The Roles of Circadian Clock Genes in Plant Temperature Stress Responses
Abstract
1. Introduction
2. The Plant Circadian Clock
2.1. Circadian Feedback Loops in Plants and Environmental-Stress-Related Clock Genes
2.2. Clock Genes Related to Temperature Compensation Mechanisms
3. Circadian Clock Gene Responses under Cold Stress
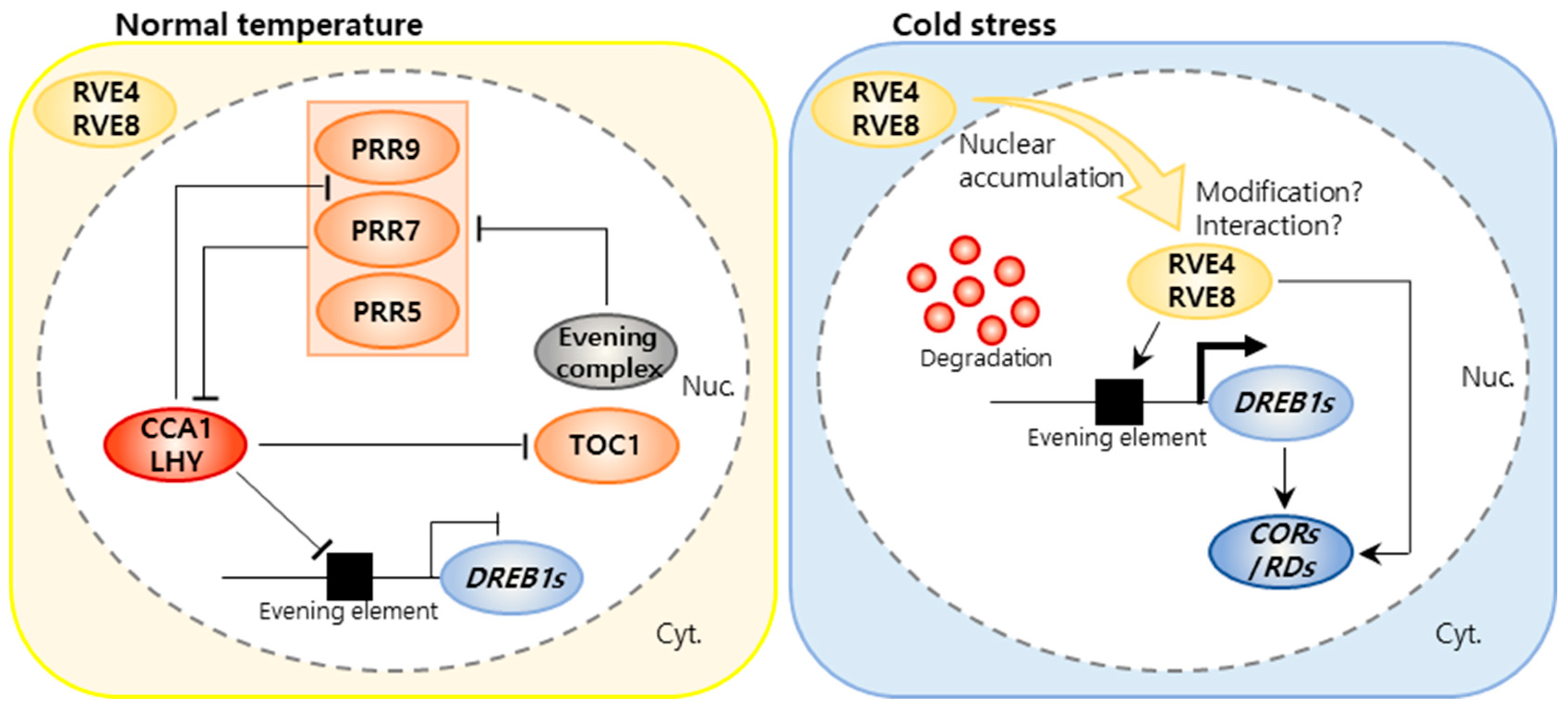
3.1. Cold Stress Response in Plants
3.2. Cold Signaling Pathways Involving Clock-Related Transcription Factors
3.3. Clock Genes Involved in Detecting Temperature Decreases
4. Circadian Clock Genes Associated with Heat Stress Responses
4.1. Responsive Transcriptomes Revealing a Diurnal Response to Heat Stress in Plants
4.2. Clock Genes Involved in Thermomorphogenesis
4.3. Circadian Clock Genes Involved in Heat Resistance
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Creux, N.; Harmer, S. Circadian rhythms in plants. Cold Spring Harb. Perspect. Biol. 2019, 11, a034611. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Shamim, M.; Kumar, M.; Mishra, A.; Maurya, R.; Sharma, D.; Pandey, P.; Singh, K.N. Role of circadian rhythm in plant system: An update from development to stress response. Environ. Exp. Bot. 2019, 162, 256–271. [Google Scholar] [CrossRef]
- Venkat, A.; Muneer, S. Role of circadian rhythms in major plant metabolic and signaling pathways. Front. Plant Sci. 2022, 13, 836244. [Google Scholar] [CrossRef] [PubMed]
- Oravec, M.W.; Greenham, K. The adaptive nature of the plant circadian clock in natural environments. Plant Physiol. 2022, 190, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Kim, H.-S.; Choi, S.-H.; Jang, J.-Y.; Jeong, M.-J.; Lee, S.I. The Importance of the Circadian Clock in Regulating Plant Metabolism. Int. J. Mol. Sci. 2017, 18, 2680. [Google Scholar] [CrossRef]
- de Barros Dantas, L.L.; Eldridge, B.M.; Dorling, J.; Dekeya, R.; Lynch, D.A.; Dodd, A.N. Circadian regulation of metabolism across photosynthetic organisms. Plant J. 2023, 116, 650–668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ma, Y.; Zhang, H.; Xu, J.; Gao, X.; Zhang, T.; Liu, X.; Guo, L.; Zhao, D. Environmental F actors coordinate circadian clock function and rhythm to regulate plant development. Plant Signal Behav. 2023, 18, e2231202. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Temperature Entrainment of the Arabidopsis Circadian Clock. Plant Cell 2005, 17, 645–647. [Google Scholar] [CrossRef]
- Park, M.-J.; Seo, P.J.; Park, C.-M. CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signal Behav. 2012, 7, 1194–1196. [Google Scholar] [CrossRef]
- Gil, K.E.; Kim, W.Y.; Lee, H.J.; Faisal, M.; Saquib, Q.; Alatar, A.A.; Park, C.M. ZEITLUPE contributes to a thermoresponsive protein quality control system in Arabidopsis. Plant Cell 2017, 29, 2882–2894. [Google Scholar] [CrossRef]
