MicroRNA Profiling Identifies Age-Associated MicroRNAs and Potential Biomarkers for Early Diagnosis of Autism
Abstract
:1. Introduction
2. Results
2.1. Study Population Characteristics
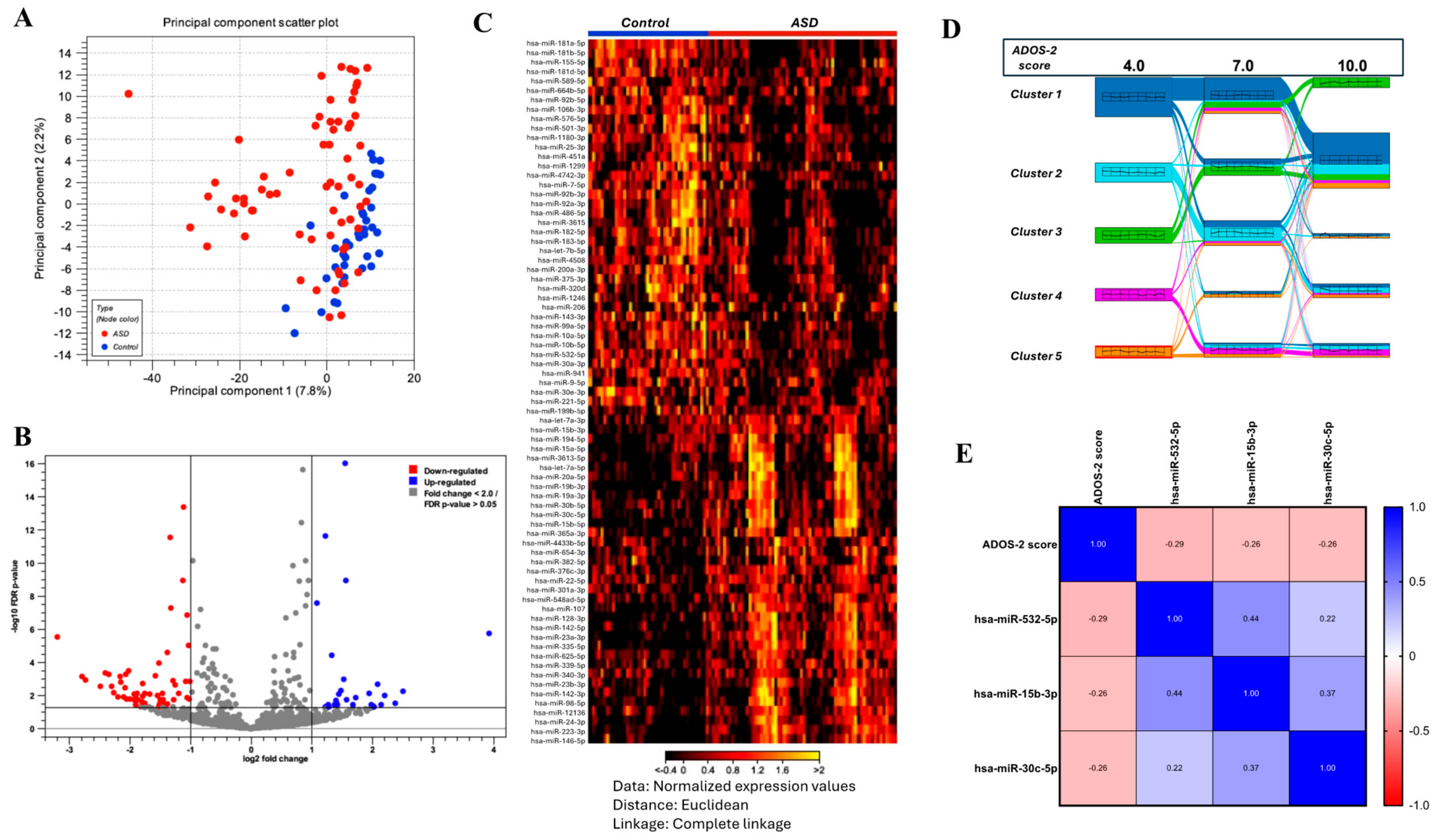
2.2. Differential Expression of Plasma miRNAs in Children with ASD
2.3. Correlation of Several miRNAs with the Clinical Severity of ASD
2.4. Correlation of Circulating miRNAs with the Age of Children with ASD
2.5. Validation of the 17-miRNA Signature in Plasma Samples of Children with ASD
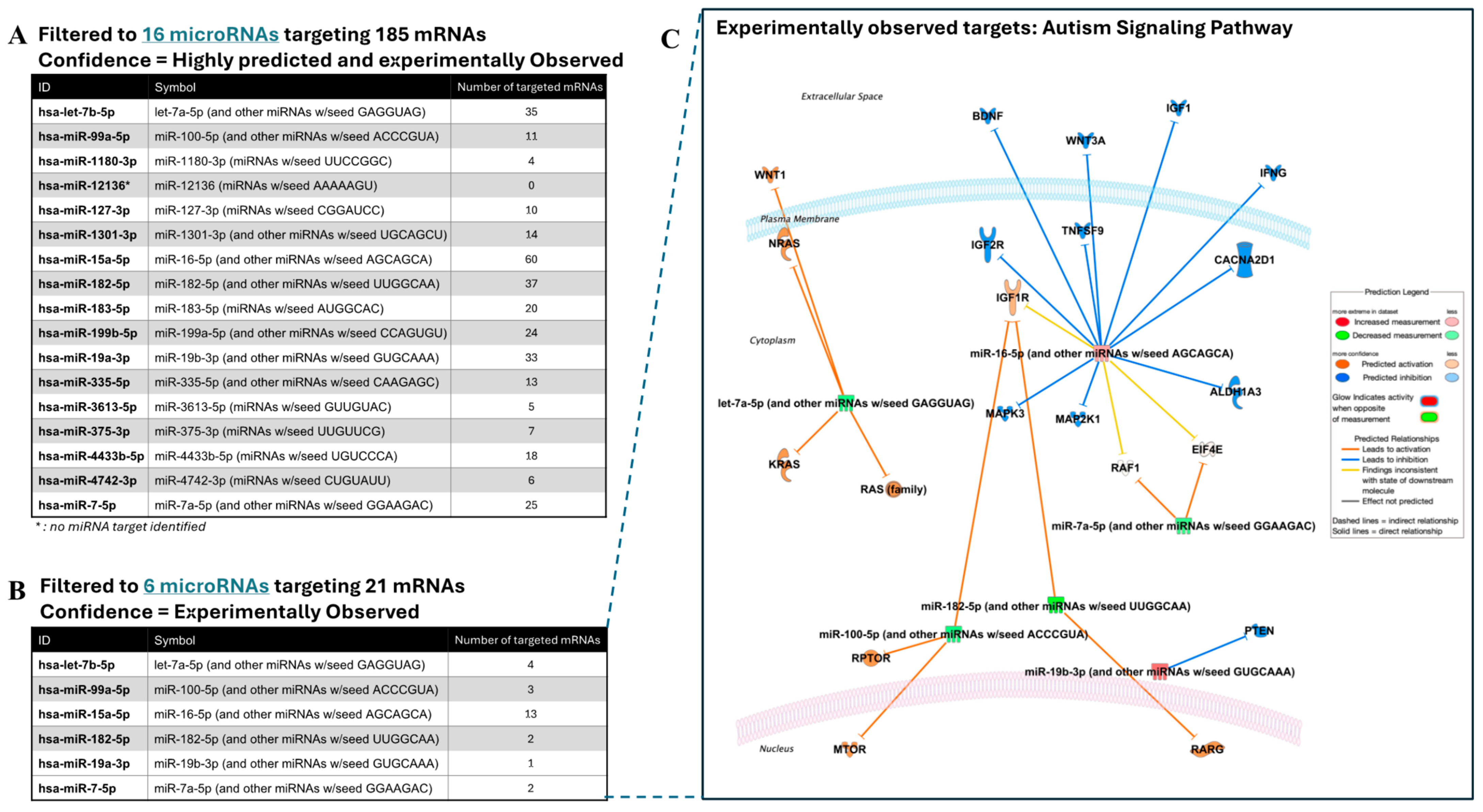
2.6. Dysregulated Expression of the Target Genes of 17-miRNA Signature in the Blood and Brain Tissues in ASD
2.7. Molecular Functions of the 17-miRNA Signature in ASD
2.8. Logistic Regression Model Revealed Predictor Markers for ASD Diagnosis
2.9. Diagnostic Accuracy of the Four-miRNA Panel in ASD Detection
3. Discussion
4. Methods
4.1. Study Participants
4.2. Plasma Isolation and Processing
4.3. Total RNA Extraction from Plasma
4.4. Quality Check of the Extracted RNA
4.5. Small RNA Library Preparation
4.6. High-Throughput Sequencing of Small RNAs
4.7. Read Mapping and Annotation of Small RNA Sequences
4.8. Quality Check of Sequencing Data and Abundance of miRNAs Within the Sample Cohort
4.9. Bioinformatics Data Analysis
4.10. Differential Expression Analysis
4.11. Advanced miRNA Pathway and Network Analysis
4.12. BaseSpace Correlation Engine
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lord, C.; Brugha, T.S.; Charman, T.; Cusack, J.; Dumas, G.; Frazier, T.; Jones, E.J.H.; Jones, R.M.; Pickles, A.; State, M.W.; et al. Autism spectrum disorder. Nat. Rev. Dis. Prim. 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Zeidan, J.; Fombonne, E.; Scorah, J.; Ibrahim, A.; Durkin, M.S.; Saxena, S.; Yusuf, A.; Shih, A.; Elsabbagh, M. Global prevalence of autism: A systematic review update. Autism Res. 2022, 15, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Fusar-Poli, L.; Brondino, N.; Politi, P.; Aguglia, E. Missed diagnoses and misdiagnoses of adults with autism spectrum disorder. Eur. Arch. Psychiatry Clin. Neurosci. 2020, 272, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Becerra, M.A. Closing the Diagnostic Gap: Early Autism Spectrum Disorder Screening for Every Child. Health Soc. Work 2022, 47, 87–91. [Google Scholar] [CrossRef]
