KLF14 and SREBF-1 Binding Site Associations with Orphan Receptor Promoters in Metabolic Syndrome
Abstract
:1. Introduction
2. Results
2.1. Alignment Sequence Similarity-Based Process
2.2. Important Region Identification and Similarity
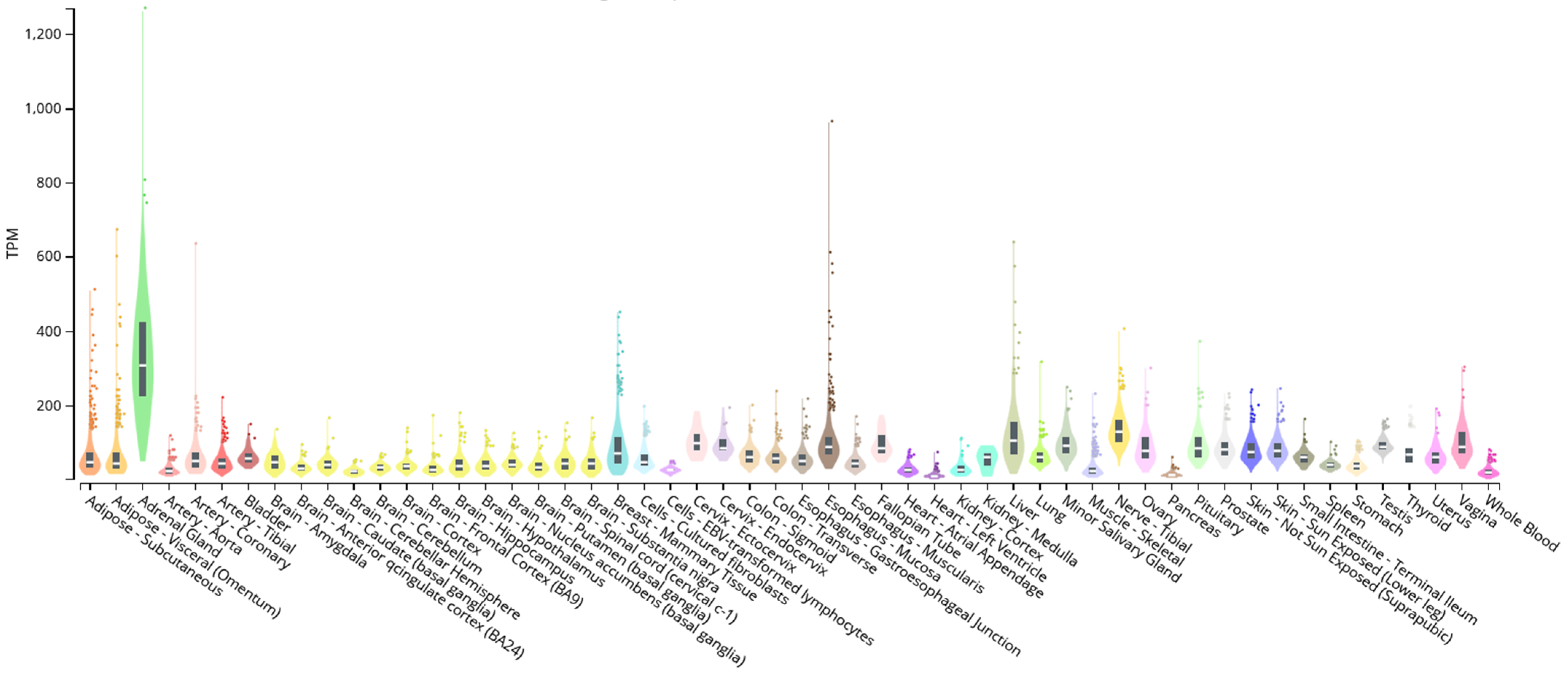
2.3. KLF14, SREBF-1, and oGPCRs Genic Expression Correlation
2.4. Identification of GPCRs Associated with Metabolic Syndrome (GPCRs-MetS)
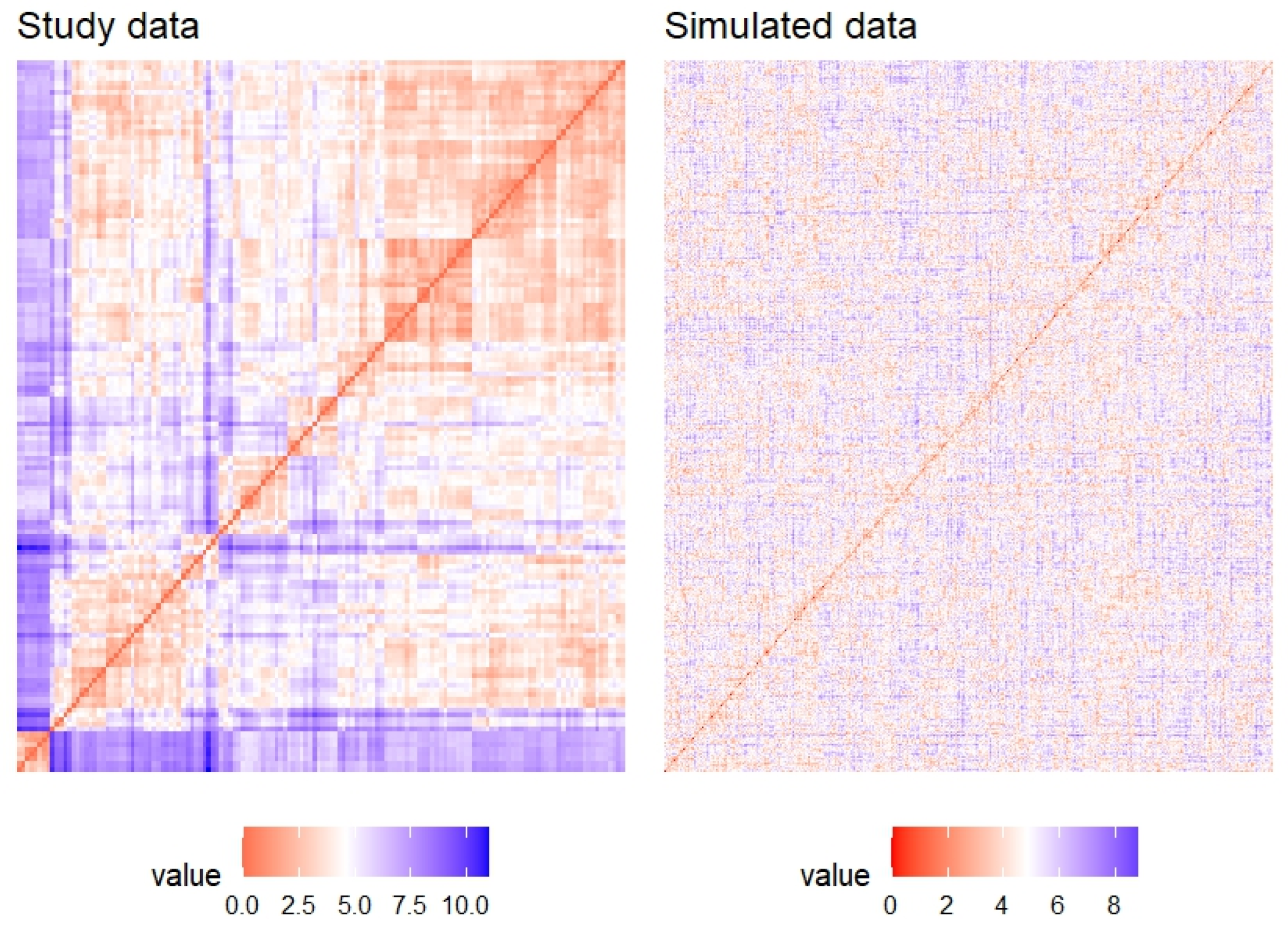
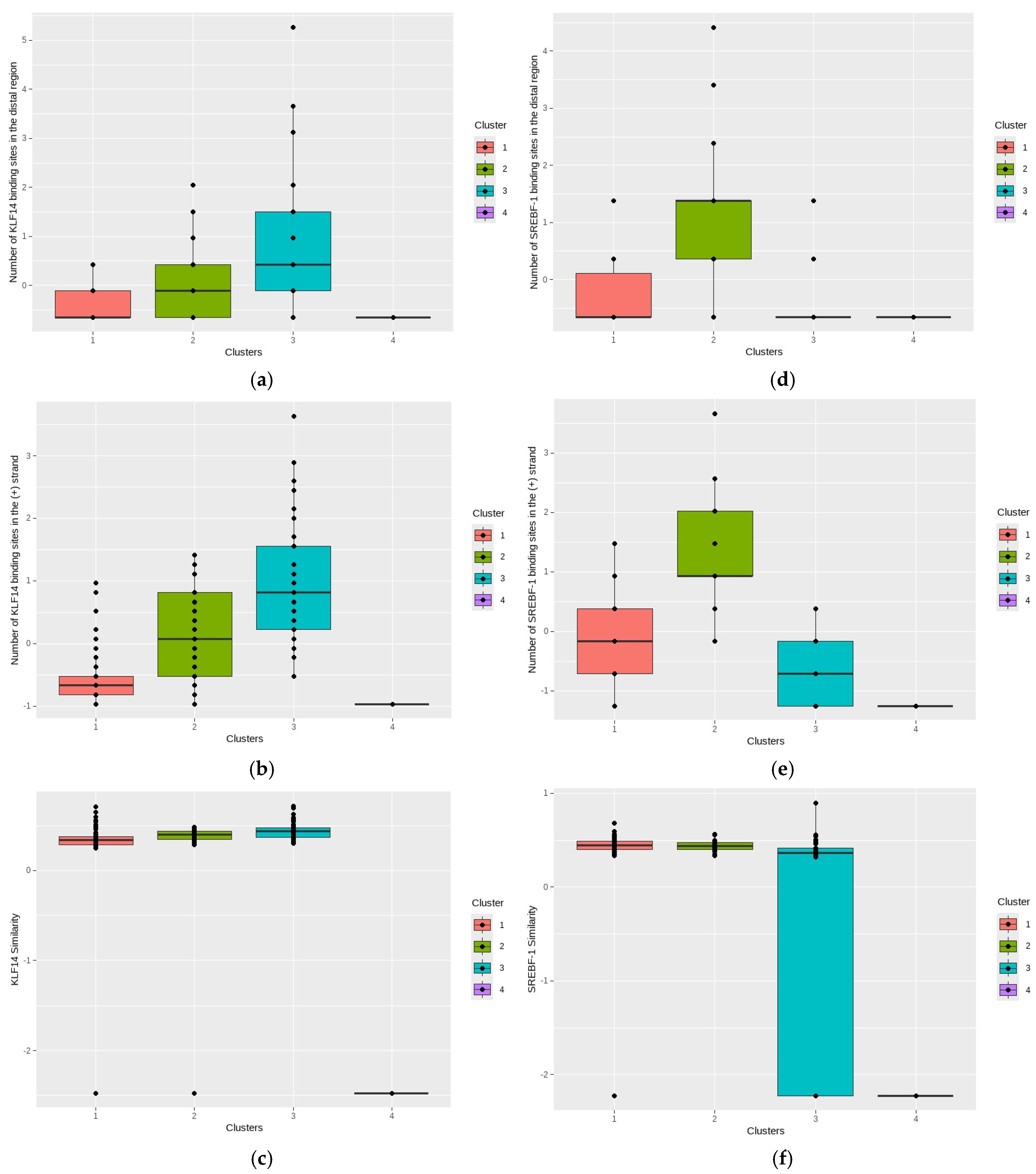
2.5. Clustering Analysis for the Bioinformatics Outcomes
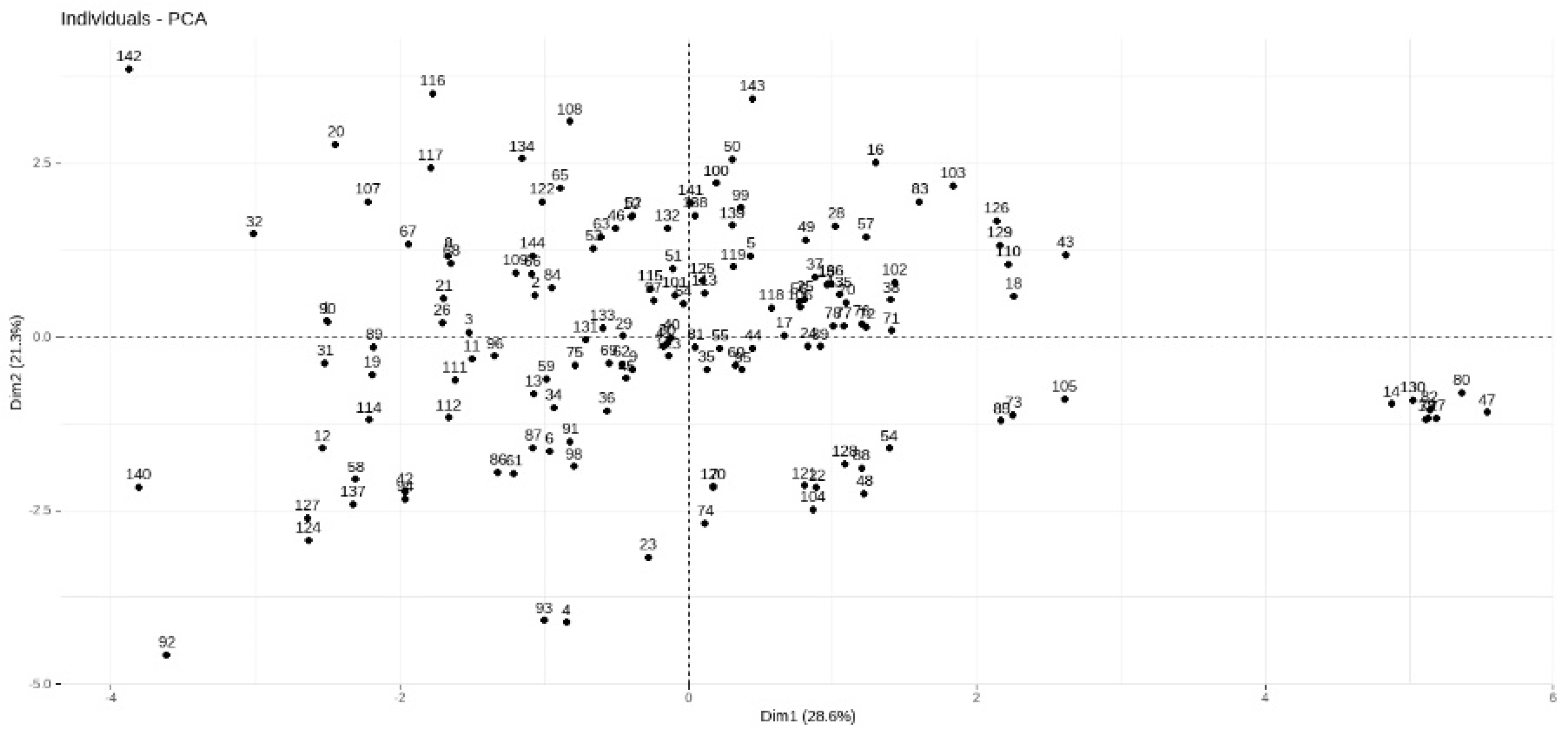
