The Second Class of Tetrahydrofolate (THF-II) Riboswitches Recognizes the Tetrahydrofolic Acid Ligand via Local Conformation Changes
Abstract
:1. Introduction
2. Results
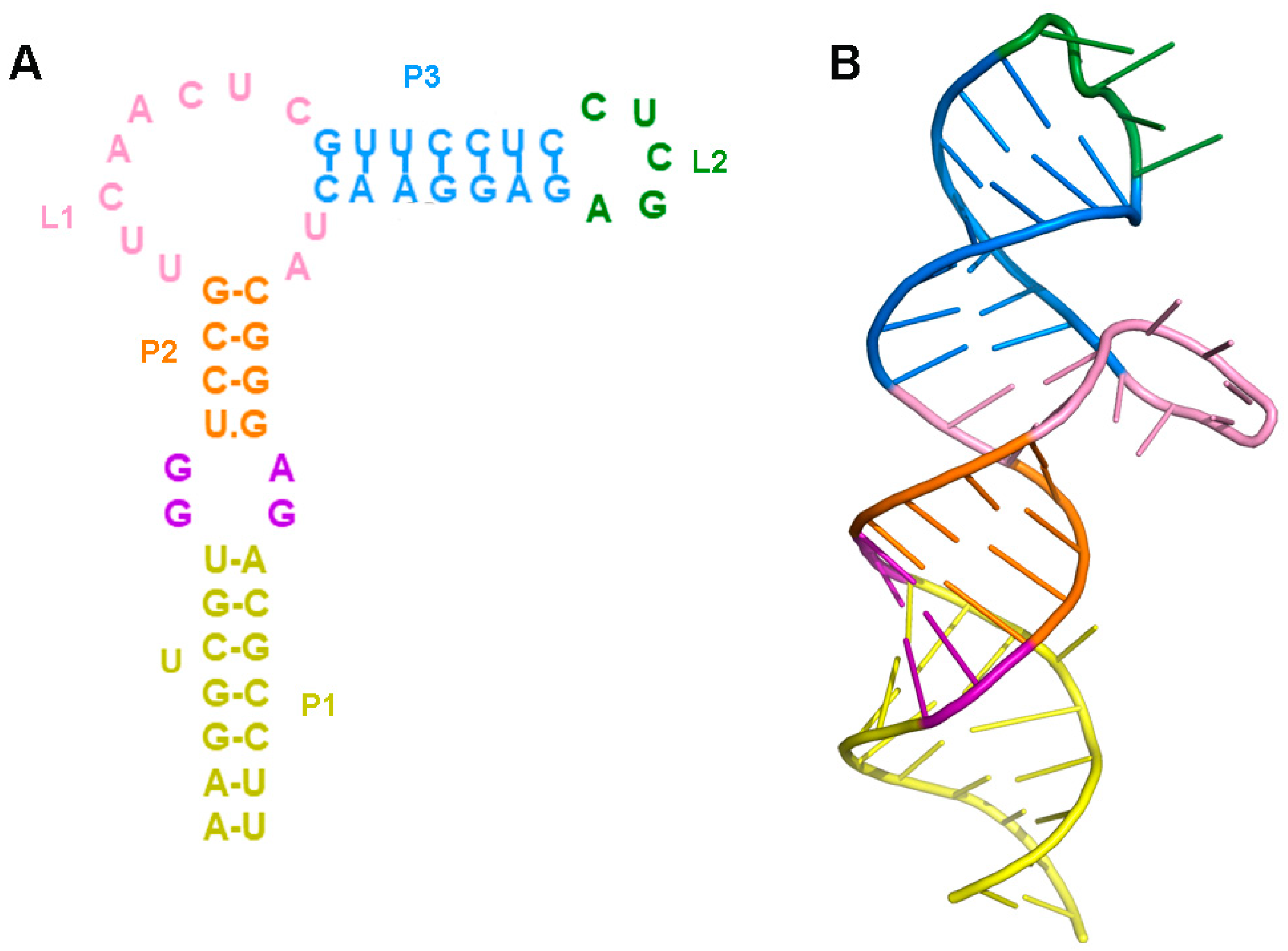
2.1. Prediction of the Three-Dimensional Structure of THF-II Riboswitch
2.2. Tetrahydrofolic Acid Recognizes the THF-II Riboswitch via Loop Regions
2.3. Tetrahydrofolic Acid Binding Induces Local Conformational Changes in the THF-II Riboswitch
2.4. Loop1 Rotates in the THF-II Riboswitch upon Ligand Binding
3. Discussion
4. Materials and Methods
4.1. RNA Preparation
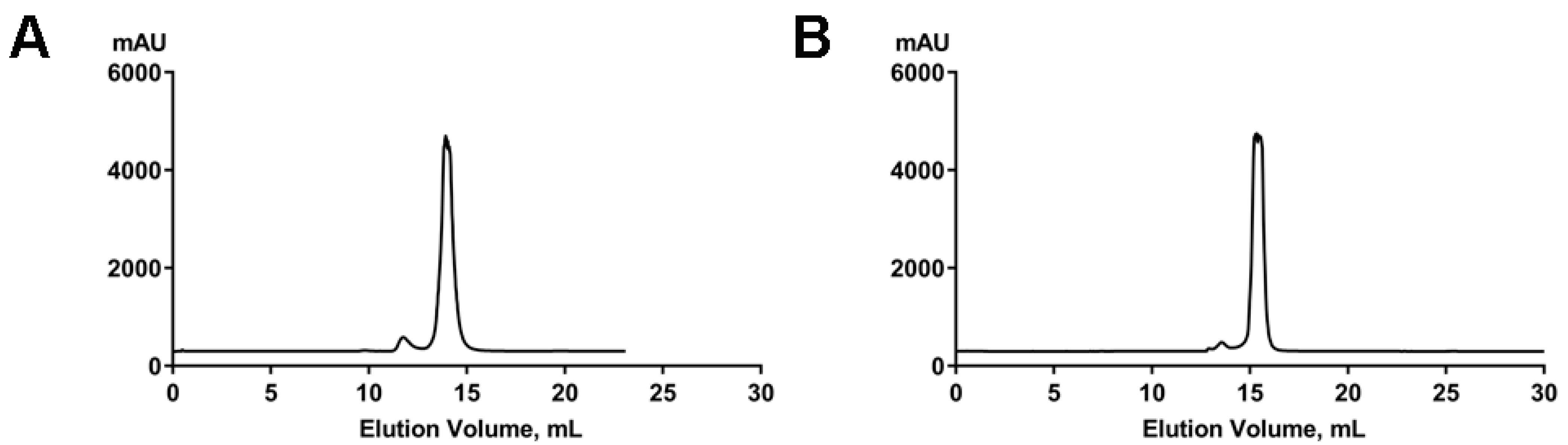
4.2. Size-Exclusion Chromatography
4.3. Small-Angle X-ray Scattering Experiments and Data Analysis
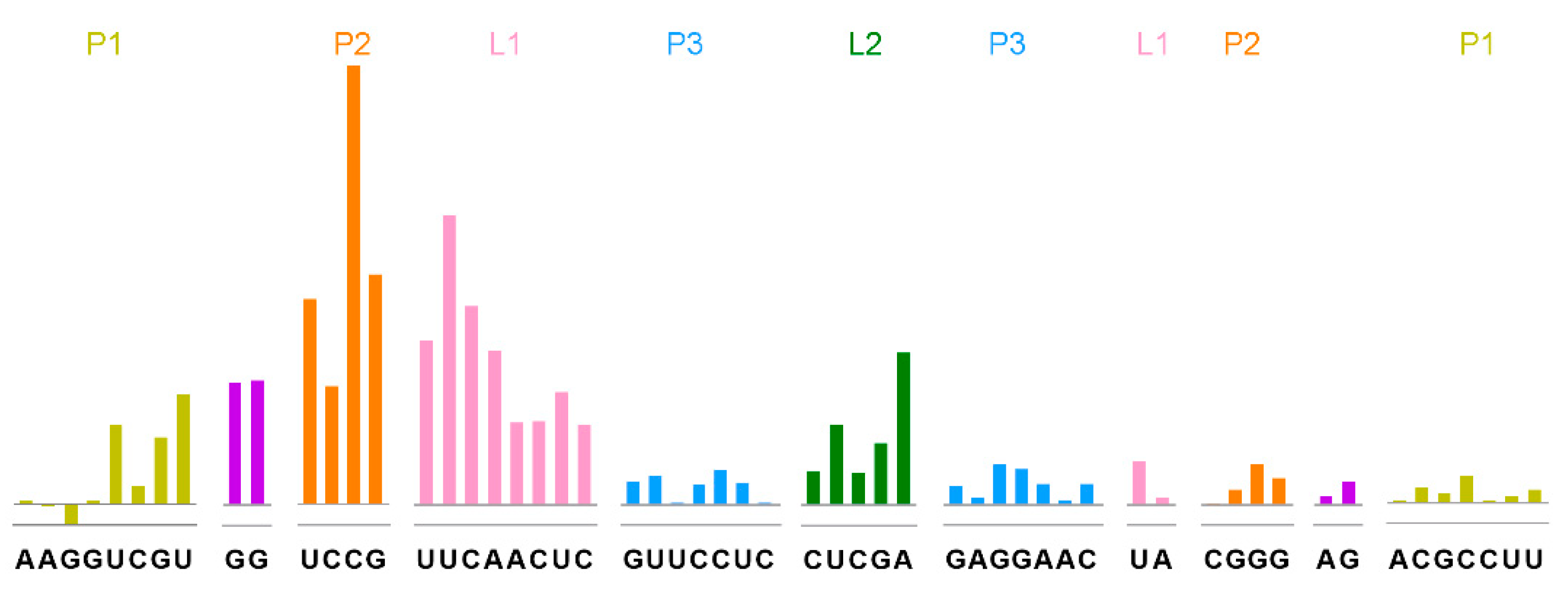
4.4. SHAPE Sample Preparation and SHAPE Experiments of THF-II RNA
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [Green Version]
- Lv, H.; Lv, G.; Han, Q.; Yang, W.; Wang, H. Noncoding RNAs in liver cancer stem cells: The big impact of little things. Cancer Lett. 2018, 418, 51–63. [Google Scholar] [CrossRef]
- Huang, T.; Alvarez, A.; Hu, B.; Cheng, S.Y. Noncoding RNAs in cancer and cancer stem cells. Chin. J. Cancer 2013, 32, 582–593. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 1–12. [Google Scholar] [CrossRef]
- Serganov, A.; Patel, D.J. Ribozymes, riboswitches and beyond: Regulation of gene expression without proteins. Nat. Rev. Genet. 2007, 8, 776–790. [Google Scholar] [CrossRef]
- Bastet, L.; Dube, A.; Masse, E.; Lafontaine, D.A. New insights into riboswitch regulation mechanisms. Mol. Microbiol. 2011, 80, 1148–1154. [Google Scholar] [CrossRef]
- Ariza-Mateos, A.; Nuthanakanti, A.; Serganov, A. Riboswitch Mechanisms: New Tricks for an Old Dog. Biochemistry 2021, 86, 962–975. [Google Scholar] [CrossRef]
- Miranda-Rios, J.; Navarro, M.; Soberon, M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. USA 2001, 98, 9736–9741. [Google Scholar] [CrossRef] [Green Version]
- Nou, X.; Kadner, R.J. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J. Bacteriol. 1998, 180, 6719–6728. [Google Scholar] [CrossRef] [Green Version]