- Salloum-Asfar, S.; Elsayed, A.K.; Abdulla, S.A. Chapter 6—Potential approaches and recent advances in biomarker discovery in autism spectrum disorders. In Neural Engineering Techniques for Autism Spectrum Disorder; El-Baz, A.S., Suri, J.S., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 121–145. [Google Scholar] [CrossRef]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531, Correction in Nat. Rev. Genet. 2004, 5, 631. [Google Scholar] [CrossRef]
- Kosik, K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006, 7, 911–920. [Google Scholar] [CrossRef]
- Kyzar, E.J.; Bohnsack, J.P.; Pandey, S.C. Current and Future Perspectives of Noncoding RNAs in Brain Function and Neuropsychiatric Disease. Biol. Psychiatry 2021, 91, 183–193. [Google Scholar] [CrossRef]
- Martins, H.C.; Schratt, G. MicroRNA-dependent control of neuroplasticity in affective disorders. Transl. Psychiatry 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Li, J.; Xu, X.; Liu, J.; Zhang, S.; Tan, X.; Li, Z.; Zhang, J.; Wang, Z. Decoding microRNAs in autism spectrum disorder. Mol. Ther. Nucleic Acids 2022, 30, 535–546. [Google Scholar] [CrossRef]
- Garrido-Torres, N.; Guzmán-Torres, K.; García-Cerro, S.; Bermúdez, G.P.; Cruz-Baquero, C.; Ochoa, H.; García-González, D.; Canal-Rivero, M.; Crespo-Facorro, B.; Ruiz-Veguilla, M. miRNAs as biomarkers of autism spectrum disorder: A systematic review and meta-analysis. Eur. Child Adolesc. Psychiatry 2023, 33, 2957–2990. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.-F.; Wu, N.; Wang, L.; Li, J. Circulating MicroRNAs: A Novel Class of Potential Biomarkers for Diagnosing and Prognosing Central Nervous System Diseases. Cell. Mol. Neurobiol. 2013, 33, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2015, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.D.; Ignacio, C.; Gentile, K.; Middleton, F.A. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016, 16, 52. [Google Scholar] [CrossRef] [PubMed]
- Vasu, M.M.; Anitha, A.; Thanseem, I.; Suzuki, K.; Yamada, K.; Takahashi, T.; Wakuda, T.; Iwata, K.; Tsujii, M.; Sugiyama, T.; et al. Serum microRNA profiles in children with autism. Mol. Autism 2014, 5, 40. [Google Scholar] [CrossRef]
- Wu, Y.E.; Parikshak, N.N.; Belgard, T.G.; Geschwind, D.H. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat. Neurosci. 2016, 19, 1463–1476. [Google Scholar] [CrossRef]
- Salloum-Asfar, S.; Elsayed, A.K.; Elhag, S.F.; Abdulla, S.A. Circulating Non-Coding RNAs as a Signature of Autism Spectrum Disorder Symptomatology. Int. J. Mol. Sci. 2021, 22, 6549. [Google Scholar] [CrossRef]