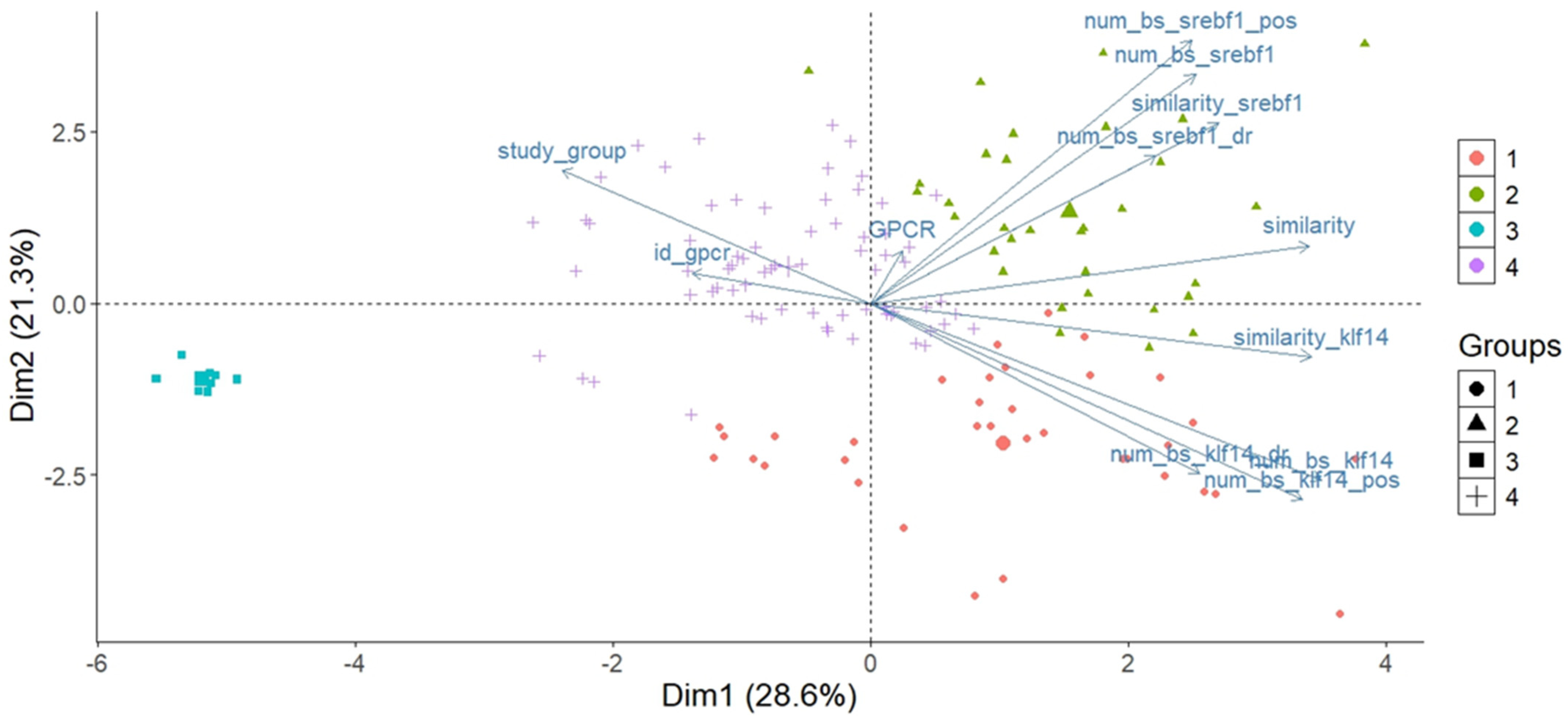
2.5.1. PCA for the Distribution of Individual Observations
2.5.2. K-Means Clustering Algorithm
- Five proposed two as the best number of clusters;
- Four proposed three as the best number of clusters;
- Two proposed four as the best number of clusters;
- One proposed five as the best number of clusters;
- Two proposed six as the best number of clusters;
- Three proposed eight as the best number of clusters;
- Five proposed nine as the best number of clusters;
- Five proposed ten as the best number of clusters.
