Interleukin-1 Induces the Release of Lubricating Phospholipids from Human Osteoarthritic Fibroblast-like Synoviocytes
Abstract
:1. Introduction
2. Results
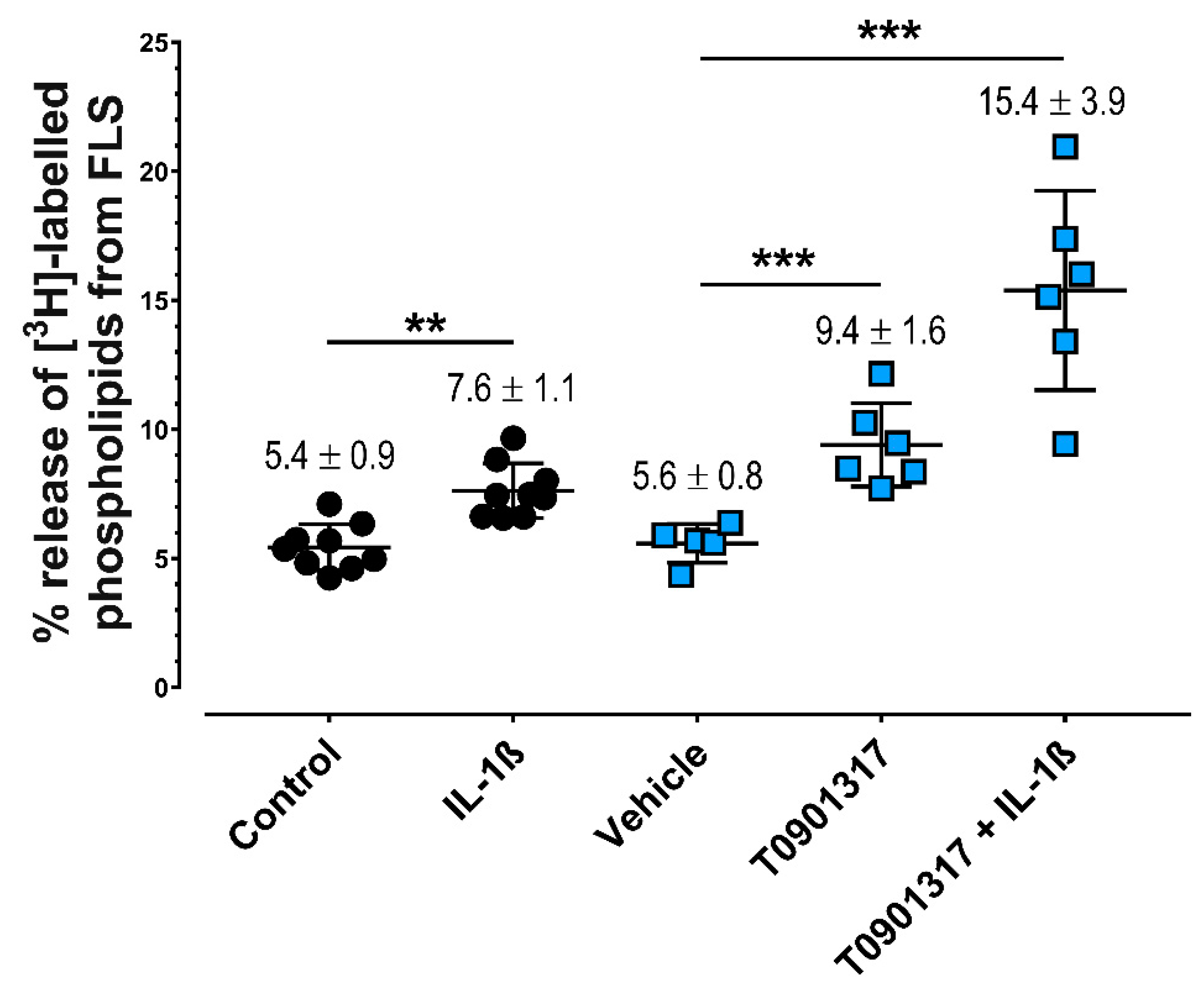
2.1. IL-1ß and LXRα Agonists Induce Greater Release of PLs from FLS
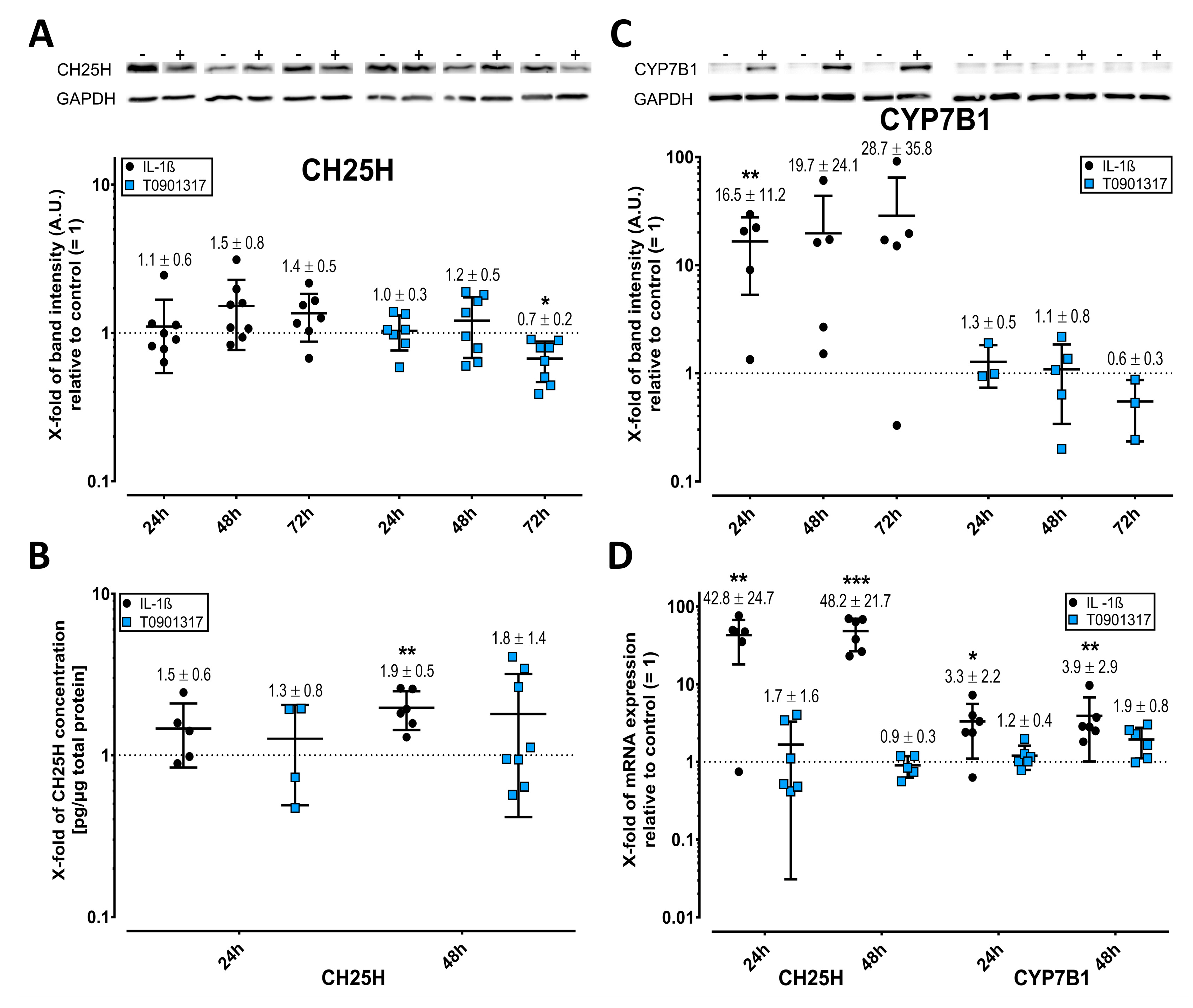
2.2. Cholesterol Hydroxylases Are Upregulated by IL-1ß
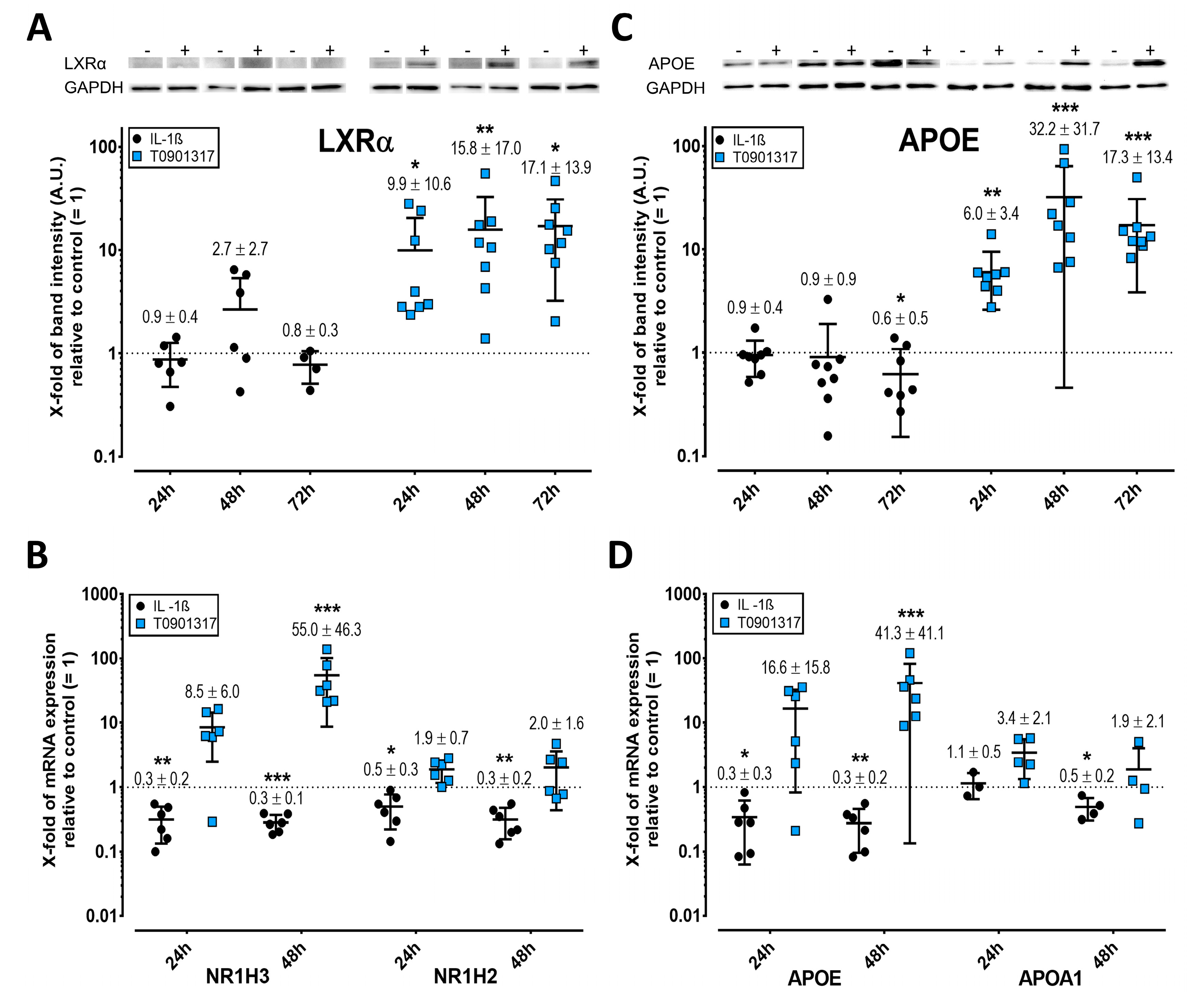
2.3. LXR Expression Is Enhanced by T0901317 but Not IL-1ß
2.4. APOE but Not APOA1 Is Upregulated by T0901317
2.5. Elevated ABC Transporter Expression after Treatment with IL-1ß and T0901317
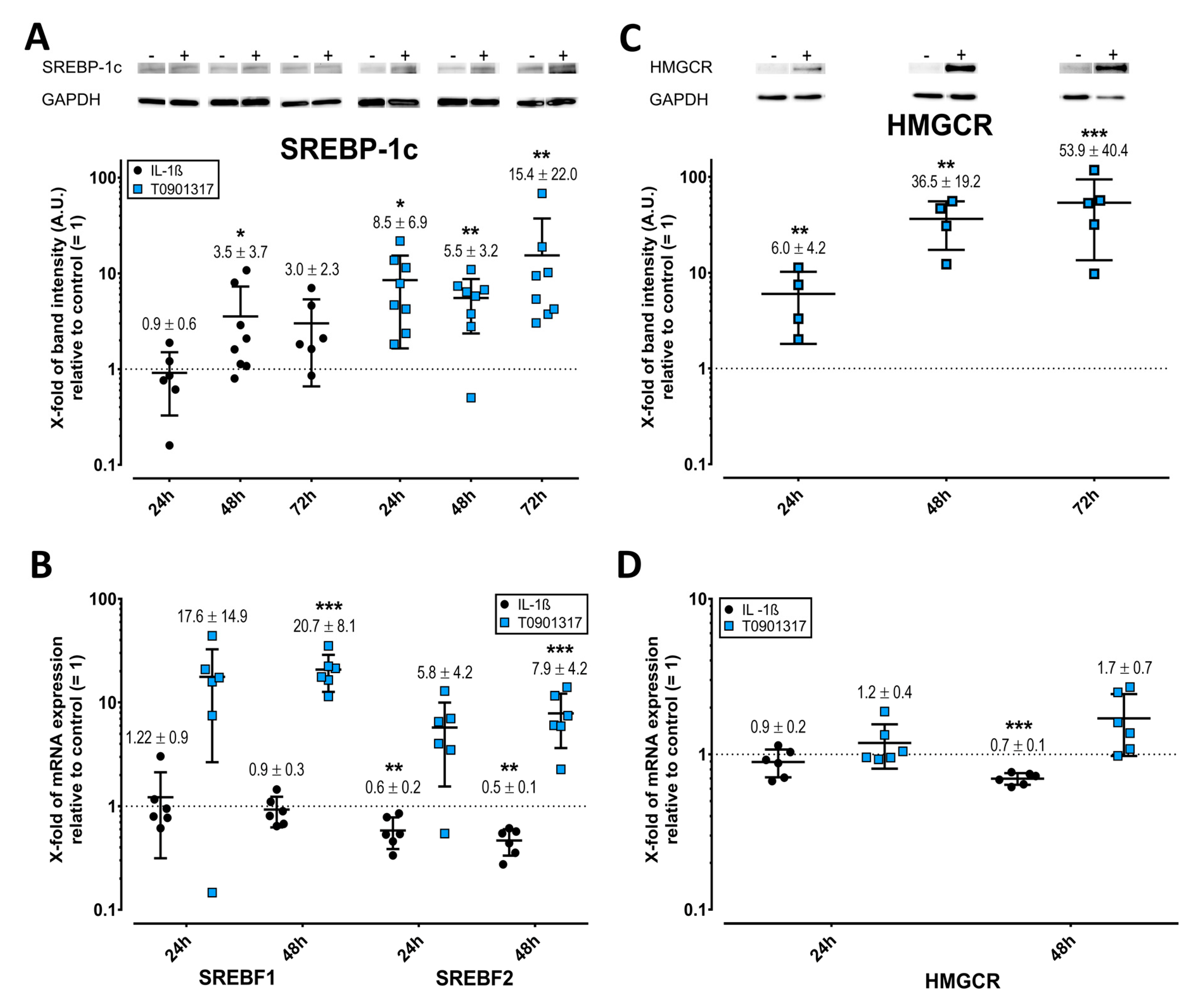
2.6. Opposing Effects of T0901317 and IL-1ß on SREBPs and HMGCR
3. Discussion
4. Materials and Methods
4.1. Source of Fibroblast-Lile Synoviocytes
4.2. Isolation, Cell Propagation, and Purity of FLS
4.3. Treatment of FLS to Study PL Release
4.4. Determination of Radiolabeled PLs
4.5. Treatment of FLS to Study PL Transport Mechanism
4.6. Analysis of mRNA Expression
4.7. ELISA of CH25H
4.8. Western Blot
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seror, J.; Zhu, L.; Goldberg, R.; Day, A.J.; Klein, J. Supramolecular synergy in the boundary lubrication of synovial joints. Nat. Commun. 2015, 6, 6497. [Google Scholar] [CrossRef]
- Kosinska, M.K.; Liebisch, G.; Lochnit, G.; Wilhelm, J.; Klein, H.; Kaesser, U.; Lasczkowski, G.; Rickert, M.; Schmitz, G.; Steinmeyer, J. A lipidomic study of phospholipid classes and species in human synovial fluid. Arthritis Rheum. 2013, 65, 2323–2333. [Google Scholar] [CrossRef]