- Kurz, A.; Kumar, R.; Northoff, B.H.; Wenk, C.; Schirra, J.; Donakonda, S.; Höglinger, G.U.; Schwarz, J.; Rozanski, V.; Hübner, R. Differential expression of gut miRNAs in idiopathic Parkinson's disease. Park. Relat. Disord. 2021, 88, 46–50. [Google Scholar] [CrossRef]
- Alter, M.D.; Kharkar, R.; Ramsey, K.E.; Craig, D.W.; Melmed, R.D.; Grebe, T.A.; Bay, R.C.; Ober-Reynolds, S.; Kirwan, J.; Jones, J.J.; et al. Autism and Increased Paternal Age Related Changes in Global Levels of Gene Expression Regulation. PLoS ONE 2011, 6, e16715. [Google Scholar] [CrossRef]
- Pramparo, T.; Lombardo, M.V.; Campbell, K.; Barnes, C.C.; Marinero, S.; Solso, S.; Young, J.; Mayo, M.; Dale, A.; Ahrens-Barbeau, C.; et al. Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers. Mol. Syst. Biol. 2015, 11, 841. [Google Scholar] [CrossRef]
- Ginsberg, M.R.; Rubin, R.A.; Natowicz, M.R. Patterning of Regional Gene Expression in Autism: New Complexity. Sci. Rep. 2013, 3, srep01831. [Google Scholar] [CrossRef] [PubMed]
- Gregg, J.P.; Lit, L.; Baron, C.A.; Hertz-Picciotto, I.; Walker, W.; Davis, R.A.; Croen, L.A.; Ozonoff, S.; Hansen, R.; Pessah, I.N.; et al. Gene expression changes in children with autism. Genomics 2007, 91, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Pramparo, T.; Pierce, K.; Lombardo, M.V.; Barnes, C.C.; Marinero, S.; Ahrens-Barbeau, C.; Murray, S.S.; Lopez, L.; Xu, R.; Courchesne, E. Prediction of Autism by Translation and Immune/Inflammation Coexpressed Genes in Toddlers From Pediatric Community Practices. JAMA Psychiatry 2015, 72, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Kaya, N.; Colak, D.; Albakheet, A.; Al-Owain, M.; Abu-Dheim, N.; Al-Younes, B.; Al-Zahrani, J.; Mukaddes, N.M.; Dervent, A.; Al-Dosari, N.; et al. A novel X-linked disorder with developmental delay and autistic features. Ann. Neurol. 2011, 71, 498–508. [Google Scholar] [CrossRef]
- Chow, M.L.; Li, H.-R.; Winn, M.E.; April, C.; Barnes, C.C.; Wynshaw-Boris, A.; Fan, J.-B.; Fu, X.-D.; Courchesne, E.; Schork, N.J. Genome-wide expression assay comparison across frozen and fixed postmortem brain tissue samples. BMC Genom. 2011, 12, 449. [Google Scholar] [CrossRef]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef]
- Ginsberg, M.R.; Rubin, R.A.; Falcone, T.; Ting, A.H.; Natowicz, M.R. Brain Transcriptional and Epigenetic Associations with Autism. PLoS ONE 2012, 7, e44736. [Google Scholar] [CrossRef]