2.6. Regression Analysis for Orphan GPCRs and KLF14 and SREBF-1
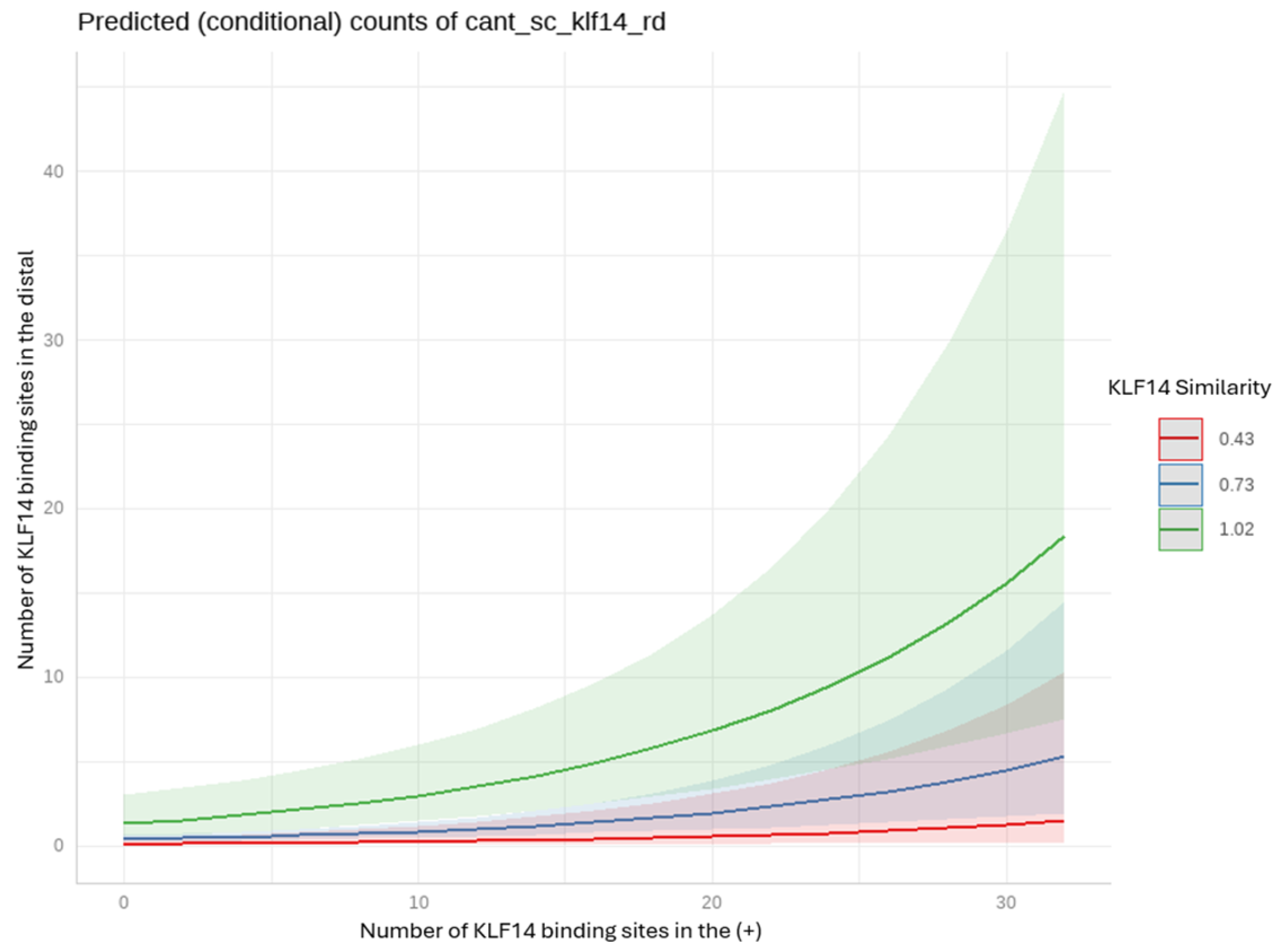
2.6.1. Negative Binomial Regression Model for KLF14
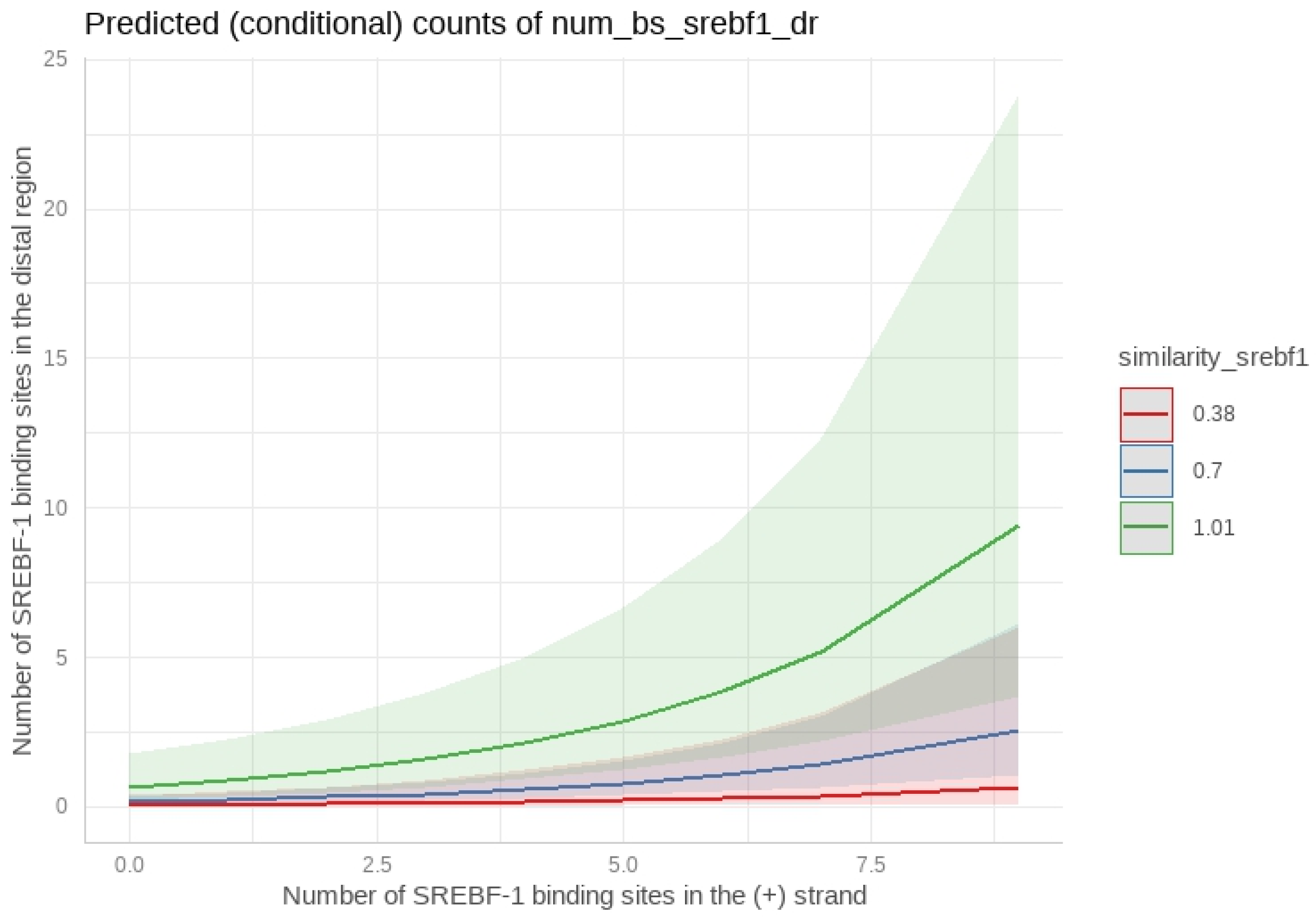
2.6.2. Poisson Regression Model for SREBF-1
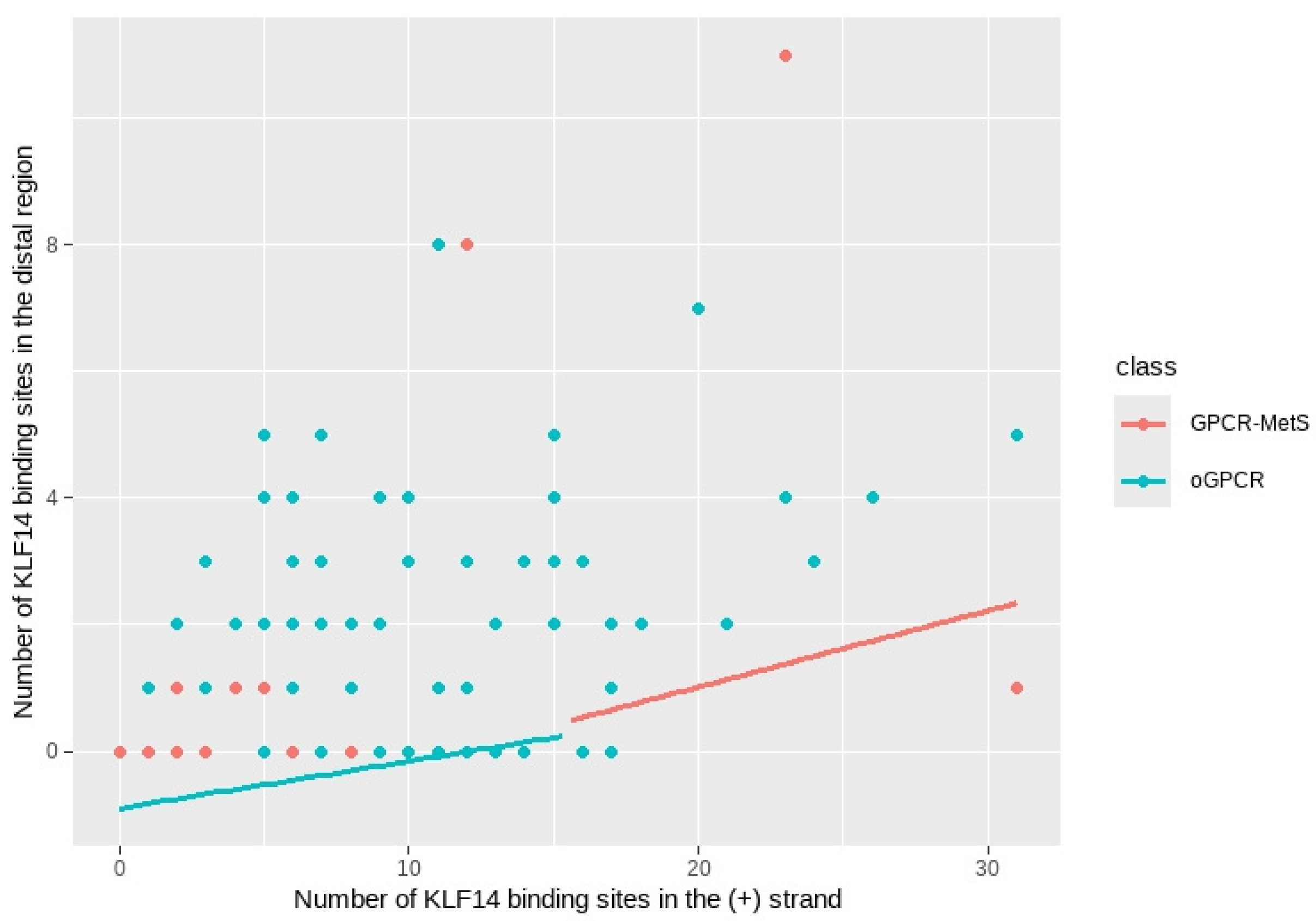
2.7. Comparison Between Receptor Groups
3. Discussion
3.1. Orphan Receptors Analysis
3.2. Analysis of GPCRs Associated with MetS
3.3. Statistical Analysis-Based
- KLF14: the density of KLF14 binding sites in the positive strand was significantly associated with the distal region binding sites density, even after adjusting for sequence similarity. This suggests that the presence of binding sites in the positive strand is a strong predictor of the number of binding sites in the distal region. Additionally, the sequence similarity of KLF14 significantly modulated the distal region binding site density, indicating that the structure and conservation of the KLF14 sequence are crucial for its regulatory function.
- SREBF-1: The density of SREBF-1 binding sites in the positive strand was also significantly associated with the distal region binding site density. While the sequence similarity of SREBF-1 showed a significant association, the confidence interval was wider, suggesting a potentially more complex relationship.
3.4. Study Implications
3.5. Study Limitations and Strengths
3.6. Future Directions
4. Materials and Methods
4.1. Alignment Sequences Similarity-Based Process
- Local alignment: the Smith–Waterman algorithm, which seeks the best local match between two sequences, regardless of their total length. This approach is useful for identifying regions of high similarity within longer sequences.
4.2. Important Region Identification and Similarity
- GenBank extraction: promoters of GPCRs were extracted from the NCBI GenBank database.
- Transcription factor binding site prediction: the JASPAR database (jaspar.genereg.net) was employed to analyze these promoters for potential binding sites of the transcription factors KLF14 and SREBF-1.
- Smith–Waterman algorithm: this dynamic programming algorithm was utilized to align the promoter sequences, identifying regions of similarity and dissimilarity.
- Pearson correlation coefficient: this metric was employed to quantify the degree of similarity between sequences.
- Unweighted pair group method with arithmetic mean (UPGMA): this clustering method was used to construct a phylogenetic tree based on sequence similarities, allowing for visualization of evolutionary relationships.
- Region classification: viable binding sites were identified and categorized based on their position relative to the transcription start site, including distal regions and regions with potential enhancer or silencer activity.
- A thorough literature review on MetS;
- Identification of orphan GPCRs;
- Extraction of gene sequences and identification of BS using NCBI GenBank and JASPAR;
- Classification of BS into distal and regulatory regions.