- Kosinska, M.K.; Liebisch, G.; Lochnit, G.; Wilhelm, J.; Klein, H.; Kaesser, U.; Lasczkowski, G.; Rickert, M.; Schmitz, G.; Steinmeyer, J. Sphingolipids in human synovial fluid—A lipidomic study. PLoS ONE 2014, 9, e91769. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Kampf, N.; Kosinska, M.K.; Steinmeyer, J.; Klein, J. Interactions between Bilayers of Phospholipids Extracted from Human Osteoarthritic Synovial Fluid. Biotribology 2021, 25, 100157. [Google Scholar] [CrossRef]
- Kosinska, M.K.; Ludwig, T.E.; Liebisch, G.; Zhang, R.; Siebert, H.-C.; Wilhelm, J.; Kaesser, U.; Dettmeyer, R.B.; Klein, H.; Ishaque, B.; et al. Articular Joint Lubricants during Osteoarthritis and Rheumatoid Arthritis Display Altered Levels and Molecular Species. PLoS ONE 2015, 10, e0125192. [Google Scholar] [CrossRef]
- Sluzalska, K.D.; Liebisch, G.; Lochnit, G.; Ishaque, B.; Hackstein, H.; Schmitz, G.; Rickert, M.; Steinmeyer, J. Interleukin-1β affects the phospholipid biosynthesis of fibroblast-like synoviocytes from human osteoarthritic knee joints. Osteoarthr. Cartil. 2017, 25, 1890–1899. [Google Scholar] [CrossRef] [Green Version]
- Sluzalska, K.D.; Liebisch, G.; Wilhelm, J.; Ishaque, B.; Hackstein, H.; Schmitz, G.; Rickert, M.; Steinmeyer, J. Growth factors regulate phospholipid biosynthesis in human fibroblast-like synoviocytes obtained from osteoarthritic knees. Sci. Rep. 2017, 7, 13469. [Google Scholar] [CrossRef] [Green Version]
- Neumann, J.; Rose-Sperling, D.; Hellmich, U.A. Diverse relations between ABC transporters and lipids: An overview. Biochim. Biophys. Acta Biomembr. 2017, 1859, 605–618. [Google Scholar] [CrossRef]