- Shalit-kaneh, A.; Kumimoto, R.W.; Filkov, V.; Harmer, S.L. Multiple feedback loops of the Arabidopsis circadian clock provide rhythmic robustness across environmental conditions. Proc. Natl. Acad. Sci. USA 2018, 115, 7147–7152. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.E.; Park, C.M. Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef]
- Fornara, F.; de Montaigu, A.; Sánchez-Villarreal, A.; Takahashi, Y.; Ver Loren van Themaat, E.; Huettel, B.; Davis, S.J.; Coupland, G. The GI–CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 2015, 81, 695–706. [Google Scholar] [CrossRef]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef]
- Keily, J.; MacGregor, D.R.; Smith, R.W.; Millar, A.J.; Halliday, K.J.; Penfield, S. Model selection reveals control of cold signalling by evening-phased components of the plant circadian clock. Plant J. 2013, 76, 247–257. [Google Scholar] [CrossRef]
- Cao, S.; Ye, M.; Jiang, S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005, 24, 683–690. [Google Scholar] [CrossRef]
- Espinoza, C.; Degenkolbe, T.; Caldana, C.; Zuther, E.; Leisse, A.; Willmitzer, L.; Hincha, D.K.; Hannah, M.A. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS ONE 2010, 5, e14101. [Google Scholar] [CrossRef]
- Dong, M.A.; Farré, E.M.; Thomashow, M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar] [CrossRef] [PubMed]
- Chow, B.Y.; Sanchez, S.E.; Breton, G.; Pruneda-Paz, J.L.; Krogan, N.T.; Kay, S.A. Transcriptional regulation of LUX by CBF1 mediates cold input to the circadian clock in Arabidopsis. Curr. Biol. 2014, 24, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Ding, Z.; Duan, M.; Xiong, Y.; Li, X.; Yuan, X.; Huang, J. OsLUX confers rice cold tolerance as a positive regulatory factor. Int. J. Mol. Sci. 2023, 24, 6727. [Google Scholar] [CrossRef] [PubMed]
- Ford, B.; Deng, W.; Clausen, J.; Oliver, S.; Boden, S.; Hemming, M.; Trevaskis, B. Barley (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an EARLY FLOWERING 3-dependent manner. J. Exp. Bot. 2016, 67, 5517–5528. [Google Scholar] [CrossRef] [PubMed]
- Ibañez, C.; Ramos, A.; Acebo, P.; Contreras, A.; Casado, R.; Allona, I.; Aragoncillo, C. Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS ONE 2008, 3, e3567. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Yi, H.; Han, C.T.; Park, J.I.; Hur, Y. Allelic variation in Brassica oleracea CIRCADIAN CLOCK ASSOCIATED 1 (BoCCA1) is associated with freezing tolerance. Hortic. Environ. Biotechnol. 2018, 59, 426–434. [Google Scholar] [CrossRef]
- Marcolino-Gomes, J.; Rodrigues, F.A.; Fuganti-Pagliarini, R.; Bendix, C.; Nakayama, T.J.; Celaya, B.; Molinari, H.B.C.; Oliveira, M.C.N.; Harmon, F.G.; Nepomuceno, A. Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS ONE 2014, 9, e86402. [Google Scholar] [CrossRef]
- Tian, L.; Zhao, X.; Liu, H.; Ku, L.; Wang, S.; Han, Z.; Wu, L.; Shi, Y.; Song, X.; Chen, Y. Alternative splicing of ZmCCA1 mediates drought response in tropical maize. PLoS ONE 2019, 14, e0211623. [Google Scholar] [CrossRef]