- Liu, X.; Campanac, E.; Cheung, H.-H.; Ziats, M.N.; Canterel-Thouennon, L.; Raygada, M.; Baxendale, V.; Pang, A.L.-Y.; Yang, L.; Swedo, S.; et al. Idiopathic Autism: Cellular and Molecular Phenotypes in Pluripotent Stem Cell-Derived Neurons. Mol. Neurobiol. 2016, 54, 4507–4523. [Google Scholar] [CrossRef]
- Qu, S.; Qiu, O.; Huang, J.; Liu, J.; Wang, H. Upregulation of hsa-miR-196a-5p is associated with MIR196A2 methylation and affects the malignant biological behaviors of glioma. Genomics 2021, 113, 1001–1010. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Bauer, A.; ElSharawy, A.; Haas, J.; Backes, C.; Wendschlag, A.; Giese, N.; Tjaden, C.; Ott, K.; et al. Toward the blood-borne miRNome of human diseases. Nat. Methods 2011, 8, 841–843. [Google Scholar] [CrossRef]
- Nour-Eldine, W.; Manaph, N.P.A.; Ltaief, S.M.; Aati, N.A.; Mansoori, M.H.; Al Abdulla, S.; Al-Shammari, A.R. Discovery of a novel cytokine signature for the diagnosis of autism spectrum disorder in young Arab children in Qatar. Front. Psychiatry 2024, 15, 1333534. [Google Scholar] [CrossRef] [PubMed]
- Ltaief, S.M.; Nour-Eldine, W.; Manaph, N.P.A.; Tan, T.; Anuar, N.D.; Bensmail, I.; George, J.; Abdesselem, H.B.; Al-Shammari, A.R. Dysregulated plasma autoantibodies are associated with B cell dysfunction in young Arab children with autism spectrum disorder in Qatar. Autism Res. 2024, 17, 1974–1993. [Google Scholar] [CrossRef] [PubMed]
- Salloum-Asfar, S.; Satheesh, N.J.; Abdulla, S.A. Circulating miRNAs, Small but Promising Biomarkers for Autism Spectrum Disorder. Front. Mol. Neurosci. 2019, 12, 253. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kupershmidt, I.; Su, Q.J.; Grewal, A.; Sundaresh, S.; Halperin, I.; Flynn, J.; Shekar, M.; Wang, H.; Park, J.; Cui, W.; et al. Ontology-Based Meta-Analysis of Global Collections of High-Throughput Public Data. PLoS ONE 2010, 5, e13066. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]

| Total (n = 108) | Control (n = 42) | ASD (n = 66) | p-Value | |
|---|---|---|---|---|
| Sex | ||||
| Male Female | 87 (80.6) 21 (19.4) | 33 (78.6) 9 (21.4) | 54 (81.8) 12 (18.2) | 0.678 |
| Age in years | 3.44 (2.99–3.75) | 3.37 (2.93–3.81) | 3.46 (3.11–3.75) | 0.504 |
| Family history of ASD | ||||
| Yes No | – – | NA NA | 13 (19.7) 53 (80.3) | – |
| Consanguinity | ||||