- Schmitz, G.; Liebisch, G.; Langmann, T. Lipidomic strategies to study structural and functional defects of ABC-transporters in cellular lipid trafficking. FEBS Lett. 2006, 580, 5597–5610. [Google Scholar] [CrossRef] [Green Version]
- Nakaya, K.; Ayaori, M.; Ikewaki, K. Role of ATP-Binding Cassette Transporters A1 and G1 in Reverse Cholesterol Transport and Atherosclerosis. In The HDL Handbook; Elsevier: Amsterdam, The Netherlands, 2017; pp. 121–151. ISBN 9780128125137. [Google Scholar]
- Fessler, M.B. The challenges and promise of targeting the Liver X Receptors for treatment of inflammatory disease. Pharmacol. Ther. 2018, 181, 1–12. [Google Scholar] [CrossRef]
- Collins-Racie, L.A.; Yang, Z.; Arai, M.; Li, N.; Majumdar, M.K.; Nagpal, S.; Mounts, W.M.; Dorner, A.J.; Morris, E.; LaVallie, E.R. Global analysis of nuclear receptor expression and dysregulation in human osteoarthritic articular cartilage: Reduced LXR signaling contributes to catabolic metabolism typical of osteoarthritis. Osteoarthr. Cartil. 2009, 17, 832–842. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Zhao, R.Q.; Parro, C.; Zhao, W.; Chou, H.-Y.; Robert, J.; Deeb, T.Z.; Raynoschek, C.; Barichievy, S.; Engkvist, O.; et al. Small molecule inducers of ABCA1 and apoE that act through indirect activation of the LXR pathway. J. Lipid Res. 2018, 59, 830–842. [Google Scholar] [CrossRef] [Green Version]
- Jakobsson, T.; Treuter, E.; Gustafsson, J.-Å.; Steffensen, K.R. Liver X receptor biology and pharmacology: New pathways, challenges and opportunities. Trends Pharmacol. Sci. 2012, 33, 394–404. [Google Scholar] [CrossRef]