4.3. GPCRs-MetS Identification
4.4. Clustering and PCA Statistical Analysis
4.5. Regression Models for Counting Data
- Information criteria: information criteria such as AIC and BIC were used to select the model that best fits the data.
- Vuong test: the Vuong test was employed to compare the fit of different models, especially non-nested models.
- Residual analysis: residuals were examined to verify if the model assumptions were met.
- Diagnostic plots: diagnostic plots were created to assess the quality of the model fit (scatter plot with smoothing lines and facets, histograms, marginal effects plots).
4.6. Software Packages and Libraries
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Clustering Analysis for the Bioinformatics Outcomes


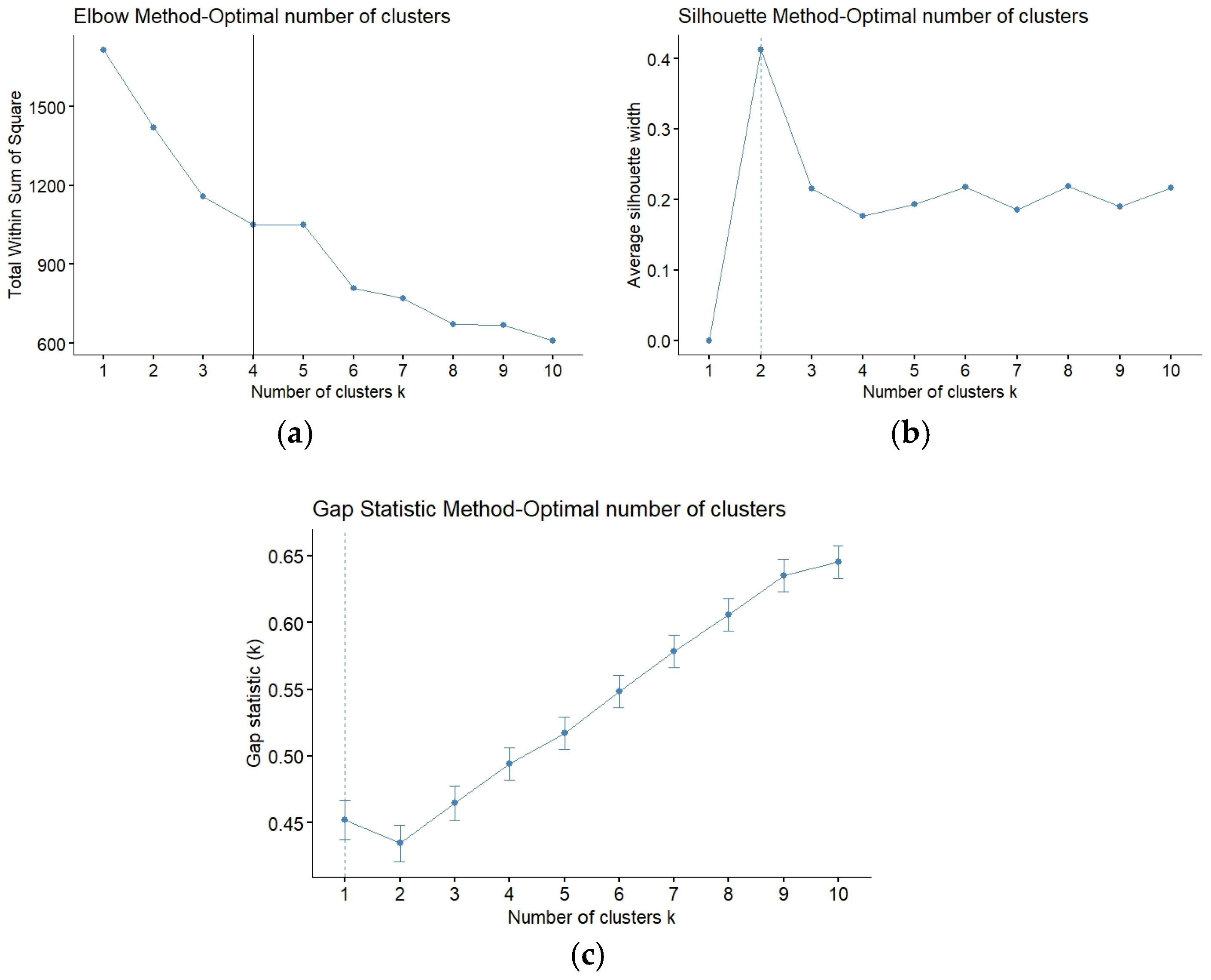
| Methods | Optimal K Value |
|---|---|
| Elbow | 4 |
| Silhouette | 2 |
| Gap statistic | - |
| Complete | 2 |
| WBSS | 6 |
| Clusterboot | 4 |
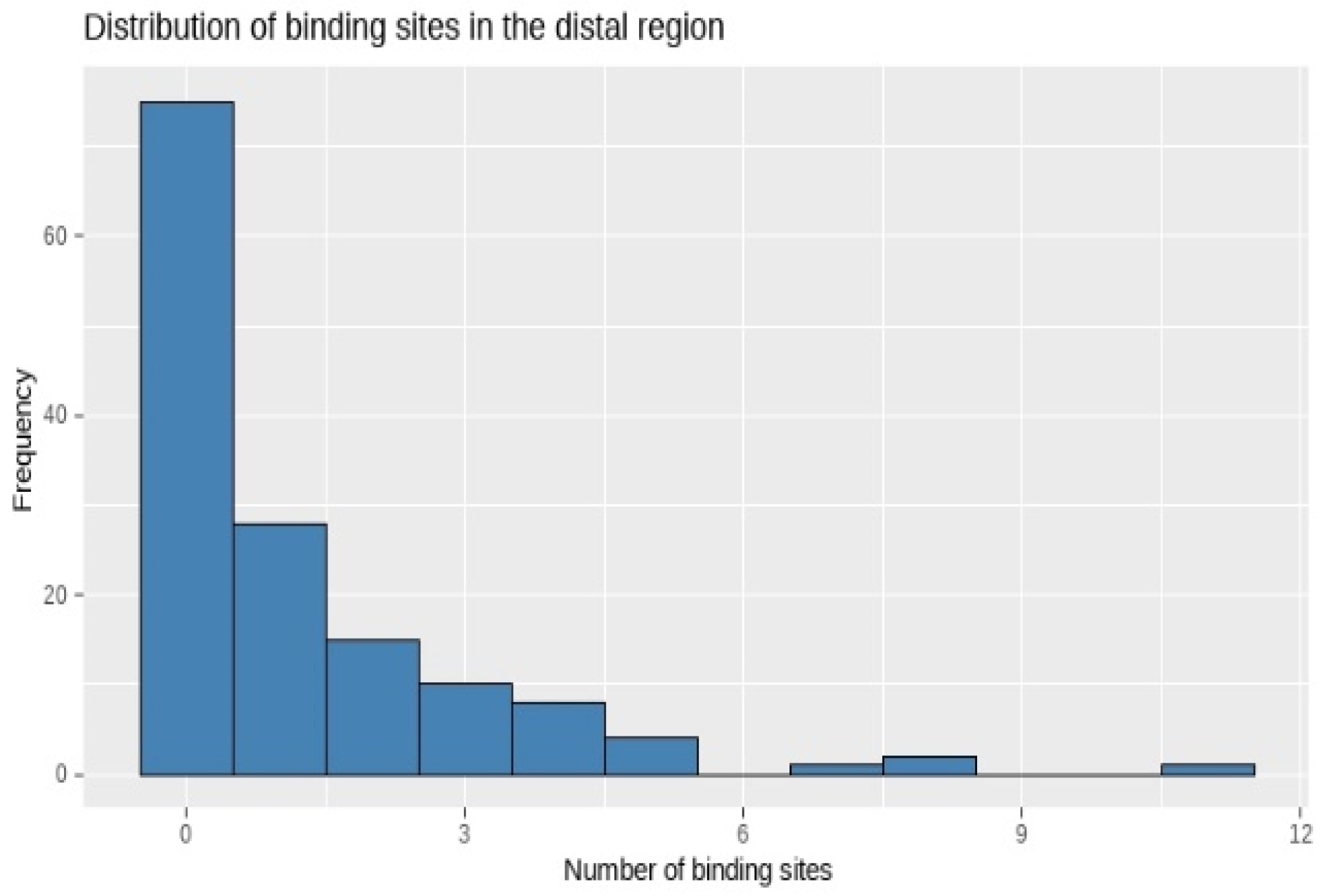
Appendix A.2. Exploring Data Distribution


Appendix A.3. Regression Analysis for Orphan GPCRs and KLF14 and SREBF-1
Negative Binomial Regression Model for KLF14

Appendix A.4. Poisson Regression Model for SREBF-1
References
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Maslov, L.N.; Naryzhnaya, N.V.; Boshchenko, A.A.; Popov, S.V.; Ivanov, V.V.; Oeltgen, P.R. Is Oxidative Stress of Adipocytes a Cause or a Consequence of the Metabolic Syndrome? J. Clin. Transl. Endocrinol. 2019, 15, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.C.; Zaidi, A.; Cao, X.; Weil, O.; Bey, P.; Patte, C.; Samudio, A.; Haddad, L.; Lam, C.G.; Moreira, C.; et al. The My Child Matters Programme: Effect of Public-Private Partnerships on Paediatric Cancer Care in Low-Income and Middle-Income Countries. Lancet Oncol. 2018, 19, e252–e266. [Google Scholar] [CrossRef] [PubMed]
- Saif-Ali, R.; Kamaruddin, N.A.; AL-Habori, M.; Al-Dubai, S.A.; Ngah, W.Z.W. Relationship of Metabolic Syndrome Defined by IDF or Revised NCEP ATP III with Glycemic Control among Malaysians with Type 2 Diabetes. Diabetol. Metab. Syndr. 2020, 12, 67. [Google Scholar] [CrossRef]
- Alberti, K.G.M.M.; Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z.; Cleeman, J.I.; Donato, K.A.; Fruchart, J.-C.; James, W.P.T.; Loria, C.M.; Smith, S.C. Harmonizing the Metabolic Syndrome. Circulation 2009, 120, 1640–1645. [Google Scholar] [CrossRef]
- Encuesta Nacional de Salud y Nutrición (ENSANUT). 2018. Available online: https://www.inegi.org.mx/programas/ensanut/2018/default.html#Tabulados (accessed on 20 September 2024).
- Rojas-Martínez, R.; Aguilar-Salinas, C.A.; Romero-Martínez, M.; Castro-Porras, L.; Gómez-Velasco, D.; Mehta, R. Trends in the prevalence of metabolic syndrome and its components in Mexican adults, 2006–2018. Salud Pública México 2021, 63, 713–724. [Google Scholar] [CrossRef]
