Metalloprotein-Specific or Critical Amino Acid Residues: Perspectives on Plant-Precise Detoxification and Recognition Mechanisms under Cadmium Stress
Abstract
:1. Introduction
2. Influx Protein
2.1. A Brief Introduction to the NRAMP Family, the Key Proteins Related to Cd Influx and the Mechanism of Action
2.2. A Brief Introduction to the ZIP Family, the Key Proteins Related to Cd Influx and the Mechanism of Action
2.3. Comparison of Core Characteristics of Cd Influx Key Proteins
3. Chelating Protein
3.1. A Brief Introduction to the MT Family, Key Proteins and Chelation Mechanism
3.2. A Brief Introduction to the PDF Family, Key Proteins and Chelation Mechanism
3.3. Comparison of the Core Features of Key Cd Chelation Proteins
4. Vacuolar Protein
4.1. Introduction to CAX Family, Functional Proteins and Vacuolar Transfer Mechanism
4.2. An Introduction to the ABCC Subfamily, Functional Proteins and Vacuolar Transfer Mechanism
4.3. Introduction to MTP Family, Functional Proteins and Vacuolar Transfer Mechanism
4.4. Comparison of Core Characteristics of Key Proteins in Vacuolar Transport
5. Long-Distance Transport Protein
5.1. Introduction to the OPT Family, the Key Proteins Involved in the Transport of Cd and the Mode of Transport
5.2. An Introduction to the HMA Family, the Key Proteins Involved in the Transport of Cd and the Mode of Transport
5.3. Comparison of Core Characteristics of Key Proteins for Long-Distance Transportation of Cd
6. Efflux Protein
6.1. A Brief Introduction to the PCR Family, Key Efflux Proteins and Efflux Mechanisms
6.2. A Brief Introduction to the ABCG Family, Key Efflux Proteins and Efflux Mechanisms
6.3. Comparison of Core Features of Key Cd Efflux Proteins
7. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| ABC | ATP-binding cassette transporters |
| At | Arabidopsis thaliana |
| Bb | Bordetella bronchiseptica |
| CAX | Cation/proton exchanger |
| Cd | cadmium |
| HMA | heavy metal ATPase |
| IRT1 | iron-regulated transporter 1 |
| JA | jasmonic acid |
| MT | metallothionei |
| MTP/CDF | metal tolerance protein/ cation diffusion facilitator |
| NRAMP | Natural resistance-associated macrophage protein |
| OPT | oligopeptide transporter |
| Os | Oryza sativa |
| PCR | plant Cd resistance |
| plant defensins | |
| SA | salicylic acid |
| TFs | transcription factor |
| ZIP | ZRT, IRT-like protein |
References
- Ratajczak, A.E.; Rychter, A.M.; Zawada, A.; Dobrowolska, A.; Krela-Kaźmierczak, I. Do Only Calcium and Vitamin D Matter? Micronutrients in the Diet of Inflammatory Bowel Diseases Patients and the Risk of Osteoporosis. Nutrients 2021, 13, 525. [Google Scholar] [CrossRef]
- Zheng, L.Y.; Sanders, A.P.; Saland, J.M.; Wright, R.O.; Arora, M. Environmental exposures and pediatric kidney function and disease: A systematic review. Env. Res. 2017, 158, 625–648. [Google Scholar] [CrossRef] [Green Version]
- Buha, A.; Wallace, D.; Matovic, V.; Schweitzer, A.; Oluic, B.; Micic, D.; Djordjevic, V. Cadmium Exposure as a Putative Risk Factor for the Development of Pancreatic Cancer: Three Different Lines of Evidence. Biomed. Res. Int. 2017, 2017, 1981837. [Google Scholar] [CrossRef] [PubMed]
- Filippini, T.; Torres, D.; Lopes, C.; Carvalho, C.; Moreira, P.; Naska, A.; Kasdagli, M.; Malavolti, M.; Orsini, N.; Vinceti, M. Cadmium exposure and risk of breast cancer: A dose-response meta-analysis of cohort studies. Env. Int. 2020, 142, 105879. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Kim, K.; Lee, Y.; Fernandes, J.; Smith, M.R.; Jung, Y.; Orr, M.; Kang, S.; Jones, D.P.; Go, Y. Environmental Cadmium Enhances Lung Injury by Respiratory Syncytial Virus Infection. Am. J. Pathol. 2019, 189, 1513–1525. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Kashifuddin, M. Adsorption studies of Cd (II) on ball clay: Comparison with other natural clays. Arab J. Chem. 2016, 9, S1233–S1241. [Google Scholar] [CrossRef] [Green Version]
- Kashif Irshad, M.; Chen, C.; Noman, A.; Ibrahim, M.; Adeel, M.; Shang, J. Goethite-modified biochar restricts the mobility and transfer of cadmium in soil-rice system. Chemosphere 2020, 242, 125152. [Google Scholar] [CrossRef]
- Sarwar, N.; Saifullah; Malhi, S.S.; Zia, M.H.; Naeem, A.; Bibi, S.; Farid, G. Role of mineral nutrition in minimizing cadmium accumulation by plants. J. Sci. Food Agr. 2010, 90, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Asad, S.A.; Farooq, M.; Afzal, A.; West, H. Integrated phytobial heavy metal remediation strategies for a sustainable clean environment—A review. Chem.osphere 2019, 217, 925–941. [Google Scholar] [CrossRef]