- Tsezou, A.; Iliopoulos, D.; Malizos, K.N.; Simopoulou, T. Impaired expression of genes regulating cholesterol efflux in human osteoarthritic chondrocytes. J. Orthop. Res. 2010, 28, 1033–1039. [Google Scholar] [CrossRef]
- Ratneswaran, A.; Sun, M.M.-G.; Dupuis, H.; Sawyez, C.; Borradaile, N.; Beier, F. Nuclear receptors regulate lipid metabolism and oxidative stress markers in chondrocytes. J. Mol. Med. 2017, 95, 431–444. [Google Scholar] [CrossRef] [Green Version]
- Joseph, S.B.; Castrillo, A.; Laffitte, B.A.; Mangelsdorf, D.J.; Tontonoz, P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 2003, 9, 213–219. [Google Scholar] [CrossRef]
- Chintalacharuvu, S.R.; Sandusky, G.E.; Burris, T.P.; Burmer, G.C.; Nagpal, S. Liver X receptor is a therapeutic target in collagen-induced arthritis. Arthritis Rheum. 2007, 56, 1365–1367. [Google Scholar] [CrossRef]
- Li, N.; Rivéra-Bermúdez, M.A.; Zhang, M.; Tejada, J.; Glasson, S.S.; Collins-Racie, L.A.; Lavallie, E.R.; Wang, Y.; Chang, K.C.N.; Nagpal, S.; et al. LXR modulation blocks prostaglandin E2 production and matrix degradation in cartilage and alleviates pain in a rat osteoarthritis model. Proc. Natl. Acad. Sci. USA 2010, 107, 3734–3739. [Google Scholar] [CrossRef] [Green Version]
- Favari, E.; Chroni, A.; Tietge, U.J.F.; Zanotti, I.; Escolà-Gil, J.C.; Bernini, F. Cholesterol efflux and reverse cholesterol transport. Handb. Exp. Pharmacol. 2015, 224, 181–206. [Google Scholar] [CrossRef] [Green Version]