- Plaza-López, M.F.; Valdez-Flores, M.A.; Herrán-Arita, A.K.d.l.; Osuna-Ramos, J.F.; Angulo-Rojo, C.E.; Llanos, A.M.G.; Camberos-Barraza, J.; Calderón-Zamora, L.; Norzagaray-Valenzuela, C.D. Prevalencia Del Síndrome Metabólico y Sus Componentes En La Población Adulta de Culiacán, Sinaloa, México. Rev. Med. UAS 2024, 13, 367–378. [Google Scholar] [CrossRef]
- Lorenzo, C.; Williams, K.; Gonzalez-Villalpando, C.; Haffner, S.M. The Prevalence of the Metabolic Syndrome Did Not Increase in Mexico City between 1990–1992 and 1997–1999 despite More Central Obesity. Diabetes Care 2005, 28, 2480–2485. [Google Scholar] [CrossRef]
- Civelek, M.; Lusis, A.J. Conducting the Metabolic Syndrome Orchestra. Nat. Genet. 2011, 43, 506–508. [Google Scholar] [CrossRef]
- McConnell, B.B.; Yang, V.W. Mammalian Krüppel-like Factors in Health and Diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef]
- Tetreault, M.-P.; Yang, Y.; Katz, J.P. Krüppel-like Factors in Cancer. Nat. Rev. Cancer 2013, 13, 701–713. [Google Scholar] [CrossRef]
- Jiang, J.; Chan, Y.-S.; Loh, Y.-H.; Cai, J.; Tong, G.-Q.; Lim, C.-A.; Robson, P.; Zhong, S.; Ng, H.-H. A Core Klf Circuitry Regulates Self-Renewal of Embryonic Stem Cells. Nat. Cell Biol. 2008, 10, 353–360. [Google Scholar] [CrossRef]
- Miranzadeh-Mahabadi, H.; Emadi-Baygi, M.; Nikpour, P.; Kelishadi, R. Association Study Between Metabolic Syndrome and rs8066560 Polymorphism in the Promoter Region of Sterol Regulatory Element-Binding Transcription Factor 1 Gene in Iranian Children and Adolescents. Int. J. Prev. Med. 2016, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yun, J.-H.; Ryoo, J.E.; Lee, K.-J.; Choi, B.-C.; Baek, K.-H. 54G/C Polymorphism of SREBF-1 Gene Is Associated with Polycystic Ovary Syndrome. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015, 188, 95–99. [Google Scholar] [CrossRef]
- Adams, L.A.; Marsh, J.A.; Ayonrinde, O.T.; Olynyk, J.K.; Ang, W.Q.; Beilin, L.J.; Mori, T.; Palmer, L.J.; Oddy, W.W.; Lye, S.J.; et al. Cholesteryl Ester Transfer Protein Gene Polymorphisms Increase the Risk of Fatty Liver in Females Independent of Adiposity. J. Gastroenterol. Hepatol. 2012, 27, 1520–1527. [Google Scholar] [CrossRef]
- Harding, A.-H.; Loos, R.J.F.; Luan, J.; O’Rahilly, S.; Wareham, N.J.; Barroso, I. Polymorphisms in the Gene Encoding Sterol Regulatory Element-Binding Factor-1c Are Associated with Type 2 Diabetes. Diabetologia 2006, 49, 2642–2648. [Google Scholar] [CrossRef]
- Alcántara-Hernández, R.; Hernández-Espinosa, D.A.; Medina, L.d.C.; García-Sáinz, J.A. Los receptores acoplados a proteínas G como blanco terapéutico. Gac. Méd. México 2022, 158, 101–107. [Google Scholar] [CrossRef]
- Mo, X.-L.; Yang, Z.; Tao, Y.-X. Chapter Four—Targeting GPR119 for the Potential Treatment of Type 2 Diabetes Mellitus. In Progress in Molecular Biology and Translational Science; Tao, Y.-X., Ed.; Glucose Homeostatis and the Pathogenesis of Diabetes Mellitus; Academic Press: Cambridge, MA, USA, 2014; Volume 121, pp. 95–131. Available online: https://www.sciencedirect.com/science/article/pii/B9780128001011000041 (accessed on 20 November 2022).