- Kim, J.A.; Jung, H.E.; Hong, J.K.; Hermand, V.; McClung, R.; Lee, Y.H.; Kim, J.Y.; Lee, S.I.; Jeong, M.J.; Kim, J.; et al. Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep. 2016, 35, 1943–1954. [Google Scholar] [CrossRef]
- Dong, L.; Hou, Z.; Li, H.; Li, Z.; Fang, C.; Kong, L.; Li, Y.; Du, H.; Li, T.; Wang, L.; et al. Agronomical selection on loss-of-function of GIGANTEA simultaneously facilitates soybean salt tolerance and early maturity. J. Integr. Plant Biol. 2022, 64, 1866–1882. [Google Scholar] [CrossRef]
- Cheng, Q.; Gan, Z.; Wang, Y.; Lu, S.; Hou, Z.; Li, H.; Xiang, H.; Liu, B.; Kong, F.; Dong, L. The soybean gene J contributes to salt stress tolerance by up-regulating salt-responsive genes. Front. Plant Sci. 2020, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Y.; Wei, H.; Wang, L. A clock regulatory module is required for salt tolerance and control of heading date in rice. Plant Cell Environ. 2021, 44, 3283–3301. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, X.; He, Y.; Xu, H.; Wang, L. Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1-mediated sodium homeostasis. EMBO J. 2020, 40, e105086. [Google Scholar] [CrossRef] [PubMed]
- Habte, E.; Müller, L.M.; Shtaya, M.; Davis, S.J.; Von Korf, M. Osmotic stress at the barley root affects expression of circadian clock genes in the shoot: Osmotic stress changes the barley circadian clock. Plant Cell Environ. 2014, 37, 1321–1337. [Google Scholar] [CrossRef]
- Tang, W.; Yan, H.; Su, Z.X.; Park, S.C.; Liu, Y.J.; Zhang, Y.G.; Wang, X.; Kou, M.; Ma, D.F.; Kwak, S.S.; et al. Cloning and characterization of a novel GIGANTEA gene in sweet potato. Plant Physiol. Biochem. 2017, 116, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Xu, H.; Su, C.; Wang, X.; Wang, L. Rice CIRCADIAN CLOCK ASSOCIATED 1 transcriptionally regulates ABA signaling to confer multiple abiotic stress tolerance. Plant Physiol. 2022, 190, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Lou, P.; Hermand, V.; McClung, C.R. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. USA 2015, 112, 3829–3834. [Google Scholar] [CrossRef]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef]
- Gould, P.D.; Locke, J.C.; Larue, C.; Southern, M.M.; Davis, S.J.; Hanano, S.; Moyle, R.; Milich, R.; Putterill, J.; Millar, A.J. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 2006, 18, 1177–1187. [Google Scholar] [CrossRef]
- Salomé, P.A.; Weigel, D.; McClung, C.R. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 2010, 22, 3650–3661. [Google Scholar] [CrossRef]
- Salome, P.A.; McClung, C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 2005, 17, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Song, S.; Xiao, Y.; Fan, F.; Yan, Z.; Jia, G.; Tang, W.; Peng, J. Circadian clock-coordinated response to chilling stress in rice. Environ. Exp. Bot. 2021, 185, 104398. [Google Scholar] [CrossRef]