- Schultz, J.R.; Tu, H.; Luk, A.; Repa, J.J.; Medina, J.C.; Li, L.; Schwendner, S.; Wang, S.; Thoolen, M.; Mangelsdorf, D.J.; et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000, 14, 2831–2838. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.-S.; Lee, G.; Song, W.-H.; Koh, J.-T.; Yang, J.; Kwak, J.-S.; Kim, H.-E.; Kim, S.K.; Son, Y.-O.; Nam, H.; et al. The CH25H-CYP7B1-RORα axis of cholesterol metabolism regulates osteoarthritis. Nature 2019, 566, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Cyster, J.G.; Dang, E.V.; Reboldi, A.; Yi, T. 25-Hydroxycholesterols in innate and adaptive immunity. Nat. Rev. Immunol. 2014, 14, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Hui, A.Y.; McCarty, W.J.; Masuda, K.; Firestein, G.S.; Sah, R.L. A systems biology approach to synovial joint lubrication in health, injury, and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 15–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.-P.; Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef]
- Glyn-Jones, S.; Palmer, A.J.R.; Agricola, R.; Price, A.J.; Vincent, T.L.; Weinans, H.; Carr, A.J. Osteoarthritis. Lancet 2015, 386, 376–387. [Google Scholar] [CrossRef]
- Jenei-Lanzl, Z.; Meurer, A.; Zaucke, F. Interleukin-1β signaling in osteoarthritis—Chondrocytes in focus. Cell. Signal. 2019, 53, 212–223. [Google Scholar] [CrossRef]
- McNulty, A.L.; Rothfusz, N.E.; Leddy, H.A.; Guilak, F. Synovial fluid concentrations and relative potency of interleukin-1 alpha and beta in cartilage and meniscus degradation. J. Orthop. Res. 2013, 31, 1039–1045. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, V.; Kim, M.-J.; Gelissen, I.C.; Brown, A.J.; Sandoval, C.; Hallab, J.C.; Kockx, M.; Traini, M.; Jessup, W.; Kritharides, L. Cellular cholesterol regulates ubiquitination and degradation of the cholesterol export proteins ABCA1 and ABCG1. J. Biol. Chem. 2014, 289, 7524–7536. [Google Scholar] [CrossRef] [Green Version]