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR Drug Discovery: New Agents, Targets and Indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Sriram, K.; Insel, P.A. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol. Pharmacol. 2018, 93, 251–258. [Google Scholar] [CrossRef]
- Fang, Y.; Kenakin, T.; Liu, C. Editorial: Orphan GPCRs as Emerging Drug Targets. Front. Pharmacol. 2015, 6, 295. [Google Scholar] [CrossRef]
- García Sáinz, J.A. Receptores acoplados a proteínas G y su desensibilización. Rev. Odontol. Mex. 2011, 15, 210–213. [Google Scholar]
- Ruiz-Hernández, A.; Sánchez-Muñoz, F.; Rodriguez, J.; Calderón-Zamora, L.; Romero-Nava, R.; Huang, F.; Hong, E.; Villafaña, S. Expression of Orphan Receptors GPR22 and GPR162 in Streptozotocin-Induced Diabetic Rats. J. Recept. Signal Transduct. 2015, 35, 46–53. [Google Scholar] [CrossRef]
- Li, Y.; Li, B.; Chen, W.-D.; Wang, Y.-D. Role of G-Protein Coupled Receptors in Cardiovascular Diseases. Front. Cardiovasc. Med. 2023, 10, 1130312. [Google Scholar] [CrossRef]
- Kolar, G.R.; Grote, S.M.; Yosten, G.L.C. Targeting Orphan G Protein-Coupled Receptors for the Treatment of Diabetes and Its Complications: C-Peptide and GPR146. J. Intern. Med. 2016, 281, 25–40. [Google Scholar] [CrossRef]
- The GTEx Consortium. The Genotype-Tissue Expression (GTEx) Project; National Institutes of Health (NIH): Bethesda, MD, USA, 2025.
- Xiong, J. Essential Bioinformatics; Cambridge University Press: Cambridge, UK, 2006; 360p, ISBN 978-1-139-45062-1. [Google Scholar]
- Harding, S.D.; Armstrong, J.F.; Faccenda, E.; Southan, C.; Alexander, S.P.H.; Davenport, A.P.; Spedding, M.; Davies, J.A. The IUPHAR/BPS Guide to PHARMACOLOGY in 2024. Nucleic Acids Res. 2024, 52, D1438–D1449. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Yang, H.; Huang, Q.; Yu, H.; Quan, Z. Associations Between Obesity-Related Gene MC4R rs17782313 Locus Polymorphism and Components of Metabolic Syndrome: A Systematic Review and Meta-Analysis. Metab. Syndr. Relat. Disord. 2024, 22, 241–250. [Google Scholar] [CrossRef]
- Rana, S.; Sultana, A.; Bhatti, A.A. Association of BDNF rs6265 and MC4R rs17782313 with Metabolic Syndrome in Pakistanis. J. Biosci. 2019, 44, 95. [Google Scholar]
- Mendes de Oliveira, E.; Keogh, J.M.; Talbot, F.; Henning, E.; Ahmed, R.; Perdikari, A.; Bounds, R.; Wasiluk, N.; Ayinampudi, V.; Barroso, I.; et al. Obesity-Associated GNAS Mutations and the Melanocortin Pathway. N. Engl. J. Med. 2021, 385, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Vanwong, N.; Puangpetch, A.; Unaharassamee, W.; Jiratjintana, N.; Na Nakorn, C.; Hongkaew, Y.; Sukasem, C. Effect of 5-HT2C Receptor Gene Polymorphism (HTR2C-759C/T) on Metabolic Adverse Effects in Thai Psychiatric Patients Treated with Risperidone. Pharmacoepidemiol. Drug Saf. 2021, 30, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Maimaitirexiati, T.; Zhang, R.; Gui, X.; Zhang, W.; Xu, G.; Hu, G. HTR2C Polymorphisms, Olanzapine-Induced Weight Gain and Antipsychotic-Induced Metabolic Syndrome in Schizophrenia Patients: A Meta-Analysis. Int. J. Psychiatry Clin. Pract. 2014, 18, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Romero-Nava, R.; Zhou, D.-S.; García, N.; Ruiz-Hernández, A.; Si, Y.-C.; Sánchez-Muñoz, F.; Huang, F.; Hong, E.; Villafaña, S. Evidence of Alterations in the Expression of Orphan Receptors GPR26 and GPR39 Due to the Etiology of the Metabolic Syndrome. J. Recept. Signal Transduct. 2017, 37, 422–429. [Google Scholar] [CrossRef]
- Bjursell, M.; Gerdin, A.-K.; Jönsson, M.; Surve, V.V.; Svensson, L.; Huang, X.-F.; Törnell, J.; Bohlooly-Y, M. G Protein-Coupled Receptor 12 Deficiency Results in Dyslipidemia and Obesity in Mice. Biochem. Biophys. Res. Commun. 2006, 348, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.; Wu, S.; Ling, L.; Danao, J.; Li, Y.; Yeh, W.-C.; Tian, H.; Baribault, H. G-Protein-Coupled Receptor GPR21 Knockout Mice Display Improved Glucose Tolerance and Increased Insulin Response. Biochem. Biophys. Res. Commun. 2012, 418, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Osborn, O.; Oh, D.Y.; McNelis, J.; Sanchez-Alavez, M.; Talukdar, S.; Lu, M.; Li, P.; Thiede, L.; Morinaga, H.; Kim, J.J.; et al. G Protein-Coupled Receptor 21 Deletion Improves Insulin Sensitivity in Diet-Induced Obese Mice. J. Clin. Investig. 2012, 122, 2444–2453. [Google Scholar] [CrossRef]
- Marazziti, D.; Golini, E.; Mandillo, S.; Magrelli, A.; Witke, W.; Matteoni, R.; Tocchini-Valentini, G.P. Altered Dopamine Signaling and MPTP Resistance in Mice Lacking the Parkinson’s Disease-Associated GPR37/Parkin-Associated Endothelin-like Receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 10189–10194. [Google Scholar] [CrossRef]
- Imai, Y.; Inoue, H.; Kataoka, A.; Hua-Qin, W.; Masuda, M.; Ikeda, T.; Tsukita, K.; Soda, M.; Kodama, T.; Fuwa, T.; et al. Pael Receptor Is Involved in Dopamine Metabolism in the Nigrostriatal System. Neurosci. Res. 2007, 59, 413–425. [Google Scholar] [CrossRef]
- Moechars, D.; Depoortere, I.; Moreaux, B.; de Smet, B.; Goris, I.; Hoskens, L.; Daneels, G.; Kass, S.; Ver Donck, L.; Peeters, T.; et al. Altered Gastrointestinal and Metabolic Function in the GPR39-Obestatin Receptor-Knockout Mouse. Gastroenterology 2006, 131, 1131–1141. [Google Scholar] [CrossRef]
- Le, L.Q.; Kabarowski, J.H.; Weng, Z.; Satterthwaite, A.B.; Harvill, E.T.; Jensen, E.R.; Miller, J.F.; Witte, O.N. Mice Lacking the Orphan G Protein-Coupled Receptor G2A Develop a Late-Onset Autoimmune Syndrome. Immunity 2001, 14, 561–571. [Google Scholar] [CrossRef]
- Venkataraman, C.; Kuo, F. The G-Protein Coupled Receptor, GPR84 Regulates IL-4 Production by T Lymphocytes in Response to CD3 Crosslinking. Immunol. Lett. 2005, 101, 144–153. [Google Scholar] [CrossRef]
- Kelly, L.M.; Pereira, J.P.; Yi, T.; Xu, Y.; Cyster, J.G. EBI2 Guides Serial Movements of Activated B Cells and Ligand Activity Is Detectable in Lymphoid and Nonlymphoid Tissues. J. Immunol. 2011, 187, 3026–3032. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Wojciech, S.; Jockers, R. Hunting for the Function of Orphan GPCRs—Beyond the Search for the Endogenous Ligand. Br. J. Pharmacol. 2015, 172, 3212–3228. [Google Scholar] [CrossRef] [PubMed]
- van Bodegom, M.; Homberg, J.R.; Henckens, M.J.A.G. Modulation of the Hypothalamic-Pituitary-Adrenal Axis by Early Life Stress Exposure. Front. Cell. Neurosci. 2017, 11, 87. [Google Scholar] [CrossRef]