| Yes No | 33 (30.6) 75 (69.4) | 15 (35.7) 27 (64.3) | 18 (27.3) 48 (72.7) | 0.353 |
| Method of reproduction | ||||
| Natural Assisted (IVF) | 103 (95.4) 5 (4.6) | 41 (97.6) 1 (2.4) | 62 (93.9) 4 (6.1) | 0.647 |
| Maternal complications | ||||
| Yes Diabetes Asthma Allergy Hypertension | 40 (37) 23 (21.3) 2 (1.9) 3 (2.8) 3 (2.8) | 15 (35.7) 11 (26.2) 1 (2.4) 0 (0) 0 (0) | 25(37.9) 12 (18.2) 1 (1.5) 3 (4.5) 3 (4.5) | 0.820 |
| Multiple conditions (asthma, allergy, diabetes, and hypertension) | 9 (8.3) | 3 (7.1) | 6 (9.1) | |
| No | 68 (63) | 27 (64.3) | 41 (62.1) | |
| Pregnancy duration | ||||
| <9 months ≥9 months | 6 (5.6) 102 (94.4) | 1 (2.4) 41 (97.6) | 5 (7.6) 61 (92.4) | 0.401 |
| Maternal age at labor | 0.570 | |||
| Age in years <35 years ≥35 years | 29 (26.00–34.00) 87 (80.6) 21 (19.4) | 30.00 (27.00–33.00) 36 (85.7) 6 (14.3) | 29.00 (26.00–34.00) 51 (77.3) 15 (22.7) | 0.280 |
| Type of delivery | ||||
| Normal C-section Induced | 65 (60.2) 39 (36.1) 4 (3.7) | 27 (64.3) 15 (35.7) 0 (0) | 38 (57.6) 24 (36.74) 4 (6.1) | 0.254 |
| Postnatal complications | ||||
| Yes Hypoxia Jaundice Hypoxia and Jaundice Others No | 11 (10.2) 4 (3.7) 4 (3.7) 1 (0.9) 2 (1.9) 97 (89.8) | 2 (4.8) 1 (2.4) 0 (0) 0 (0) 1 (2.4) 40 (95.2) | 9 (13.6) 3 (4.5) 4 (6.1) 1 (1.5) 1 (1.5) 57 (86.4) | 0.094 |
| Birth weight | ||||
| Weight in kg | 3.01 (2.80–3.50) | 3.00 (3.00–3.52) | 3.03 (2.57–3.50) | |
| <2.5 kg ≥2.5 kg | 12 (11.1) 96 (88.9) | 2 (4.8) 40 (95.2) | 10 (15.2) 56 (84.8) | 0.262 0.094 |
| Nationality | ||||
| Egyptian Qatari Syrian Yemeni Sudanese Jordanian Others | 26 (24.1) 21 (19.4) 19 (17.6) 16 (14.8) 13(12.0) 2 (1.9) 11 (10.2) | 12 (28.6) 5 (11.9) 11 (26.2) 4 (9.5) 7 (16.7) 1 (2.4) 2 (4.8) | 14 (21.2) 16 (24.2) 8 (12.1) 12 (18.2) 6 (9.1) 1 (1.5) 9 (13.6) | 0.121 |
| Cohort age 2–4 years | |||
| Group | miRNA | r | p-Value |
| ASD | hsa-miR-182-5p | 0.254 | 0.040 |
| hsa-miR-183-5p | 0.246 | 0.046 | |
| hsa-miR-335-5p | −0.250 | 0.043 | |
| hsa-let-7b-5p | 0.244 | 0.048 | |
| hsa-miR-92b-3p | 0.270 | 0.028 | |
| hsa-miR-486-5p | 0.243 | 0.049 | |
| hsa-miR-146a-5p | −0.245 | 0.047 | |
| hsa-miR-532-5p | 0.256 | 0.038 | |
| hsa-miR-10b-5p | 0.318 | 0.009 | |
| Control | hsa-miR-92b-3p | −0.352 | 0.022 |
| hsa-miR-365a-3p | 0.476 | 0.025 | |
| hsa-miR-9-5p | 0.413 | 0.009 | |
| Cohort age 5–12 years | |||
| Group | miRNA | r | p-value |
| ASD | hsa-miR-182-5p | −0.355 | 0.005 |
| hsa-miR-183-5p | −0.341 | 0.008 | |