- Jończyk, M.; Sobkowiak, A.; Trzcinska-Danielewicz, J.; Skoneczny, M.; Solecka, D.; Fronk, J.; Sowiński, P. Global analysis of gene expression in maize leaves treated with low temperature. II. Combined effect of severe cold (8 °C) and circadian rhythm. Plant Mol. Biol. 2017, 95, 279–302. [Google Scholar] [CrossRef]
- Nagel, D.H.; Pruneda-Paz, J.L.; Kay, S.A. FBH1 affects warm temperature responses in the Arabidopsis circadian clock. Proc. Natl Acad. Sci. USA 2014, 111, 14595–14600. [Google Scholar] [CrossRef] [PubMed]
- Kolmos, E.; Chow, B.Y.; Pruneda-Paz, J.L.; Kay, S.A. HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 16172–16177. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.-S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef]
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004, 38, 982–993. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2013, 2, e00473. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyama, T.; Kinoshita, T.; Nakamichi, N. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Aditya Nayaka, A.; Laia, X.; Hutina, S.; Hugouvieuxa, V.; Jung, J.-H.; López-Vidrieroc, I.; Franco-Zorrillac, J.M.; Panigrahid, K.C.S.; Nanaoe, M.H.; Wiggef, P.A.; et al. Molecular mechanisms of Evening Complex activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 6901–6909. [Google Scholar]
- Jung, J.H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Wigge, P.A. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef]
- Karayekov, E.; Sellaro, R.; Legris, M.; Yanovsky, M.J.; Casal, J.J. Heat shock–induced fluctuations in clock and light signaling enhance phytochrome B–mediated Arabidopsis deetiolation. Plant Cell 2013, 25, 2892–2906. [Google Scholar] [CrossRef]
- Grundy, J.; Stoker, C.; Carré, I.A. Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 2015, 6, 648. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Oh, E.; Wang, T.; Wang, Z.Y. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 2016, 7, 13692. [Google Scholar] [CrossRef]
- Park, Y.J.; Kim, J.Y.; Lee, J.H.; Lee, B.D.; Paek, N.C.; Park, C.M. GIGANTEA shapes the photoperiodic rhythms of thermomorphogenic growth in Arabidopsis. Mol. Plant 2020, 13, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wang, J.; Huang, S.; Xie, Y.; Zhu, T.; Liu, L.; Li, L. HSP90s are required for hypocotyl elongation during skotomorphogenesis and thermomorphogenesis via the COP1–ELF3–PIF4 pathway in Arabidopsis. New Phytol. 2023, 239, 1157–1159. [Google Scholar] [CrossRef] [PubMed]
- Balcerowicz, M. Thermomorphogenesis goes like clockwork: How the circadian clock fine-tunes temperature responses through competing transcriptional repressors. New Phytol. 2023, 237, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Blair, E.J.; Bonnot, T.; Hummel, M.; Hay, E.; Marzolino, J.M.; Quijada, I.A.; Nagel, D.H. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci. Rep. 2019, 9, 4814. [Google Scholar] [CrossRef]