- Wellington, C.L.; Walker, E.K.Y.; Suarez, A.; Kwok, A.; Bissada, N.; Singaraja, R.; Yang, Y.-Z.; Zhang, L.-H.; James, E.; Wilson, J.E.; et al. ABCA1 mRNA and protein distribution patterns predict multiple different roles and levels of regulation. Lab. Investig. 2002, 82, 273–283. [Google Scholar] [CrossRef] [Green Version]
- Hozoji, M.; Munehira, Y.; Ikeda, Y.; Makishima, M.; Matsuo, M.; Kioka, N.; Ueda, K. Direct interaction of nuclear liver X receptor-beta with ABCA1 modulates cholesterol efflux. J. Biol. Chem. 2008, 283, 30057–30063. [Google Scholar] [CrossRef] [Green Version]
- Hozoji-Inada, M.; Munehira, Y.; Nagao, K.; Kioka, N.; Ueda, K. Liver X receptor beta (LXRbeta) interacts directly with ATP-binding cassette A1 (ABCA1) to promote high density lipoprotein formation during acute cholesterol accumulation. J. Biol. Chem. 2011, 286, 20117–20124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janowski, B.A.; Grogan, M.J.; Jones, S.A.; Wisely, G.B.; Kliewer, S.A.; Corey, E.J.; Mangelsdorf, D.J. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. USA 1999, 96, 266–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laffitte, B.A.; Repa, J.J.; Joseph, S.B.; Wilpitz, D.C.; Kast, H.R.; Mangelsdorf, D.J.; Tontonoz, P. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. USA 2001, 98, 507–512. [Google Scholar] [CrossRef]
- Wójcicka, G.; Jamroz-Wiśniewska, A.; Horoszewicz, K.; Bełtowski, J. Liver X receptors (LXRs). Part I: Structure, function, regulation of activity, and role in lipid metabolism. Postepy Hig. Med. Dosw. 2007, 61, 736–759. [Google Scholar]