- Liu, B.; Song, S.; Jones, P.M.; Persaud, S.J. GPR55: From Orphan to Metabolic Regulator? Pharmacol. Ther. 2015, 145, 35–42. [Google Scholar] [CrossRef]
- Wong, T.; Gao, W.; Chen, G.; Qiu, C.; He, G.; Ye, F.; Wu, Z.; Zeng, Z.; Du, Y. Cryo-EM Structure of Orphan G Protein-coupled Receptor GPR21. MedComm 2023, 4, e205. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.P.; Kelly, L.M.; Xu, Y.; Cyster, J.G. EBI2 Mediates B Cell Segregation between the Outer and Centre Follicle. Nature 2009, 460, 1122–1126. [Google Scholar] [CrossRef]
- Kaczmarek, I.; Suchý, T.; Prömel, S.; Schöneberg, T.; Liebscher, I.; Thor, D. The Relevance of Adhesion G Protein-Coupled Receptors in Metabolic Functions. Biol. Chem. 2022, 403, 195–209. [Google Scholar] [CrossRef]
- Aljorani, R.H.; Saleh, E.S.; Al Mohammadawi, K.G. Association between CNR1 Gene Polymorphisms and Susceptibility to Diabetic Nephropathy in Iraqi Patients with T2DM. J. Med. Life 2023, 16, 1663–1669. [Google Scholar] [CrossRef]
- Messeha, S.S.; Agarwal, M.; Gendy, S.G.; Mehboob, S.B.; Soliman, K.F.A. The Anti-Obesogenic Effects of Muscadine Grapes through Ciliary Neurotrophic Factor Receptor (Cntfr) and Histamine Receptor H1 (Hrh1) Genes in 3T3-L1 Differentiated Mouse Cells. Nutrients 2024, 16, 1817. [Google Scholar] [CrossRef]
- Strassheim, D.; Sullivan, T.; Irwin, D.C.; Gerasimovskaya, E.; Lahm, T.; Klemm, D.J.; Dempsey, E.C.; Stenmark, K.R.; Karoor, V. Metabolite G-Protein Coupled Receptors in Cardio-Metabolic Diseases. Cells 2021, 10, 3347. [Google Scholar] [CrossRef]
- Lan, H.; Hoos, L.M.; Liu, L.; Tetzloff, G.; Hu, W.; Abbondanzo, S.J.; Vassileva, G.; Gustafson, E.L.; Hedrick, J.A.; Davis, H.R. Lack of FFAR1/GPR40 Does Not Protect Mice from High-Fat Diet-Induced Metabolic Disease. Diabetes 2008, 57, 2999–3006. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, M.B.; Fridriksson, J.; Madsen, L.; Rishi, V.; Vinson, C.; Holmsen, H.; Berge, R.K.; Mandrup, S. Glucose-Induced Lipogenesis in Pancreatic β-Cells Is Dependent on SREBP-1. Mol. Cell. Endocrinol. 2005, 240, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Fan, Y.; Zhang, J.; Lomberk, G.A.; Zhou, Z.; Sun, L.; Mathison, A.J.; Garcia-Barrio, M.T.; Zhang, J.; Zeng, L.; et al. Perhexiline Activates KLF14 and Reduces Atherosclerosis by Modulating ApoA-I Production. J. Clin. Investig. 2015, 125, 3819–3830. [Google Scholar] [CrossRef]
- Akash, M.S.H.; Rasheed, S.; Rehman, K.; Ibrahim, M.; Imran, M.; Assiri, M.A. Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms. Metabolites 2023, 13, 199. [Google Scholar] [CrossRef]
- Yerra, V.G.; Drosatos, K. Specificity Proteins (SP) and Krüppel-like Factors (KLF) in Liver Physiology and Pathology. Int. J. Mol. Sci. 2023, 24, 4682. [Google Scholar] [CrossRef]
- Ferré, P.; Phan, F.; Foufelle, F. SREBP-1c and Lipogenesis in the Liver: An Update. Biochem. J. 2021, 478, 3723–3739. [Google Scholar] [CrossRef]
- Shahvazian, E.; Mahmoudi, M.B.; Farashahi Yazd, E.; Gharibi, S.; Moghimi, B.; HosseinNia, P.; Mirzaei, M. The KLF14 Variant Is Associated with Type 2 Diabetes and HbA1C Level. Biochem. Genet. 2021, 59, 574–588. [Google Scholar] [CrossRef]
- Haghipanah, M.; Safarbalou, A.; Saadat, M.; Rostami Mehr, S. Relationship between SREBF-1 Polymorphism and Insulin Resistance in Polycystic Ovary Syndrome Women. GMJ Med. 2024, 3, 97–101. Available online: http://gmedicine.de/article-2-372-en.html (accessed on 13 December 2022).
- R Foundation. R: The R Project for Statistical Computing. 2022. Available online: https://www.r-project.org/ (accessed on 13 December 2022).
- Rinker, T.W.; Kurkiewicz, D. Pacman: Package Management for R. 2018. Available online: http://github.com/trinker/pacman (accessed on 24 September 2024).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. Available online: https://www.stats.ox.ac.uk/pub/MASS4/ (accessed on 24 September 2024).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models. Biom. J. 2008, 50, 346–363. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Zeileis, A.; Kleiber, C.; Jackman, S. Regression Models for Count Data in R. J. Stat. Softw. 2018, 27, 1–25. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 24 September 2024).
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2023. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 24 September 2024).
- Forbes, S.H. PupillometryR: An R package for preparing and analysing pupillometry data. J. Open Source Softw. 2020, 5, 2285. [Google Scholar]
- Schloerke, B.; Cook, D.; Larmarange, J.; Briatte, F.; Marbach, M.; Edwin Thoen, E.T. GGally: Extension to “ggplot2”. 2024. Available online: https://CRAN.R-project.org/package=GGally (accessed on 24 September 2024).
- Zeileis, A.; Hothorn, T. Diagnostic Checking in Regression Relationships. R News 2002, 2, 7–10. [Google Scholar]
- Williams, G.J. Data Mining with Rattle and R: The Art of Excavating Data for Knowledge Discovery; Use R! Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar] [CrossRef]
- Galili, T. Dendextend: An R Package for Visualizing, Adjusting, and Comparing Trees of Hierarchical Clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Hadley Wickham, H.W.; Henry, L. purrr: Functional Programming Tools. 2023. Available online: https://CRAN.R-project.org/package=purrr (accessed on 24 September 2024).
- Murdoch, D.; Adler, D. rgl: 3D Visualization Using OpenGL. 2024. Available online: https://CRAN.R-project.org/package=rgl (accessed on 24 September 2024).
- Alboukadel Kassambara, A.K.; Mundt, F. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 24 September 2024).
- Kassambara, A. ggpubr: “ggplot2” Based Publication Ready Plots. 2023. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 24 September 2024).
- Brock, G.; Pihur, V.; Datta, S.; Datta, S. clValid: An R Package for Cluster Validation. J. Stat. Softw. 2008, 25, 1–22. [Google Scholar]
- Hahsler, M.; Piekenbrock, M. Density-Based Spatial Clustering of Applications with Noise (DBSCAN) and Related Algorithms. 2024. Available online: https://CRAN.R-project.org/package=dbscan (accessed on 24 September 2024).
- Scrucca, L.; Fraley, C.; Murphy, T.B.; Raftery, A.E. Model-Based Clustering, Classification, and Density Estimation Using Mclust in R; Chapman & Hall/CRC The R Series; CRC Press: Boca Raton, FL, USA, 2023; ISBN 978-1-032-23495-3. Available online: https://mclust-org.github.io/book/ (accessed on 24 September 2024).