- Driedonks, N.; Xu, J.; Peters, J.L.; Park, S.; Rieu, I. Multi-level interactions between heat shock factors, heat shock proteins, and the redox system regulate acclimation to heat. Front. Plant Sci. 2015, 6, 999. [Google Scholar] [CrossRef]
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; Van Zanten, M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2016, 2, 15190. [Google Scholar] [CrossRef]
- Crawford, A.J.; McLachlan, D.H.; Hetherington, A.M.; Franklin, K.A. High temperature exposure increases plant cooling capacity. Curr. Biol. 2012, 22, R396–R397. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, H.J.; Gil, K.E.; Kim, J.Y.; Lee, J.H.; Lee, H.; Cho, H.T.; Vu, L.D.; De Smet, I.; Park, C.M. Developmental programming of thermonastic leaf movement. Plant Physiol. 2019, 180, 1185–1197. [Google Scholar] [CrossRef]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef]
- Sun, J.; Qi, L.; Li, Y.; Chu, J.; Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012, 8, e1002594. [Google Scholar] [CrossRef]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Gray, W.M. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Stavang, J.A.; Gallego-Bartolomé, J.; Gómez, M.D.; Yoshida, S.; Asami, T.; Olsen, J.E.; García-Martínez, J.L.; Alabadí, D.; Blázquez, M.A. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009, 60, 589–601. [Google Scholar] [CrossRef]
- Koini, M.A.; Alvey, L.; Allen, T.; Harberb, N.P.; Whitelam, G.C.; Franklin, K.A. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009, 19, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Yamashino, T.; Matsushika, A.; Fujimori, T.; Sato, S.; Kato, T.; Tabata, S.; Mizuno, T. A Link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.; Park, J.; Kim, S.; Park, J.; Seo, D.; Oh, E. Overexpression of BBX18 promotes thermomorphogenesis through the PRR5-PIF4 pathway. Front. Plant Sci. 2021, 12, 782352. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 21893–21899. [Google Scholar] [CrossRef]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.S.; Kim, W.Y.; Fujiwara, S.; Kim, J.; Cha, J.Y.; Park, J.H.; Lee, S.Y.; Somers, D.E. HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc. Natl. Acad. Sci. USA 2011, 108, 16843–16848. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, J.; Kim, T.S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.Y.; Somers, D.E. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, Y.; Kieffer, M.; Yu, H.; Kepinski, S.; Estelle, M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 2016, 7, 10269. [Google Scholar] [CrossRef]
- Seo, D.; Park, J.; Park, J.; Hwang, G.; Seo, P.J.; Oh, E. ZTL regulates thermomorphogenesis through TOC1 and PRR5. Plant Cell Environ. 2023, 46, 1442–1452. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, J.Y.; Kim, J.I.; Park, Y.J.; Park, C.M. Plant thermomorphogenic adaptation to global warming. J. Plant Biol. 2020, 63, 1–9. [Google Scholar] [CrossRef]