- Kockx, M.; Jessup, W.; Kritharides, L. Regulation of endogenous apolipoprotein E secretion by macrophages. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.; Chow, R.; Brown, A.J. Sterol regulators of cholesterol homeostasis and beyond: The oxysterol hypothesis revisited and revised. Prog. Lipid Res. 2008, 47, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Jeon, T.-I.; Osborne, T.F. SREBPs: Metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 2012, 23, 65–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, E.; Riepl, B.; Knedla, A.; Lefèvre, S.; Tarner, I.H.; Grifka, J.; Steinmeyer, J.; Schölmerich, J.; Gay, S.; Müller-Ladner, U. Cell culture and passaging alters gene expression pattern and proliferation rate in rheumatoid arthritis synovial fibroblasts. Arthritis Res. Ther. 2010, 12, R83. [Google Scholar] [CrossRef] [Green Version]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. 2020. Available online: https://www.R-project.org/ (accessed on 7 October 2021).
- Sarkar, D. Lattice: Trellis Graphics for R. Available online: https://CRAN.R-project.org/package=lattice (accessed on 7 October 2021).
- Fox, J.; Weisberg, W.S. Car: Companion to Applied Regression. Available online: https://CRAN.R-project.org/package=car (accessed on 12 August 2020).
- Pinheiro, J.; Bates, D.; R Core Team. Nlme: Linear and Nonlinear Mixed Effects Models. Available online: https://svn.r-project.org/R-packages/trunk/nlme/ (accessed on 7 October 2021).