| hsa-miR-335-5p | 0.311 | 0.016 | |
| hsa-let-7b-5p | −0.413 | 0.001 | |
| hsa-miR-7-5p | −0.331 | 0.010 | |
| hsa-miR-4742-3p | −0.367 | 0.007 | |
| hsa-miR-141-3p | −0.369 | 0.004 | |
| hsa-miR-196a-5p | −0.332 | 0.012 | |
| hsa-miR-148b-5p | −0.412 | 0.003 | |
| hsa-miR-328-3p | 0.329 | 0.010 | |
| hsa-miR-224-5p | 0.260 | 0.045 | |
| hsa-miR-744-5p | 0.276 | 0.033 | |
| hsa-miR-221-3p | 0.396 | 0.002 | |
| Control | hsa-miR-182-5p | −0.857 | 0.011 |
| Gene Name | Fold Change | Adjusted p-Value | Role in Disease | Implications in ASD | Reference |
|---|---|---|---|---|---|
| miR-335-5p | 2.945 | 1.993 × 10−17 | Brain development, neural differentiation, migration; involved in cancer metastasis. | Contributes to abnormal brain development in ASD. | [19] |
| miR-3613-5p | 2.962 | 1.194 × 10−9 | Involved in gene regulation; less studied. | Role in ASD unclear. | [20] |
| miR-182-5p | −2.171 | 1.194 × 10−9 | Sensory processing, neural development; implicated in cancer. | Relevant to sensory processing abnormalities in ASD. | [12] |
| miR-19a-3p | 2.121 | 2.631 × 10−8 | Part of miR-17-92 cluster; involved in cancer and immune regulation, cell proliferation, apoptosis. | Relevant to immune regulation and apoptosis in ASD. | [19] |
| miR-1180-3p | −2.508 | 5.482 × 10−8 | Linked to cancer; involved in cell cycle regulation, apoptosis. | Relevant to cell cycle regulation and apoptosis in ASD. | [21] |
| miR-375-3p | −2.041 | 9.618 × 10−6 | Pancreatic function, insulin secretion; implicated in various cancers. | Role in ASD unclear. | [8] |
| miR-15a-5p | 1.631 | 2.703 × 10−5 | Apoptosis, cell cycle regulation (cancer, cardiovascular diseases). | Relevant to neuronal death and abnormal cell cycles in ASD. | [20] |
| miR-7-5p | −1.560 | 1.247 × 10−4 | Neuroprotection, synaptic plasticity; implicated in Parkinson’s disease, insulin signaling. | Influences synaptic and neurodevelopmental abnormalities in ASD. | [22] |
| let-7b-5p | −1.620 | 2.439 × 10−4 | Regulates cell differentiation, maintains stem cell populations; involved in cancers and development. | Regulates developmental timing, cellular differentiation; potentially contributing to ASD. | [14] |
| miR-183-5p | −1.717 | 5.641 × 10−4 | Sensory organ development, neuronal function, plasticity; implicated in various cancers. | Important for sensory processing and neural connectivity in ASD. | [10] |