- Lu, L.; Tian, S.; Yang, X.; Wang, X.; Brown, P.; Li, T.; He, Z. Enhanced root-to-shoot translocation of cadmium in the hyperaccumulating ecotype of Sedum alfredii. J. Exp. Bot. 2008, 59, 3203–3213. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; He, G.; Tian, W.; Li, D.; Meng, L.; Wu, D.; He, T. Genome-Wide Identification of MATE Gene Family in Potato (Solanum tuberosum L.) and Expression Analysis in Heavy Metal Stress. Front. Genet. 2021, 12, 650500. [Google Scholar] [CrossRef] [PubMed]
- Van der Vliet, L.; Peterson, C.; Hale, B. Cd accumulation in roots and shoots of durum wheat: The roles of transpiration rate and apoplastic bypass. J. Exp. Bot. 2007, 58, 2939–2947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, Q.; Jupa, R.; Liu, Y.; Luo, J.; Li, J.; Kováč, J.; Li, B.; Li, Q.; Wu, K.; Liang, Y.; et al. Abscisic acid-mediated modifications of radial apoplastic transport pathway play a key role in cadmium uptake in hyperaccumulator Sedum alfredii. Plant Cell Environ. 2019, 42, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Asgher, M.; Khan, M.I.R.; Anjum, N.A.; Khan, N.A. Minimising toxicity of cadmium in plants—role of plant growth regulators. Protoplasma 2015, 252, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, Y.; Kuşvuran, A.; Alharby, H.F.; Kuşvuran, S.; Rady, M.M. The defensive role of silicon in wheat against stress conditions induced by drought, salinity or cadmium. Ecotox Environ Safe. 2018, 154, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; You, Y.; Shentu, J.; Weng, Y.; Wang, S.; Xu, Q.; Liu, H.; Du, S. Abscisic acid (ABA)-importing transporter 1 (AIT1) contributes to the inhibition of Cd accumulation via exogenous ABA application in Arabidopsis. J. Hazard. Mater. 2020, 391, 122189. [Google Scholar] [CrossRef]
- Wang, F.; Tan, H.; Huang, L.; Cai, C.; Ding, Y.; Bao, H.; Chen, Z.; Zhu, C. Application of exogenous salicylic acid reduces Cd toxicity and Cd accumulation in rice. Ecotox. Env. Safe 2021, 207, 111198. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Pazmiño, D.M.; Testillano, P.S.; Risueño, M.C.; Del Río, L.A.; Sandalio, L.M. Cellular response of pea plants to cadmium toxicity: Cross talk between reactive oxygen species, nitric oxide, and calcium. Plant Physiol. 2009, 150, 229–243. [Google Scholar] [CrossRef] [Green Version]
- Feng Shao, J.; Che, J.; Yamaji, N.; Fang Shen, R.; Feng Ma, J. Silicon reduces cadmium accumulation by suppressing expression of transporter genes involved in cadmium uptake and translocation in rice. J. Exp. Bot. 2017, 68, 5641–5651. [Google Scholar] [CrossRef] [Green Version]
- Dai, S.; Wang, B.; Song, Y.; Xie, Z.; Li, C.; Li, S.; Huang, Y.; Jiang, M. Astaxanthin and its gold nanoparticles mitigate cadmium toxicity in rice by inhibiting cadmium translocation and uptake. Sci. Total Env. 2021, 786, 147496. [Google Scholar] [CrossRef]
- Sun, W.; Zhan, J.; Zheng, T.; Wu, G.; Xu, H.; Chen, Y.; Yao, M.; Zeng, J.; Yan, J.; Chen, H. Involvement of several putative transporters of different families in β-cyclocitral-induced alleviation of cadmium toxicity in quinoa (Chenopodium quinoa) seedlings. J. Hazard. Mater. 2021, 419, 126474. [Google Scholar] [CrossRef]
- Jiang, M.; Jiang, J.; Li, S.; Li, M.; Tan, Y.; Song, S.; Shu, Q.; Huang, J. Glutamate alleviates cadmium toxicity in rice via suppressing cadmium uptake and translocation. J. Hazard. Mater. 2020, 384, 121319. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Nie, W.; Yan, Y.; Gao, Z.; Shi, Q. Unravelling cadmium toxicity and nitric oxide induced tolerance in Cucumis sativus: Insight into regulatory mechanisms using proteomics. J. Hazard. Mater. 2017, 336, 202–213. [Google Scholar] [CrossRef]
- Schueler-Furman, O.; Baker, D. Conserved residue clustering and protein structure prediction. Proteins-Struct. Funct. Bioinform. 2010, 52, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Sloof, J.E.; Viragh, A.; Van Der Veer, B. Kinetics of cadmium uptake by green algae. Water Air Soil Pollut. 1995, 83, 105–122. [Google Scholar] [CrossRef]
- Nevo, Y.; Nelson, N. The NRAMP family of metal-ion transporters. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2006, 1763, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; He, G.; Qin, L.; Li, D.; Meng, L.; Huang, Y.; He, T. Genome-wide analysis of the NRAMP gene family in potato (Solanum tuberosum): Identification, expression analysis and response to five heavy metals stress. Ecotox. Env. Safe 2021, 208, 111661. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [Green Version]