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002, 9, 1043. [Google Scholar] [CrossRef] [Green Version]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Mironov, A.S.; Gusarov, I.; Rafikov, R.; Lopez, L.E.; Shatalin, K.; Kreneva, R.A.; Perumov, D.A.; Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 2002, 111, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Nudler, E.; Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004, 29, 11–17. [Google Scholar] [CrossRef]
- Zimmermann, G.R.; Jenison, R.D.; Wick, C.L.; Simorre, J.P.; Pardi, A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol. 1997, 4, 644–649. [Google Scholar] [CrossRef]
- Ritchey, L.E.; Tack, D.C.; Yakhnin, H.; Jolley, E.A.; Assmann, S.M.; Bevilacqua, P.C.; Babitzke, P. Structure-seq2 probing of RNA structure upon amino acid starvation reveals both known and novel RNA switches in Bacillus subtilis. RNA 2020, 26, 1431–1447. [Google Scholar] [CrossRef]
- Tollerson, R., 2nd; Ibba, M. Translational regulation of environmental adaptation in bacteria. J. Biol. Chem. 2020, 295, 10434–10445. [Google Scholar] [CrossRef]
- Sanders, T.J.; Wenck, B.R.; Selan, J.N.; Barker, M.P.; Trimmer, S.A.; Walker, J.E.; Santangelo, T.J. FttA is a CPSF73 homologue that terminates transcription in Archaea. Nat. Microbiol. 2020, 5, 545–553. [Google Scholar] [CrossRef]
- Roth, A.; Breaker, R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009, 78, 305–334. [Google Scholar] [CrossRef] [Green Version]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef] [Green Version]
- Richards, J.; Belasco, J.G. Riboswitch control of bacterial RNA stability. Mol. Microbiol. 2021, 116, 361–365. [Google Scholar] [CrossRef]
- Pavlova, N.; Kaloudas, D.; Penchovsky, R. Riboswitch distribution, structure, and function in bacteria. Gene 2019, 708, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Atilho, R.M.; Mirihana Arachchilage, G.; Greenlee, E.B.; Knecht, K.M.; Breaker, R.R. A bacterial riboswitch class for the thiamin precursor HMP-PP employs a terminator-embedded aptamer. Elife 2019, 8, e45210. [Google Scholar] [CrossRef] [PubMed]