- Kim, H.Y.; Coté, G.G.; Crain, R.C. Potassium channels in Samanea saman protoplasts controlled by phytochrome and the biological clock. Science 1993, 260, 960–962. [Google Scholar] [CrossRef]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian clock genes universally control key agricultural traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef]
- Cheng, Q.; Dong, L.; Su, T.; Li, T.; Gan, Z.; Nan, H.; Lu, S.; Fang, C.; Kong, L.; Li, H.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 2019, 19, 562. [Google Scholar] [CrossRef]
- Kim, N.-S.; Yu, J.; Bae, S.; Kim, H.S.; Park, S.; Lee, K.; Lee, S.I.; Kim, J.A. Identification and Characterization of PSEUDO-RESPONSE REGULATOR (PRR) 1a and 1b Genes by CRISPR/Cas9-Targeted Mutagenesis in Chinese Cabbage (Brassica rapa L.). Int. J. Mol. Sci. 2022, 23, 6963. [Google Scholar] [CrossRef]

| Stress Conditions | Plant Species | Responsible Genes | References |
|---|---|---|---|
| Cold | Oryza sativa | LUX | Huang et al., 2023 [23] |
| Hordeum vulgare | CCA1, GI, PRR59, PRR73, PRR95, LUX | Ford et al., 2016 [24] | |
| Castanea sativa | TOC1, LHY, PRR5, PRR7, PRR9 | Ibañez et al., 2008 [25] | |
| Brassica oleracea | CCA1 | Song et al., 2018 [26] | |
| Drought | Glycine max | LCL1, GmELF4, PRR-like, TOC1-like, LUX-like, PRR7-like genes | Marcolino-Gomes et al., 2014 [27] |
| Zea mays | CCA1 | Tian et al., 2019 [28] | |
| Salt | Brassica rapa | GI | Kim et al., 2016 [29] |
| G. max | E2 (an ortholog of GI) | Dong et al., 2022 [30] | |
| G. max | J (an ortholog of ELF3) | Cheng et al., 2020 [31] | |
| O. sativa | GI, EC1 | Wang et al., 2021 [32] | |
| O. sativa | PRR73 | Wei et al., 2020 [33] | |
| Osmotic | H. vulgare | ELF3 | Habte et al., 2014 [34] |
| Cold, drought, heat, salt | Ipomoea batatas | GI | Tang et al., 2017 [35] |
| Salinity, osmotic, drought | O. sativa | CCA1 | Wei et al., 2022 [36] |
| Salt, cold | B. rapa | GI | Xie et al., 2015 [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, J.; Lee, S.; Kim, J.-I.; Lee, S.; Kim, J.A. The Roles of Circadian Clock Genes in Plant Temperature Stress Responses. Int. J. Mol. Sci. 2024, 25, 918. https://doi.org/10.3390/ijms25020918
Jang J, Lee S, Kim J-I, Lee S, Kim JA. The Roles of Circadian Clock Genes in Plant Temperature Stress Responses. International Journal of Molecular Sciences. 2024; 25(2):918. https://doi.org/10.3390/ijms25020918
Chicago/Turabian StyleJang, Juna, Sora Lee, Jeong-Il Kim, Sichul Lee, and Jin A. Kim. 2024. "The Roles of Circadian Clock Genes in Plant Temperature Stress Responses" International Journal of Molecular Sciences 25, no. 2: 918. https://doi.org/10.3390/ijms25020918
APA StyleJang, J., Lee, S., Kim, J.-I., Lee, S., & Kim, J. A. (2024). The Roles of Circadian Clock Genes in Plant Temperature Stress Responses. International Journal of Molecular Sciences, 25(2), 918. https://doi.org/10.3390/ijms25020918