- Courville, P.; Urbankova, E.; Rensing, C.; Chaloupka, R.; Quick, M.; Cellier, M.F.M. Solute Carrier 11 Cation Symport Requires Distinct Residues in Transmembrane Helices 1 and 6*. J. Biol. Chem. 2008, 283, 9651–9658. [Google Scholar] [CrossRef] [Green Version]
- Thomine, S.; Wang, R.; Ward, J.M.; Crawford, N.M.; Schroeder, J.I. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. USA 2000, 97, 4991–4996. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, R.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. Role of the iron transporter OsNRAMP1 in cadmium uptake and accumulation in rice. Plant Signal. Behav. 2011, 6, 1813–1816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrnstorfer, I.A.; Manatschal, C.; Arnold, F.M.; Laederach, J.; Dutzler, R. Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family. Nat. Commun. 2017, 8, 14033. [Google Scholar] [CrossRef] [Green Version]
- Pottier, M.; Oomen, R.; Picco, C.; Giraudat, J.; Scholz-Starke, J.; Richaud, P.; Carpaneto, A.; Thomine, S. Identification of mutations allowing Natural Resistance Associated Macrophage Proteins (NRAMP) to discriminate against cadmium. Plant J. 2015, 83, 625–637. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, L.; Zheng, L.; Wang, Y.; Chen, X.; Zhang, W. A Functional Study Identifying Critical Residues Involving Metal Transport Activity and Selectivity in Natural Resistance-Associated Macrophage Protein 3 in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramesh, S.A.; Shin, R.; Eide, D.J.; Schachtman, D.P. Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 2003, 133, 126–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedas, P.; Husted, S. Zinc transport mediated by barley ZIP proteins are induced by low pH. Plant Signal. Behav. 2009, 4, 842–845. [Google Scholar] [CrossRef] [Green Version]
- Barberon, M.; Dubeaux, G.; Kolb, C.; Isono, E.; Zelazny, E.; Vert, G. Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl Acad Sci. USA 2014, 111, 8293–8298. [Google Scholar] [CrossRef] [Green Version]
- Cohen, C.K.; Garvin, D.F.; Kochian, L.V. Kinetic properties of a micronutrient transporter from Pisum sativum indicate a primary function in Fe uptake from the soil. Planta 2004, 218, 784–792. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Chen, L.; Li, X. Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress. Bot. Stud. 2018, 59, 22. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, L.; Yang, A. The Molecular Mechanisms Underlying Iron Deficiency Responses in Rice. Int. J. Mol. Sci. 2019, 21, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavitha, P.G.; Kuruvilla, S.; Mathew, M.K. Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol. Bioch. 2015, 97, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Et Biophys. Acta (BBA) Biomembr. 2000, 1465, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Kuliyev, E.; Sui, D.; Hu, J. The histidine-rich loop in the extracellular domain of ZIP4 binds zinc and plays a role in zinc transport. BioChem. J. 2019, 476, 1791–1803. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, J.; Long, C.; He, B.; Hu, Z.; Jiang, C.; Li, Y.; Ma, L.; Wen, J.; Zou, X.; et al. Identification and characterization of the ZRT, IRT-like protein (ZIP) family genes reveal their involvement in growth and kojic acid production in Aspergillus oryzae. J. Ind. Microbiol. Biotechnol. 2019, 46, 1769–1780. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, J.; Fellner, M.; Zhang, C.; Sui, D.; Hu, J. Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 2017, 3, e1700344. [Google Scholar] [CrossRef] [Green Version]
- Hu, J. Toward unzipping the ZIP metal transporters: Structure, evolution, and implications on drug discovery against cancer. FEBS J. 2021, 288, 5805–5825. [Google Scholar] [CrossRef]
- Ajeesh Krishna, T.P.; Maharajan, T.; Victor Roch, G.; Ignacimuthu, S.; Antony Ceasar, S. Structure, Function, Regulation and Phylogenetic Relationship of ZIP Family Transporters of Plants. Front. Plant Sci. 2020, 11, 662. [Google Scholar] [CrossRef]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef]
- Kawachi, M.; Kobae, Y.; Mimura, T.; Maeshima, M. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn(2+)/H(+) antiporter of Arabidopsis thaliana, stimulates the transport activity. J. Biol. Chem. 2008, 283, 8374–8383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Duan, S.; Wu, Q.; Yu, M.; Shabala, S. Reducing Cadmium Accumulation in Plants: Structure-Function Relations and Tissue-Specific Operation of Transporters in the Spotlight. Plants-Basel 2020, 9, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cailliatte, R.; Schikora, A.; Briat, J.; Mari, S.; Curie, C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 2010, 22, 904–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihnatowicz, A.; Siwinska, J.; Meharg, A.A.; Carey, M.; Koornneef, M.; Reymond, M. Conserved histidine of metal transporter AtNRAMP1 is crucial for optimal plant growth under manganese deficiency at chilling temperatures. New Phytol. 2014, 202, 1173–1183. [Google Scholar] [CrossRef]