- Mirihana Arachchilage, G.; Sherlock, M.E.; Weinberg, Z.; Breaker, R.R. SAM-VI RNAs selectively bind S-adenosylmethionine and exhibit similarities to SAM-III riboswitches. RNA Biol. 2018, 15, 371–378. [Google Scholar] [CrossRef] [Green Version]
- Kanwal, F.; Chen, T.; Zhang, Y.; Simair, A.; Lu, C. A Modified In Vitro Transcription Approach to Improve RNA Synthesis and Ribozyme Cleavage Efficiency. Mol. Biotechnol. 2019, 61, 469–476. [Google Scholar] [CrossRef]
- Weinberg, Z.; Lunse, C.E.; Corbino, K.A.; Ames, T.D.; Nelson, J.W.; Roth, A.; Perkins, K.R.; Sherlock, M.E.; Breaker, R.R. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 2017, 45, 10811–10823. [Google Scholar] [CrossRef] [Green Version]
- Chandrangsu, P.; Huang, X.; Gaballa, A.; Helmann, J.D. Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol. Microbiol. 2019, 112, 751–765. [Google Scholar] [CrossRef]
- Bodnar, J.; Fitch, S.; Sanchez, J.; Lesser, M.; Baston, D.S.; Zhong, J. GTP cyclohydrolase I activity from Rickettsia monacensis strain Humboldt, a rickettsial endosymbiont of Ixodes pacificus. Ticks Tick-Borne Dis. 2020, 11, 101434. [Google Scholar] [CrossRef]
- Ames, T.D.; Rodionov, D.A.; Weinberg, Z.; Breaker, R.R. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem. Biol. 2010, 17, 681–685. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Mirihana Arachchilage, G.; Breaker, R.R. Biochemical validation of a second class of tetrahydrofolate riboswitches in bacteria. RNA 2019, 25, 1091–1097. [Google Scholar] [CrossRef] [Green Version]
- Sudarsan, N.; Wickiser, J.K.; Nakamura, S.; Ebert, M.S.; Breaker, R.R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003, 17, 2688–2697. [Google Scholar] [CrossRef] [Green Version]
- Ren, A.; Rajashankar, K.R.; Patel, D.J. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 2012, 486, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Trausch, J.J.; Ceres, P.; Reyes, F.E.; Batey, R.T. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure 2011, 19, 1413–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coppen, A.; Bolander-Gouaille, C. Treatment of depression: Time to consider folic acid and vitamin B12. J. Psychopharmacol. 2005, 19, 59–65. [Google Scholar] [CrossRef]