| Class A | Class B/Adhesion | Class C | |
|---|---|---|---|
| Total of orphan GPCRs | 85 | 33 | 8 |
| # Binding sites | 966 | 520 | 103 |
| # Binding sites [+] | 501 | 276 | 56 |
| # Binding sites [−] | 465 | 244 | 47 |
| # Viable binding sites | 501 | 276 | 56 |
| # important regions | |||
| Silencer Enhancer | |||
| 2 | 20 | 0 | |
| 64 | 34 | 0 | |
| Binding sites in the distal region [−750 −1000] | |||
| 90 | 49 | 11 | |
| Distal Region [−750 to −1000] Upstream of the Gene Promoters Class A | |||||
|---|---|---|---|---|---|
| Number of Binding Sites | |||||
| 1 | 2 | 3 | 4 | 5–10 | |
| Genes | GPR21 | GPR4 | GPR45 | GPR3 | GPR12 (8 *) |
| GPR22 | GPR19 | GPR78 | GPR17 | GPR37 (5 *) | |
| GPR25 | GPR68 | LGR4 | GPR26 | GPR85 (5 *) | |
| GPR39 | GPR132 | MRGPRD | GPR176 | ||
| GPR50 | GPR139 | MRGPRE | P2RY10 | ||
| GPR83 | GPR182 | MRGPRG | |||
| GPR84 | MAS1 | ||||
| GPR135 | P2RY8 | ||||
| GPR142 | TAAR8 | ||||
| GPR150 | GPR55 | ||||
| GPR153 | |||||
| GPR171 | |||||
| GPR183 | |||||
| MAS1L | |||||
| Class B/Adhesion | |||||
| ADGRD1 | ADGRA1 | ADGRB1 | ADGRB2 | CELSR1 (5 *) | |
| ADGRE1 | CELSR3 | ADGRB3 | ADGRD2 | ADGRE2 (5 *) | |
| ADGRF3 | ADGRG3 | CELSR2 | ADGRG1 | ||
| ADGRF5 | ADGRG6 | ADGRG5 | |||
| ADGRL2 | ADGRL1 | ||||
| Class C | |||||
| GPR56 | GPRC5C (7 *) | ||||
| GPR158 | |||||
| GPR179 | |||||
| GPRC5A | |||||
| GPCR | KLF14 (p = 0.01) | SREBF-1 (p = 0.01) | DR-KLF14 (p = 0.01) | DR-SREBF-1 (p = 0.01) |
|---|---|---|---|---|
| LEPR | 34 | 18 | 5 | 4 |
| ADIPOR1 | 29 | 21 | 3 | 3 |
| ADIPOR2 | 20 | 14 | 3 | 0 |
| GHSR | 33 | 9 | 7 | 4 |
| AGTR1 | 14 | 19 | 2 | 5 |
| CNR1 | 47 | 19 | 10 | 5 |
| H1R/HRH1 | 36 | 12 | 7 | 2 |
| X1R/HCRTR1 | 37 | 22 | 10 | 9 |
| X2R/HCRTR2 | 14 | 11 | 4 | 4 |
| FFAR3/GPR41 | 14 | 23 | 1 | 6 |
| S1PR1 | 33 | 22 | 5 | 7 |
| P2Y6/P2RY6 | 15 | 23 | 5 | 4 |
| Transcription Factor KLF14 | |||||
|---|---|---|---|---|---|
| GPCRs-MetS | #BS | #BS+ | #BS− | #IR+ | #BS-DR |
| LEPR | 50 | 31 | 19 | 30 | 1 |
| ADIPOR1 | 6 | 5 | 1 | 0 | 1 |
| ADIPOR2 | 1 | 0 | 1 | 0 | 0 |
| MC4R | 0 | 0 | 0 | 0 | 0 |
| GHSR | 17 | 4 | 13 | 0 | 1 |
| AGTR1 | 2 | 1 | 1 | 0 | 0 |
| 5-HT2C | 12 | 4 | 8 | 0 | 1 |
| CB1/CNR1 | 9 | 2 | 7 | 0 | 1 |
| MC3R | 4 | 1 | 3 | 0 | 0 |
| H1R/HRH1 | 3 | 1 | 2 | 0 | 0 |
| OX1R/HCRTR1 | 27 | 12 | 15 | 6 | 8 |
| OX2R/HCRHTR2 | 8 | 2 | 6 | 0 | 0 |
| FFAR3/GPR41 | 5 | 2 | 3 | 0 | 0 |
| S1PR1 | 32 | 23 | 9 | 0 | 11 |
| S1PR2 | 5 | 3 | 2 | 0 | 0 |
| FFAR1/GPR40 | 31 | 8 | 23 | 0 | 0 |
| HCAR2/GPR109A | 0 | 0 | 0 | 0 | 0 |
| P2Y6 | 14 | 6 | 8 | 0 | 3 |
| Transcription Factor SREBF-1 | |||||
| LEPR | 1 | 1 | 0 | 0 | 1 |
| ADIPOR1 | 1 | 0 | 1 | 0 | 0 |
| ADIPOR2 | 3 | 2 | 1 | 0 | 0 |
| MC4R | 0 | 0 | 0 | 0 | 0 |
| GHSR | 4 | 3 | 1 | 0 | 0 |
| AGTR1 | 5 | 4 | 1 | 0 | 1 |
| 5-HT2C | 4 | 3 | 1 | 0 | 0 |
| CB1/CNR1 | 10 | 6 | 4 | 0 | 1 |
| MC3R | 3 | 1 | 2 | 0 | 0 |
| H1R/HRH1 | 2 | 2 | 0 | 0 | 0 |
| OX1R/HCRTR1 | 3 | 1 | 2 | 0 | 0 |
| OX2R/HCRHTR2 | 7 | 4 | 3 | 0 | 0 |
| FFAR3/GPR41 | 5 | 4 | 1 | 0 | 0 |
| S1PR1 | 8 | 2 | 6 | 0 | 1 |
| S1PR2 | 8 | 4 | 4 | 0 | 0 |
| FFAR1/GPR40 | 15 | 9 | 6 | 0 | 4 |
| HCAR2/GPR109A | 7 | 6 | 1 | 0 | 3 |
| P2Y6 | 6 | 3 | 3 | 0 | 2 |
| Model (Neg. Binomial) | Coefficient (Estimate) | CI (95%) | Pr (>|z|) |
|---|---|---|---|
| (Intercept) | −0.80158 | (−1.1724, −0.4455) | <0.001 |
| The number of BS in the positive strand | 0.10986 | (0.0776, 0.1443) | <0.001 |
| Dispersion parameter (θ) | 1.274 | ||
| Null deviance: 188.11 | |||
| Residual deviance: 136.71 | |||
| AIC: 397.39 | |||
| (Intercept) | −8.872 | (−16.8515, −3.1697) | 0.01247 * |
| klf14 similarity | 10.831 | (4.1175, 20.2363) | 0.00951 ** |
| Dispersion parameter (θ) | 0.9107 | ||
| Null deviance: 164.37 | |||
| Residual deviance: 126.29 | |||
| AIC: 404.48 | |||
| (Intercept) | −4.10798 | (−8.8290, −1.9167) | 0.00946 ** |
| The number of BS in the positive strand | 0.08269 | (0.0500, 0.1169) | <0.001 |
| klf14 similarity | 4.28924 | (1.6102, 9.9563) | 0.02437 * |
| Dispersion parameter (θ) | 1.4771 | ||
| Null deviance: 198.51 | |||
| Residual deviance: 128.27 | |||
| AIC: 384.08 |
| Model (Poisson) | Coefficient (Estimate) | CI (95%) | Pr (>|z|) |
|---|---|---|---|
| (Intercept) | −1.4903 | (−1.8965, −1.1109) | <0.001 |
| The number of srebf1 BS in the positive strand | 0.3513 | (0.2620, 0.4381) | <0.001 |
| Null deviance: 196.92 | |||
| Residual deviance: 143.36 | |||
| AIC: 280.15 | |||
| (Intercept) | −4.245 | (−8.2293, −2.1601) | 0.00458 ** |
| srebf1 similarity | 4.738 | (2.2484, 9.4511) | 0.00771 ** |
| Null deviance: 196.92 | |||
| Residual deviance: 164.94 | |||
| AIC: 301.74 | |||
| (Intercept) | −4.5524 | (−9.5663, −2.2291) | 0.0102 * |
| The number of srebf1 BS in the positive strand | 0.2995 | (0.2011, 0.3939) | <0.001 |
| srebf1 similarity | 3.9496 | (1.1221, 9.8782) | 0.0618 · |
| Null deviance: 196.92 | |||
| Residual deviance: 132.60 | |||
| AIC: 271.39 | |||
| Variable | Coefficient | CI (95%) | p-Value |
|---|---|---|---|
| Intercept | −4.0266 | (−8.7006, −1.8430) | 0.0106 * |
| The number of klf14 BS in the positive strand | 0.0744 | (0.0389, 0.1119) | <0.001 |
| Group GPCR-MetS | −0.50047 | (−1.7248, 0.5975) | 0.3579 |
| Klf14 similarity | 4.28399 | (1.6204, 9.8874) | 0.0235 * |
| The number of klf14 BS in the positive strand for group GPCR-MetS | −0.04647 | (−0.0367, 0.1439) | 0.191019 |
| Variable | Coefficient | CI (95%) | p-Value |
|---|---|---|---|
| Intercept | 0.02943 | (−0.3070, 0.3658) | 0.864 |
| The number of srebf1 BS in the positive strand | 0.28852 | (0.1866, 0.3903) | <0.001 *** |
| Group orphan GPCR-MetS | −0.40235 | (−1.0964, 0.2917) | 0.258 |
| srebf1 similarity | −0.03833 | (−0.5684, 0.4918) | 0.888 |
| The number of srebf1 BS in the positive strand for group orphan GPCR-MetS | 0.07942 | (−0.1134, 0.2722) | 0.421 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Coste, J.J.; Villafaña-Rauda, S.; Aguayo-Cerón, K.A.; Vargas-De-León, C.; Romero-Nava, R. KLF14 and SREBF-1 Binding Site Associations with Orphan Receptor Promoters in Metabolic Syndrome. Int. J. Mol. Sci. 2025, 26, 2849. https://doi.org/10.3390/ijms26072849
Garcia-Coste JJ, Villafaña-Rauda S, Aguayo-Cerón KA, Vargas-De-León C, Romero-Nava R. KLF14 and SREBF-1 Binding Site Associations with Orphan Receptor Promoters in Metabolic Syndrome. International Journal of Molecular Sciences. 2025; 26(7):2849. https://doi.org/10.3390/ijms26072849
Chicago/Turabian StyleGarcia-Coste, Julio Jesús, Santiago Villafaña-Rauda, Karla Aidee Aguayo-Cerón, Cruz Vargas-De-León, and Rodrigo Romero-Nava. 2025. "KLF14 and SREBF-1 Binding Site Associations with Orphan Receptor Promoters in Metabolic Syndrome" International Journal of Molecular Sciences 26, no. 7: 2849. https://doi.org/10.3390/ijms26072849
APA StyleGarcia-Coste, J. J., Villafaña-Rauda, S., Aguayo-Cerón, K. A., Vargas-De-León, C., & Romero-Nava, R. (2025). KLF14 and SREBF-1 Binding Site Associations with Orphan Receptor Promoters in Metabolic Syndrome. International Journal of Molecular Sciences, 26(7), 2849. https://doi.org/10.3390/ijms26072849