- Chang, J.; Huang, S.; Yamaji, N.; Zhang, W.; Ma, J.F.; Zhao, F. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020, 43, 2476–2491. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Ogawa, I.; Ishimaru, Y.; Mori, S.; Nishizawa, N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006, 52, 464–469. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Ueno, N.M.; Kashino, M.; Ma, J.F. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 2015, 84, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Capdevila, M.; Atrian, S. Metallothionein protein evolution: A miniassay. JBIC J. Biol. Inorg. Chem. 2011, 16, 977–989. [Google Scholar] [CrossRef]
- Mekawy, A.M.M.; Assaha, D.V.M.; Ueda, A. Constitutive overexpression of rice metallothionein-like gene OsMT-3a enhances growth and tolerance of Arabidopsis plants to a combination of various abiotic stresses. J. Plant Res. 2020, 133, 429–440. [Google Scholar] [CrossRef]
- Nezhad, R.M.; Shahpiri, A.; Mirlohi, A. Discrimination between two rice metallothionein isoforms belonging to type 1 and type 4 in metal-binding ability. Biotechnol. Appl. Bioc. 2013, 60, 275–282. [Google Scholar] [CrossRef]
- Zimeri, A.M.; Dhankher, O.P.; McCaig, B.; Meagher, R.B. The Plant MT1 Metallothioneins are Stabilized by Binding Cadmiums and are Required for Cadmium Tolerance and Accumulation. Plant Mol. Biol. 2005, 58, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Malekzadeh, R.; Shahpiri, A.; Siapoush, S. Metalation of a rice type 1 metallothionein isoform (OsMTI-1b). Protein Expres. Purif. 2020, 175, 105719. [Google Scholar] [CrossRef]
- Zhong, S.; Li, X.; Li, F.; Huang, Y.; Liu, T.; Yin, H.; Qiao, J.; Chen, G.; Huang, F. Cadmium uptake and transport processes in rice revealed by stable isotope fractionation and Cd-related gene expression. Sci Total Environ. 2022, 806, 150633. [Google Scholar] [CrossRef] [PubMed]
- Mekawy, A.M.M.; Assaha, D.V.M.; Munehiro, R.; Kohnishi, E.; Nagaoka, T.; Ueda, A.; Saneoka, H. Characterization of type 3 metallothionein-like gene (OsMT-3a) from rice, revealed its ability to confer tolerance to salinity and heavy metal stresses. Env. Exp. Bot. 2018, 147, 157–166. [Google Scholar] [CrossRef]
- Leszczyszyn, O.I.; Imam, H.T.; Blindauer, C.A. Diversity and distribution of plant metallothioneins: A review of structure, properties and functions†. Metallomics 2013, 5, 1146–1169. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qin, Y.; Xu, W.; Chai, Y.; Li, T.; Zhang, C.; Yang, M.; He, Z.; Feng, D. Differences of Cd uptake and expression of MT family genes and NRAMP2 in two varieties of ryegrasses. Env. Sci. Pollut. R 2019, 26, 13738–13745. [Google Scholar] [CrossRef]
- Freisinger, E. Structural features specific to plant metallothioneins. JBIC J. Biol. Inorg. Chem. 2011, 16, 1035–1045. [Google Scholar] [CrossRef] [Green Version]
- Stürzenbaum, S.R.; Georgiev, O.; Morgan, A.J.; Kille, P. Cadmium Detoxification in Earthworms: From Genes to Cells. Env. Sci. Technol. 2004, 38, 6283–6289. [Google Scholar] [CrossRef]
- Shahzad, Z.; Ranwez, V.; Fizames, C.; Marquès, L.; Le Martret, B.; Alassimone, J.; Godé, C.; Lacombe, E.; Castillo, T.; Saumitou-Laprade, P.; et al. Plant Defensin type 1 (PDF1): Protein promiscuity and expression variation within the Arabidopsis genus shed light on zinc tolerance acquisition in Arabidopsis halleri. New Phytol. 2013, 200, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Yang, Y.; Gu, T.; Wu, Z.; Zhang, Z. The Arabidopsis defensin gene AtPDF2.5 mediates cadmium tolerance and accumulation. Plant Cell Environ. 2019, 42, 2681–2695. [Google Scholar] [CrossRef]
- Luo, J.; Gu, T.; Yang, Y.; Zhang, Z. A non-secreted plant defensin AtPDF2.6 conferred cadmium tolerance via its chelation in Arabidopsis. Plant Mol. Biol. 2019, 100, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, D.; Yue, N.; Song, H.; Luo, J.; Zhang, Z. PDF1.5 Enhances Adaptation to Low Nitrogen Levels and Cadmium Stress. Int. J. Mol. Sci. 2021, 22, 10455. [Google Scholar] [CrossRef] [PubMed]