- Morscher, R.J.; Ducker, G.S.; Li, S.H.; Mayer, J.A.; Gitai, Z.; Sperl, W.; Rabinowitz, J.D. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 2018, 554, 128–132. [Google Scholar] [CrossRef]
- Parker, J.L.; Deme, J.C.; Kuteyi, G.; Wu, Z.; Huo, J.; Goldman, I.D.; Owens, R.J.; Biggin, P.C.; Lea, S.M.; Newstead, S. Structural basis of antifolate recognition and transport by PCFT. Nature 2021, 595, 130–134. [Google Scholar] [CrossRef]
- Akbar, S.; Anwar, A.; Kanwal, Q. Electrochemical determination of folic acid: A short review. Anal. Biochem. 2016, 510, 98–105. [Google Scholar] [CrossRef]
- Malara, N.; Coluccio, M.L.; Limongi, T.; Asande, M.; Trunzo, V.; Cojoc, G.; Raso, C.; Candeloro, P.; Perozziello, G.; Raimondo, R.; et al. Folic acid functionalized surface highlights 5-methylcytosine-genomic content within circulating tumor cells. Small 2014, 10, 4324–4331. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.H.; Seo, H.; Kim, K.J. Biochemical properties and crystal structure of formate-tetrahydrofolate ligase from Methylobacterium extorquens CM4. Biochem. Biophys. Res. Commun. 2020, 528, 426–431. [Google Scholar] [CrossRef]
- Tjong, E.; Dimri, M.; Mohiuddin, S.S. Biochemistry, Tetrahydrofolate. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Maden, B.E. Tetrahydrofolate and tetrahydromethanopterin compared: Functionally distinct carriers in C1 metabolism. Biochem. J. 2000, 350 Pt 3, 609–629. [Google Scholar] [CrossRef]
- Gonen, N.; Assaraf, Y.G. Antifolates in cancer therapy: Structure, activity and mechanisms of drug resistance. Drug Resist. Updates 2012, 15, 183–210. [Google Scholar] [CrossRef]
- Trausch, J.J.; Batey, R.T. A disconnect between high-affinity binding and efficient regulation by antifolates and purines in the tetrahydrofolate riboswitch. Chem. Biol. 2014, 21, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H.; Han, S.R.; Lee, S.W. Therapeutic Applications of Aptamer-Based Riboswitches. Nucleic Acid Ther. 2016, 26, 44–51. [Google Scholar] [CrossRef]
