S.; Ahmad, H.; Benkő, S.; Jansson, M.; Tőzsér, J.; Mahdi, M. Transcriptomic Analysis Reveals Key Pathways Influenced by HIV-2 Vpx. Int. J. Mol. Sci. 2025, 26, 3460. https://doi.org/10.3390/ijms26083460
Szojka ZI, Kunkli B, Kiarie IW, Linkner TR, Al-Muffti AS, Ahmad H, Benkő S, Jansson M, Tőzsér J, Mahdi M. Transcriptomic Analysis Reveals Key Pathways Influenced by HIV-2 Vpx. International Journal of Molecular Sciences. 2025; 26(8):3460. https://doi.org/10.3390/ijms26083460
Chicago/Turabian StyleSzojka, Zsófia Ilona, Balázs Kunkli, Irene Wanjiru Kiarie, Tamás Richárd Linkner, Aya Shamal Al-Muffti, Hala Ahmad, Szilvia Benkő, Marianne Jansson, József Tőzsér, and Mohamed Mahdi. 2025. "Transcriptomic Analysis Reveals Key Pathways Influenced by HIV-2 Vpx" International Journal of Molecular Sciences 26, no. 8: 3460. https://doi.org/10.3390/ijms26083460
APA StyleSzojka, Z. I., Kunkli, B., Kiarie, I. W., Linkner, T. R., Al-Muffti, A. S., Ahmad, H., Benkő, S., Jansson, M., Tőzsér, J., & Mahdi, M. (2025). Transcriptomic Analysis Reveals Key Pathways Influenced by HIV-2 Vpx. International Journal of Molecular Sciences, 26(8), 3460. https://doi.org/10.3390/ijms26083460