- Peroza, E.A.; Schmucki, R.; Güntert, P.; Freisinger, E.; Zerbe, O. The βE-Domain of Wheat Ec-1 Metallothionein: A Metal-Binding Domain with a Distinctive Structure. J. Mol. Biol. 2009, 387, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hu, H.; Song, M.; Tan, J.; Huang, G.; Zuo, J. Preparation, characterization, and Cd (II) sorption of/on cysteine-montmorillonite composites synthesized at various pH. Env. Sci. Pollut. R 2020, 27, 10599–10606. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Donghwan, S.; Song, W.Y.; Inhwan, H.; Youngsook, L. Arabidopsis metallothioneins 2a and 3 enhance resistance to cadmium when expressed in Vicia faba guard cells. Plant Mol. Biol. 2004, 54, 805–815. [Google Scholar] [CrossRef]
- Hegelund, J.N.; Schiller, M.; Kichey, T.; Hansen, T.H.; Pedas, P.; Husted, S.; Schjoerring, J.K. Barley metallothioneins: MT3 and MT4 are localized in the grain aleurone layer and show differential zinc binding. Plant Physiol. 2012, 159, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Pirzadeh, S.; Shahpiri, A. Functional characterization of a type 2 metallothionein isoform (OsMTI-2b) from rice. Int. J. Biol. Macromol. 2016, 88, 491–496. [Google Scholar] [CrossRef] [Green Version]
- .Rono, J.K.; Wang, L.L.; Wu, X.C.; Cao, H.W.; Zhao, Y.N.; Khan, I.U.; Yang, Z.M. Identification of a new function of metallothionein-like gene OsMT1e for cadmium detoxification and potential phytoremediation. Chemosphere 2021, 265, 129136. [Google Scholar] [CrossRef]
- Blumwald, E.; Poole, R.J. Kinetics of Ca/H Antiport in Isolated Tonoplast Vesicles from Storage Tissue of Beta vulgaris L. Plant Physiol. 1986, 80, 727–731. [Google Scholar] [CrossRef] [Green Version]
- Mei, H.; Zhao, J.; Pittman, J.K.; Lachmansingh, J.; Park, S.; Hirschi, K.D. In planta regulation of the Arabidopsis Ca2+/H+ antiporter CAX1. J. Exp. Bot. 2007, 58, 3419–3427. [Google Scholar] [CrossRef]
- Hirschi, K.D.; Korenkov, V.D.; Wilganowski, N.L.; Wagner, G.J. Expression of arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 2000, 124, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Korenkov, V.; Hirschi, K.; Crutchfield, J.D.; Wagner, G.J. Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 2007, 226, 1379–1387. [Google Scholar] [CrossRef]
- Zou, W.; Chen, J.; Meng, L.; Chen, D.; He, H.; Ye, G. The Rice Cation/H(+) Exchanger Family Involved in Cd Tolerance and Transport. Int. J. Mol. Sci. 2021, 22, 8186. [Google Scholar] [CrossRef]
- Shigaki, T.; Barkla, B.J.; Miranda-Vergara, M.C.; Zhao, J.; Pantoja, O.; Hirschi, K.D. Identification of a Crucial Histidine Involved in Metal Transport Activity in the Arabidopsis Cation/H+ Exchanger CAX1*. J. Biol. Chem. 2005, 280, 30136–30142. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, T.; Maeshima, M. Residues in Internal Repeats of the Rice Cation/H+ Exchanger Are Involved in the Transport and Selection of Cations*. J. Biol. Chem. 2004, 279, 812–819. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Song, W.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Fu, S.; Huang, J.; Li, L.; Long, Y.; Wei, Q.; Wang, Z.; Chen, Z.; Xia, J. The tonoplast-localized transporter OsABCC9 is involved in cadmium tolerance and accumulation in rice. Plant Sci. 2021, 307, 110894. [Google Scholar] [CrossRef]
- He, G.; Tian, W.; Qin, L.; Meng, L.; Wu, D.; Huang, Y.; Li, D.; Zhao, D.; He, T. Identification of novel heavy metal detoxification proteins in Solanum tuberosum: Insights to improve food security protection from metal ion stress. Sci. Total Env. 2021, 779, 146197. [Google Scholar] [CrossRef]
- Oldham, M.L.; Davidson, A.L.; Chen, J. Structural insights into ABC transporter mechanism. Curr. Opin. Struc. Biol. 2008, 18, 726–733. [Google Scholar] [CrossRef] [Green Version]
- Dawson, R.J.P.; Hollenstein, K.; Locher, K.P. Uptake or extrusion: Crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 2007, 65, 250–257. [Google Scholar] [CrossRef]
- Hofmann, S.; Januliene, D.; Mehdipour, A.R.; Thomas, C.; Stefan, E.; Brüchert, S.; Kuhn, B.T.; Geertsma, E.R.; Hummer, G.; Tampé, R.; et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 2019, 571, 580–583. [Google Scholar] [CrossRef]
- Stefan, E.; Hofmann, S.; Tampé, R. A single power stroke by ATP binding drives substrate translocation in a heterodimeric ABC transporter. Elife 2020, 9, e55943. [Google Scholar] [CrossRef]
- Kim, D.; Gustin, J.L.; Lahner, B.; Persans, M.W.; Baek, D.; Yun, D.; Salt, D.E. The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J. 2004, 39, 237–251. [Google Scholar] [CrossRef]