- Kazantsev, A.V.; Rambo, R.P.; Karimpour, S.; Santalucia, J., Jr.; Tainer, J.A.; Pace, N.R. Solution structure of RNase P RNA. RNA 2011, 17, 1159–1171. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.; Price, I.R.; Ding, F.; Wu, F.; Chen, T.; Zhang, Y.; Liu, G.; Jardine, P.J.; Lu, C.; Ke, A. ATP/ADP modulates gp16-pRNA conformational change in the Phi29 DNA packaging motor. Nucleic Acids Res. 2019, 47, 9818–9828. [Google Scholar] [CrossRef]
- Lu, C.; Ding, F.; Chowdhury, A.; Pradhan, V.; Tomsic, J.; Holmes, W.M.; Henkin, T.M.; Ke, A. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J. Mol. Biol. 2010, 404, 803–818. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Smith, A.M.; Ding, F.; Chowdhury, A.; Henkin, T.M.; Ke, A. Variable sequences outside the SAM-binding core critically influence the conformational dynamics of the SAM-III/SMK box riboswitch. J. Mol. Biol. 2011, 409, 786–799. [Google Scholar] [CrossRef] [Green Version]
- Baird, N.J.; Ferre-D’Amare, A.R. Analysis of riboswitch structure and ligand binding using small-angle X-ray scattering (SAXS). Methods Mol. Biol. 2014, 1103, 211–225. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Lipfert, J.; Seifert, S.; Herschlag, D.; Doniach, S. The ligand-free state of the TPP riboswitch: A partially folded RNA structure. J. Mol. Biol. 2010, 396, 153–165. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Zuo, X.; Wang, Y.X.; Dayie, T.K. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res. 2012, 40, 3117–3130. [Google Scholar] [CrossRef]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011, 3, a003533. [Google Scholar] [CrossRef]
- Blouin, S.; Lafontaine, D.A. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 2007, 13, 1256–1267. [Google Scholar] [CrossRef] [Green Version]
- Wickiser, J.K.; Winkler, W.C.; Breaker, R.R.; Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 2005, 18, 49–60. [Google Scholar] [CrossRef]
- Tomsic, J.; McDaniel, B.A.; Grundy, F.J.; Henkin, T.M. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J. Bacteriol. 2008, 190, 823–833. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Smith, A.M.; Fuchs, R.T.; Ding, F.; Rajashankar, K.; Henkin, T.M.; Ke, A. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat. Struct. Mol. Biol. 2008, 15, 1076–1083. [Google Scholar] [CrossRef]