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Statist. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Hothorn, T.; Bretz, F.; Westfall, P. Multcomp: Simultaneous Inference in General Parametric Models. Available online: https://CRAN.R-project.org/package=multcomp (accessed on 12 August 2020).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thottakkattumana Parameswaran, V.; Hild, C.; Eichner, G.; Ishaque, B.; Rickert, M.; Steinmeyer, J. Interleukin-1 Induces the Release of Lubricating Phospholipids from Human Osteoarthritic Fibroblast-like Synoviocytes. Int. J. Mol. Sci. 2022, 23, 2409. https://doi.org/10.3390/ijms23052409
Thottakkattumana Parameswaran V, Hild C, Eichner G, Ishaque B, Rickert M, Steinmeyer J. Interleukin-1 Induces the Release of Lubricating Phospholipids from Human Osteoarthritic Fibroblast-like Synoviocytes. International Journal of Molecular Sciences. 2022; 23(5):2409. https://doi.org/10.3390/ijms23052409
Chicago/Turabian StyleThottakkattumana Parameswaran, Vishnu, Christiane Hild, Gerrit Eichner, Bernd Ishaque, Markus Rickert, and Juergen Steinmeyer. 2022. "Interleukin-1 Induces the Release of Lubricating Phospholipids from Human Osteoarthritic Fibroblast-like Synoviocytes" International Journal of Molecular Sciences 23, no. 5: 2409. https://doi.org/10.3390/ijms23052409
APA StyleThottakkattumana Parameswaran, V., Hild, C., Eichner, G., Ishaque, B., Rickert, M., & Steinmeyer, J. (2022). Interleukin-1 Induces the Release of Lubricating Phospholipids from Human Osteoarthritic Fibroblast-like Synoviocytes. International Journal of Molecular Sciences, 23(5), 2409. https://doi.org/10.3390/ijms23052409