- Li, D.; He, G.; Tian, W.; Saleem, M.; Huang, Y.; Meng, L.; Wu, D.; He, T. Comparative and Systematic Omics Revealed Low Cd Accumulation of Potato StMTP9 in Yeast: Suggesting a New Mechanism for Heavy Metal Detoxification. Int. J. Mol. Sci. 2021, 22, 10478. [Google Scholar] [CrossRef]
- Das, N.; Bhattacharya, S.; Maiti, M.K. Enhanced cadmium accumulation and tolerance in transgenic tobacco overexpressing rice metal tolerance protein gene OsMTP1 is promising for phytoremediation. Plant Physiol. Bioch. 2016, 105, 297–309. [Google Scholar] [CrossRef]
- Ram, H.; Kaur, A.; Gandass, N.; Singh, S.; Deshmukh, R.; Sonah, H.; Sharma, T.R. Molecular characterization and expression dynamics of MTP genes under various spatio-temporal stages and metal stress conditions in rice. PLoS ONE 2019, 14, e217360. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Saier, J.M.H. A Novel Family of Ubiquitous Heavy Metal Ion Transport Proteins. J. Membr. Biol. 1997, 156, 99–103. [Google Scholar] [CrossRef]
- Keren-Khadmy, N.; Zeytuni, N.; Kutnowski, N.; Perriere, G.; Monteil, C.; Zarivach, R. From conservation to structure, studies of magnetosome associated cation diffusion facilitators (CDF) proteins in Proteobacteria. PLoS ONE 2020, 15, e231839. [Google Scholar] [CrossRef]
- Zeytuni, N.; Uebe, R.; Maes, M.; Davidov, G.; Baram, M.; Raschdorf, O.; Nadav-Tsubery, M.; Kolusheva, S.; Bitton, R.; Goobes, G.; et al. Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain. PLoS ONE 2014, 9, e92141. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Chai, J.; Fu, D. Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct Mol. Biol. 2009, 16, 1063–1067. [Google Scholar] [CrossRef]
- Uebe, R.; Junge, K.; Henn, V.; Poxleitner, G.; Katzmann, E.; Plitzko, J.M.; Zarivach, R.; Kasama, T.; Wanner, G.; Pósfai, M.; et al. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol. Microbiol. 2011, 82, 818–835. [Google Scholar] [CrossRef]
- Erbasol, I.; Bozdag, G.O.; Koc, A.; Pedas, P.; Karakaya, H.C. Characterization of two genes encoding metal tolerance proteins from Beta vulgaris subspecies maritima that confers manganese tolerance in yeast. Biometals 2013, 26, 795–804. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Fu, D. Selective Metal Binding to a Membrane-embedded Aspartate in the Escherichia coli Metal Transporter YiiP (FieF)*. J. Biol. Chem. 2005, 280, 33716–33724. [Google Scholar] [CrossRef] [Green Version]
- Migeon, A.; Blaudez, D.; Wilkins, O.; Montanini, B.; Campbell, M.M.; Richaud, P.; Thomine, S.; Chalot, M. Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cell Mol. Life Sci. 2010, 67, 3763–3784. [Google Scholar] [CrossRef]
- Qin, Q.; Li, X.; Zhuang, J.; Weng, L.; Liu, W.; Tai, P. Long-distance transport of cadmium from roots to leaves of Solanum melongena. Ecotoxicology 2015, 24, 2224–2232. [Google Scholar] [CrossRef]
- Lubkowitz, M.A.; Hauser, L.; Breslav, M.; Naider, F.; Becker, J.M. An oligopeptide transport gene from Candida albicans. Microbiology 1997, 143, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; He, G.; Tian, W.; Saleem, M.; Li, D.; Huang, Y.; Meng, L.; He, Y.; Liu, Y.; He, T. OPT gene family analysis of potato (Solanum tuberosum) responding to heavy metal stress: Comparative omics and co-expression networks revealed the underlying core templates and specific response patterns. Int. J. Biol. Macromol. 2021, 188, 892–903. [Google Scholar] [CrossRef]
- Lubkowitz, M. The Oligopeptide Transporters: A Small Gene Family with a Diverse Group of Substrates and Functions? Mol. Plant 2011, 4, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Wongkaew, A.; Asayama, K.; Kitaiwa, T.; Nakamura, S.; Kojima, K.; Stacey, G.; Sekimoto, H.; Yokoyama, T.; Ohkama-Ohtsu, N. AtOPT6 Protein Functions in Long-Distance Transport of Glutathione in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1443–1451. [Google Scholar] [CrossRef] [Green Version]
- Cagnac, O.; Bourbouloux, A.; Chakrabarty, D.; Zhang, M.; Delrot, S. AtOPT6 transports glutathione derivatives and is induced by primisulfuron. Plant Physiol. 2004, 135, 1378–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Xie, Q.; Jobe, T.O.; Kau, A.R.; Wang, C.; Li, Y.; Qiu, B.; Wang, Q.; Mendoza-Cózatl, D.G.; Schroeder, J.I. Identification of AtOPT4 as a Plant Glutathione Transporter. Mol. Plant 2016, 9, 481–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osawa, H.; Stacey, G.; Gassmann, W. ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochem. J. 2006, 393, 267–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, S.; Wiles, A.M.; Sharp, J.S.; Naider, F.R.; Becker, J.M.; Stacey, G. An oligopeptide transporter gene family in Arabidopsis. Plant Physiol. 2002, 128, 21–29. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Qin, L.; Tian, W.; Meng, L.; He, T.; Zhao, D. Heavy Metal Transporters-Associated Proteins in S. tuberosum: Genome-Wide Identification, Comprehensive Gene Feature, Evolution and Expression Analysis. Genes-Basel 2020, 11, 1269. [Google Scholar] [CrossRef]