| miR-4742-3p | −2.111 | 1.487 × 10−3 | Less characterized; possibly involved in gene regulation. | Role in ASD unclear. | [13] |
| miR-12136 | 1.850 | 1.863 × 10−3 | Linked to gene expression regulation; less studied. | Role in ASD unclear. | [21] |
| miR-199b-5p | −2.376 | 2.457 × 10−3 | Cardiac and cancer biology; regulates HIF-1α, mTOR pathways; involved in neural plasticity. | Implicated in neurodevelopmental abnormalities. | [22] |
| miR-99a-5p | −1.635 | 3.233 × 10−3 | Involved in cancers (prostate, breast); regulates cell proliferation and apoptosis. | May regulate immune responses and cell differentiation. | [8] |
| miR-1301-3p | 1.502 | 2.118 × 10−2 | Implicated in cancer, regulates cell proliferation and migration. | Could be linked to neurodevelopmental processes disrupted in ASD. | [10] |
| miR-4433b-5p | 1.516 | 2.423 × 10−2 | Gene expression regulation; less studied in neurodevelopment. | Potential role in gene expression modulation in the brain. | [11] |
| miR-127-3p | −1.508 | 2.428 × 10−2 | Regulates cell proliferation and differentiation, studied in cancer and liver fibrosis. | Potential regulatory role in neurodevelopment. | [12] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salloum-Asfar, S.; Ltaief, S.M.; Taha, R.Z.; Nour-Eldine, W.; Abdulla, S.A.; Al-Shammari, A.R. MicroRNA Profiling Identifies Age-Associated MicroRNAs and Potential Biomarkers for Early Diagnosis of Autism. Int. J. Mol. Sci. 2025, 26, 2044. https://doi.org/10.3390/ijms26052044
Salloum-Asfar S, Ltaief SM, Taha RZ, Nour-Eldine W, Abdulla SA, Al-Shammari AR. MicroRNA Profiling Identifies Age-Associated MicroRNAs and Potential Biomarkers for Early Diagnosis of Autism. International Journal of Molecular Sciences. 2025; 26(5):2044. https://doi.org/10.3390/ijms26052044
Chicago/Turabian StyleSalloum-Asfar, Salam, Samia M. Ltaief, Rowaida Z. Taha, Wared Nour-Eldine, Sara A. Abdulla, and Abeer R. Al-Shammari. 2025. "MicroRNA Profiling Identifies Age-Associated MicroRNAs and Potential Biomarkers for Early Diagnosis of Autism" International Journal of Molecular Sciences 26, no. 5: 2044. https://doi.org/10.3390/ijms26052044
APA StyleSalloum-Asfar, S., Ltaief, S. M., Taha, R. Z., Nour-Eldine, W., Abdulla, S. A., & Al-Shammari, A. R. (2025). MicroRNA Profiling Identifies Age-Associated MicroRNAs and Potential Biomarkers for Early Diagnosis of Autism. International Journal of Molecular Sciences, 26(5), 2044. https://doi.org/10.3390/ijms26052044