- Ketterer, S.; Gladis, L.; Kozica, A.; Meier, M. Engineering and characterization of fluorogenic glycine riboswitches. Nucleic Acids Res. 2016, 44, 5983–5992. [Google Scholar] [CrossRef]
- Wedekind, J.E.; Dutta, D.; Belashov, I.A.; Jenkins, J.L. Metalloriboswitches: RNA-based inorganic ion sensors that regulate genes. J. Biol. Chem. 2017, 292, 9441–9450. [Google Scholar] [CrossRef] [Green Version]
- Emadpour, M.; Karcher, D.; Bock, R. Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res. 2015, 43, e66. [Google Scholar] [CrossRef] [Green Version]
- Irla, M.; Hakvag, S.; Brautaset, T. Developing a Riboswitch-Mediated Regulatory System for Metabolic Flux Control in Thermophilic Bacillus methanolicus. Int. J. Mol. Sci. 2021, 22, 4686. [Google Scholar] [CrossRef]
- Wilson, R.A.; Fernandez, J.; Quispe, C.F.; Gradnigo, J.; Seng, A.; Moriyama, E.; Wright, J.D. Towards defining nutrient conditions encountered by the rice blast fungus during host infection. PLoS ONE 2012, 7, e47392. [Google Scholar] [CrossRef]
- Carvalho, F.M.; Souza, R.C.; Barcellos, F.G.; Hungria, M.; Vasconcelos, A.T. Genomic and evolutionary comparisons of diazotrophic and pathogenic bacteria of the order Rhizobiales. BMC Microbiol. 2010, 10, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, M.A.; Aliashkevich, A.; Krol, E.; Kuru, E.; Bouchier, J.M.; Rittichier, J.; Brun, Y.V.; VanNieuwenhze, M.S.; Becker, A.; Cava, F.; et al. Unipolar Peptidoglycan Synthesis in the Rhizobiales Requires an Essential Class A Penicillin-Binding Protein. Mbio 2021, 12, e0234621. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.B.; Gillilan, R.E.; Skou, S. BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 2017, 50, 1545–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panjkovich, A.; Svergun, D.I. CHROMIXS: Automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics 2018, 34, 1944–1946. [Google Scholar] [CrossRef] [PubMed]