- Fang, X.; Wang, L.; Deng, X.; Wang, P.; Ma, Q.; Nian, H.; Wang, Y.; Yang, C. Genome-wide characterization of soybean P 1B -ATPases gene family provides functional implications in cadmium responses. BMC Genom. 2016, 17, 376. [Google Scholar] [CrossRef] [Green Version]
- Verret, F.; Gravot, A.; Auroy, P.; Leonhardt, N.; David, P.; Nussaume, L.; Vavasseur, A.; Richaud, P. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. Febs. Lett. 2004, 576, 306–312. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.K.E.; Cobbett, C.S. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 2009, 181, 71–78. [Google Scholar] [CrossRef]
- Takahashi, R.; Bashir, K.; Ishimaru, Y.; Nishizawa, N.K.; Nakanishi, H. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal. Behav. 2012, 7, 1605–1607. [Google Scholar] [CrossRef]
- Smith, A.T.; Barupala, D.; Stemmler, T.L.; Rosenzweig, A.C. A new metal binding domain involved in cadmium, cobalt and zinc transport. Nat. Chem. Biol. 2015, 11, 678–684. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.T.; Smith, K.P.; Rosenzweig, A.C. Diversity of the metal-transporting P1B-type ATPases. J. Biol. Inorg. Chem. JBIC A Publ. Soc. Biol. Inorg. Chem. 2014, 19, 947–960. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.T.; Ross, M.O.; Hoffman, B.M.; Rosenzweig, A.C. Metal Selectivity of a Cd-, Co-, and Zn-Transporting P(1B)-type ATPase. Biochemistry-Us 2017, 56, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Daghino, S.; Di Vietro, L.; Petiti, L.; Martino, E.; Dallabona, C.; Lodi, T.; Perotto, S. Yeast expression of mammalian Onzin and fungal FCR1 suggests ancestral functions of PLAC8 proteins in mitochondrial metabolism and DNA repair. Sci. Rep-Uk 2019, 9, 6629. [Google Scholar] [CrossRef]
- Song, W.; Hörtensteiner, S.; Tomioka, R.; Lee, Y.; Martinoia, E. Common functions or only phylogenetically related? The large family of PLAC8 motif-containing/PCR genes. Mol. Cells 2011, 31, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Choi, K.; Alexis, D.A.; Martinoia, E.; Lee, Y. Brassica juncea plant cadmium resistance 1 protein (BjPCR1) facilitates the radial transport of calcium in the root. Proc. Natl. Acad. Sci. USA 2011, 108, 19808–19813. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Simmons, C.R. Cell number counts—The fw2.2 and CNR genes and implications for controlling plant fruit and organ size. Plant Sci. 2011, 181, 1–7. [Google Scholar] [CrossRef]
- Song, W.; Choi, K.S.; Kim, D.Y.; Geisler, M.; Park, J.; Vincenzetti, V.; Schellenberg, M.; Kim, S.H.; Lim, Y.P.; Noh, E.W.; et al. Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 2010, 22, 2237–2252. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Gao, X.; Zhao, J.; Zhang, J.; Chen, S.; Lu, L. Plant Cadmium Resistance 2 (SaPCR2) Facilitates Cadmium Efflux in the Roots of Hyperaccumulator Sedum alfredii Hance. Front. Plant Sci. 2020, 11, 568887. [Google Scholar] [CrossRef]
- Song, W.; Martinoia, E.; Lee, J.; Kim, D.; Kim, D.; Vogt, E.; Shim, D.; Choi, K.S.; Hwang, I.; Lee, Y. A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 2004, 135, 1027–1039. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Tan, H.; Han, J.; Zhang, Y.; He, X.; Ding, Y.; Chen, Z.; Zhu, C. A novel family of PLAC8 motif-containing/PCR genes mediates Cd tolerance and Cd accumulation in rice. Env. Sci. Eur. 2019, 31, 82. [Google Scholar] [CrossRef]
- Dhara, A.; Raichaudhuri, A. ABCG transporter proteins with beneficial activity on plants. Phytochemistry 2021, 184, 112663. [Google Scholar] [CrossRef]
- Kim, D.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.; Li, R.; et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef]
- Song, W.; Lee, H.; Jin, S.; Ko, D.; Martinoia, E.; Lee, Y.; An, G.; Ahn, S. Rice PCR1 influences grain weight and Zn accumulation in grains. Plant Cell Environ. 2015, 38, 2327–2339. [Google Scholar] [CrossRef]
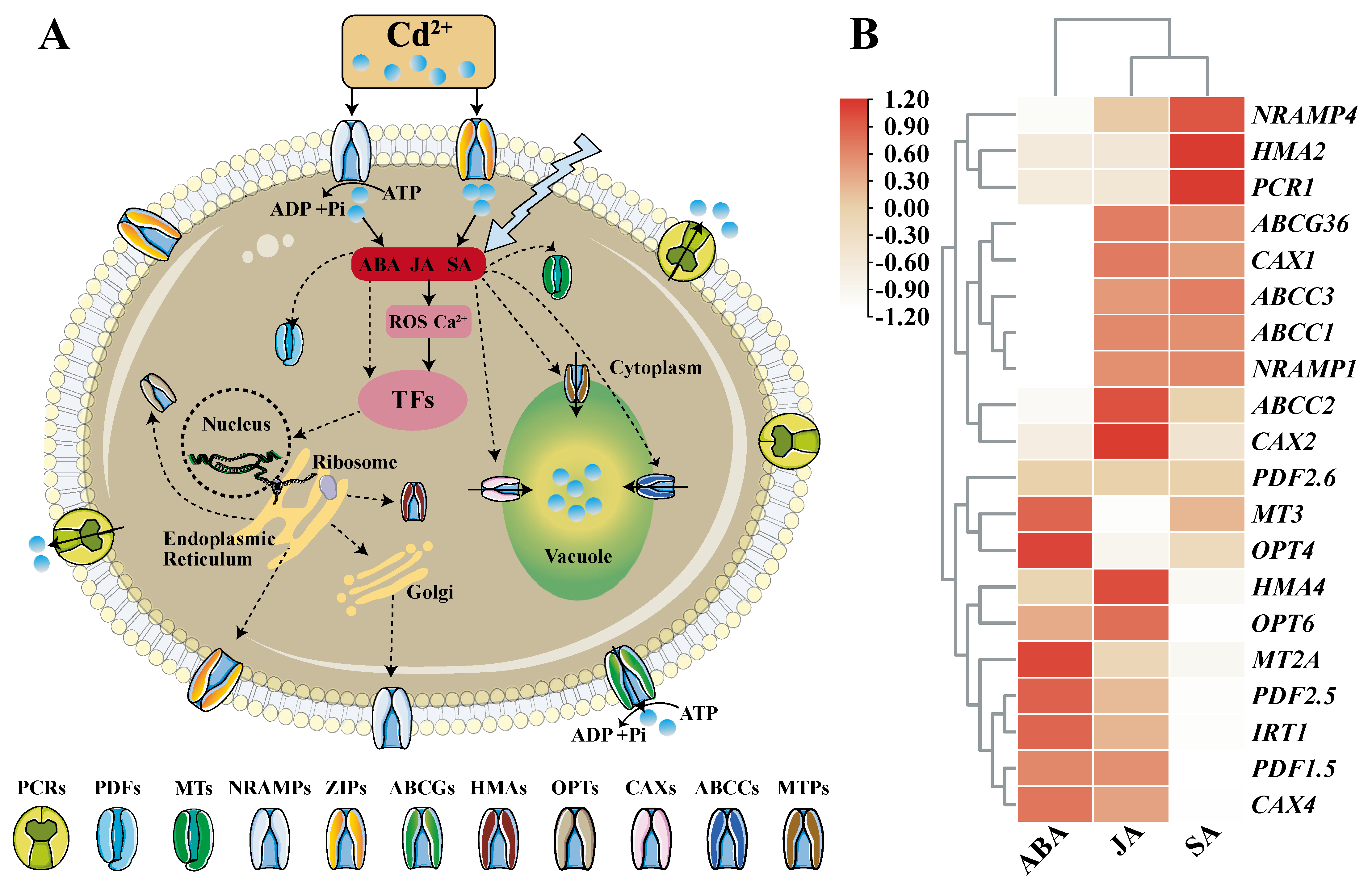
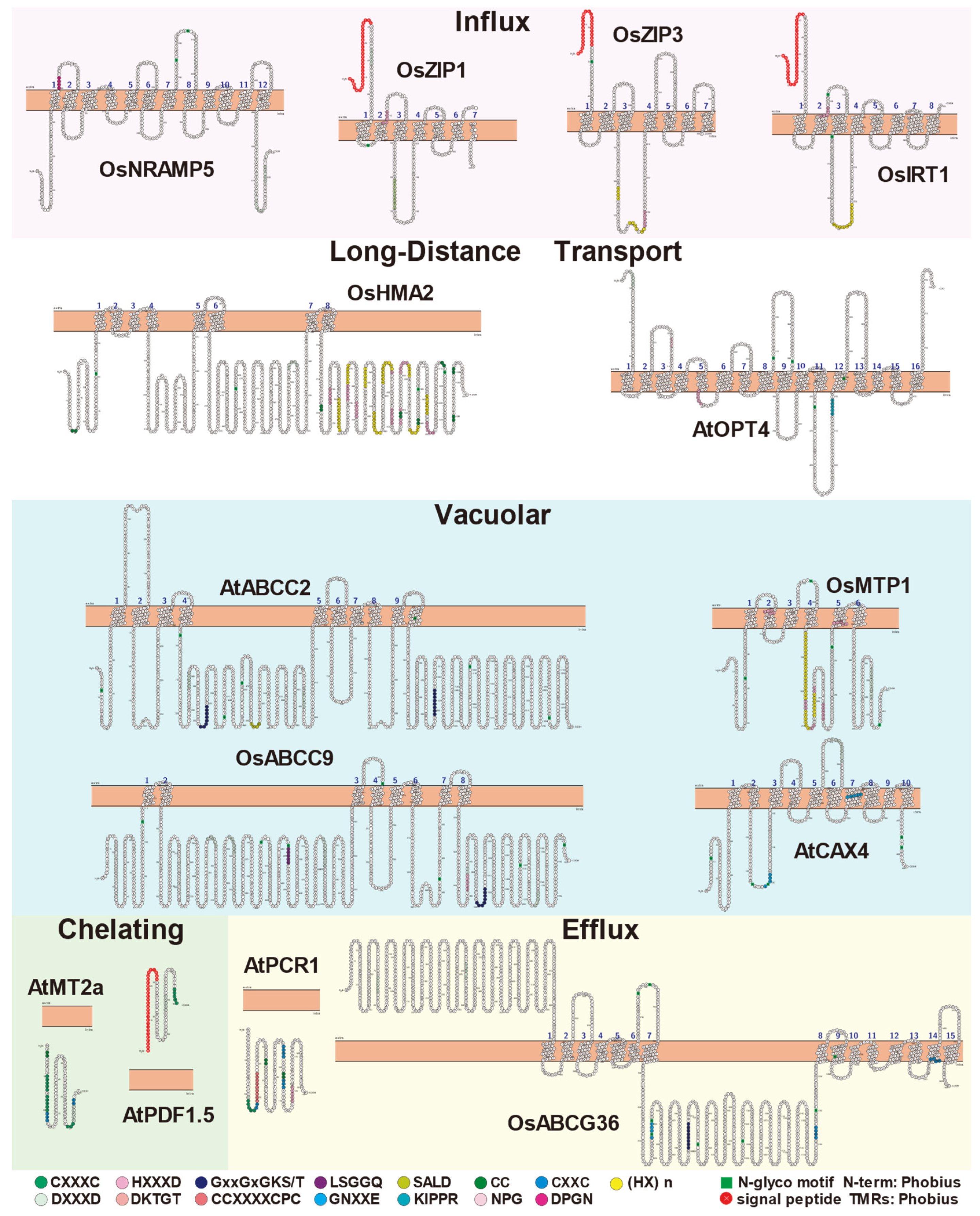
| The Key Proteins | Tissue Specific Expression | Subcellular Localization | Structural Features | References |
|---|---|---|---|---|
| AtNRAMP1 | Mainly expressed in the root | Cell membrane | 12 TMDs, DPGN near to TMD1. | [53,54] |