- ManalastaS-Cantos, K.; Konarev, P.V.; Hajizadeh, N.R.; Kikhney, A.G.; Petoukhov, M.V.; Molodenskiy, D.S.; Panjkovich, A.; Mertens, H.D.T.; Gruzinov, A.; Borges, C.; et al. ATSAS 3.0: Expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 2021, 54, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Svergun, D.I. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Franke, D.; Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009, 42, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.; Konarev, P.V.; Svergun, D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012, 45, 342–350. [Google Scholar] [CrossRef] [Green Version]
- Volkov, V.V.; Svergun, D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003, 36, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Kozin, M.B.; Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001, 34, 33–41. [Google Scholar] [CrossRef]
- Svergun, D.I.B.C.; Barberato, C.; Koch, M.H. CRYSOL—A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 25 March 2022).
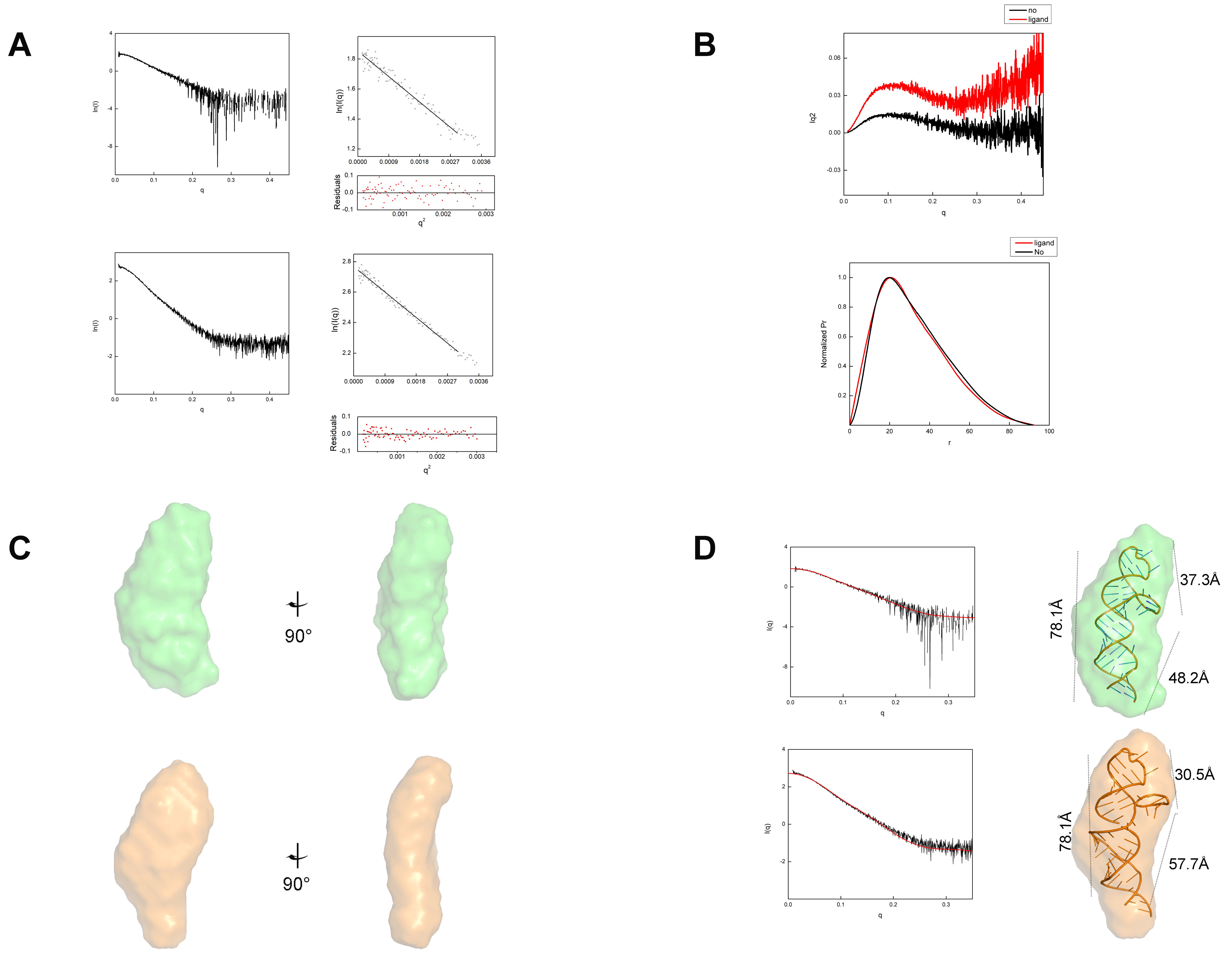

| Structural Parameters | Ligand-Free | Ligand-Bound |
|---|---|---|
| I(0) (cm−1) from Guinier fit | 6.44 ± 0.048 | 15.95 ± 0.065 |
| Rg (Å) from Guinier fit | 24.01 ± 1.21 | 23.64 ± 1.35 |
| Rg (Å) from P(r) | 25.15 | 25.08 |
| Dmax (Å) from P(r) | 89.46 | 92.5 |
| I(0) (cm−1) from P(r) | 25.18 | 25.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Liu, G.; Zhang, Y.; Chen, T.; Feng, S.; Cai, R.; Lu, C. The Second Class of Tetrahydrofolate (THF-II) Riboswitches Recognizes the Tetrahydrofolic Acid Ligand via Local Conformation Changes. Int. J. Mol. Sci. 2022, 23, 5903. https://doi.org/10.3390/ijms23115903
Zhang M, Liu G, Zhang Y, Chen T, Feng S, Cai R, Lu C. The Second Class of Tetrahydrofolate (THF-II) Riboswitches Recognizes the Tetrahydrofolic Acid Ligand via Local Conformation Changes. International Journal of Molecular Sciences. 2022; 23(11):5903. https://doi.org/10.3390/ijms23115903
Chicago/Turabian StyleZhang, Minmin, Guangfeng Liu, Yunlong Zhang, Ting Chen, Shanshan Feng, Rujie Cai, and Changrui Lu. 2022. "The Second Class of Tetrahydrofolate (THF-II) Riboswitches Recognizes the Tetrahydrofolic Acid Ligand via Local Conformation Changes" International Journal of Molecular Sciences 23, no. 11: 5903. https://doi.org/10.3390/ijms23115903