| OsNRAMP1 | Mainly expressed in the mature zone of the root | Cell membrane | 12 TMDs, DPGN near to TMD1. | [32,55] |
| OsNRAMP5 | Mainly expressed in the root | Cell membrane | 10 TMDs, DPGN is near to TMD1. There is a Asparticacid residue (DXXXD, X= any amino acid) at the C-terminus. | [31] |
| AtIRT1 | Root epidermal cells | Cell membrane | 8 TMDs; there is a signal peptide at the N-terminus, histidine, Asparticacid residues (HXXXD, X= any amino acid) on TMD2, and (HX)n between TMD3 and TMD4. | [40] |
| OsIRT1 | Mainly expressed in the root | Cell membrane | 8 TMDs; there is a signal peptide at the N-terminus, HXXXD residues on TMD2, and the (HX)n residues between TMD3 and TMD4. | [56] |
| OsIRT2 | Mainly expressed in the root | Cell membrane | There are 8 TMDs with signal peptide and DXXXD residues at the N-terminus; the (HX)n residues between TMD3-TMD4. | [56] |
| OsZIP1 | Mainly expressed in the root | Cell membrane | 8 TMDs, with signal peptide and DXXXD residues at the N-terminus; HXXXD residues on TMD2, and the (HX)n residues between TMD3-TMD4. | [41] |
| OsZIP3 | Xylem parenchyma cells | Cell membrane | There are 7 TMDs, there is a signal peptide at the N-terminus, and there are (HX)n residues and HXXXD conserved residues between TMD3-TMD4. | [41,57] |
| The Key Proteins | Tissue Specific Expression | Subcellular Localization | Structural Features | References |
|---|---|---|---|---|
| AtMT2a | / | Cytoplasm | CXCXXXCXC and DXXXD are distributed at the C end, without TMDs. | [75] |
| AtMT3 | / | Cytoplasm | CXCXXXCXCXXXCXC is distributed at the C-terminus and has no TMDs. | [75,76] |
| OsMTI-1b | / | Cytoplasm, Nucleus | CXCXXXCXCXXXCXC is distributed at the C-terminus and has no TMDs. | [62] |
| OsMTI-2b | Highly expressed in rice stems | Cytoplasm | N-terminal has 8 cysteine residues arranged in CC, CXC and CXXC residues. | [77] |
| OsMT1e | Expressed in roots at all developmental stages | Nucleus | CXCXXXCXCXXCXCXCXCX is distributed at the N-terminal and has no TMDs. | [78] |
| OsMT-3a | / | Cytoplasm | CXCXXXCXCXXCXCXCXCX is distributed at the C-terminus and has no TMDs. | [59,64] |
| AtPDF1.5 | Mainly expressed in knots and peels | Cell wall and cytoplasm | There is a signal peptide at the N-terminus, a cysteine-rich domain, and a DXXXD residue. | [72] |
| AtPDF2.5 | mainly expressed in root vascular bundle | Cell wall | Cysteine-rich domain and secretion signal peptide. | [70] |
| AtPDF2.6 | mainly expressed in root vascular bundle | Cell wall | There is a signal peptide at the N-terminus, a cysteine-rich domain. | [71] |
| The Key Proteins | Tissue Specific Expression | Subcellular Localization | Structural Features | References | |
|---|---|---|---|---|---|
| AtCAX2 | / | Tonoplast | 8 TMDs; with GNXXE residue on TMD2 and TMD7. | [81] | |
| AtCAX4 | Mainly expressed in the root | Tonoplast | 10 TMDs; a GNXXE residue on TMD7, and the DXXXD residues between TMD5-6. | [82] | |
| OsCAX1a | expressed at a high level in flowering spikelet | Vacuole | 10 TMDs; GNXXE residues on TMD3 and TMD8, and the DXXXD residues between TMD6-TMD7. | [83] | |
| OsCAX1c | Strongly expressed in leaves | Tonoplast | 10 TMDs; GNXXE residues exist on TMD2, TMD, and DXXXD residues near TMD6. | [83] | |
| AtABCC1 | Mainly expressed in the root | Vacuole | 15 TMDs, nucleotide binding site at the C-terminus, cysteine-rich loop, Q-loop, Walker B residue. | [86] | |
| AtABCC2 | Mainly expressed in the root | Vacuole | 10 TMDs, cysteine-rich loop, Walker B residue, DXXXD residues exist between TMD4-TMD5. | [86] | |
| AtABCC3 | / | Tonoplast | 15 TMDs, cysteine-rich loop, Walker B residue, HXXXD residues near TMD3, and DXXXD residues between TMD9-TMD10. | [87] | |
| OsABCC9 | Significantly induced by Cd treatment in roots | Tonoplast | 8 TMDs, cysteine-rich loop, Q-loop, Walker B residue, and HXXXD and DXXXD residues at the N-terminus. | [88] | |
| OsMTP1 | Significantly induced by Cd treatment in roots | Vacuole | The characteristic residue of CDF (SLAILTDAAHLLSDVAA), 6 TMDs and histidine-rich regions, HXXXD residues exist on and/or near TMD2, TMD5. | [96] | |
| The Key Proteins | Tissue Specific Expression | Subcellular Localization | Structural Features | References |
|---|---|---|---|---|
| AtOPT6 | It is preferentially expressed in the vascular system of rosette leaves, stems and roots | Cell membrane | 15 TMDs, including NPG (near TMD3), KIPPR (near TMD12) residues, HXXXD residues near TMD5, and DXXXD residues at the N-terminal. | [110,111] |
| AtOPT4 | Mainly expressed in the vascular bundles | Cell membrane | 14 TMDs, including NPG (tMD2-TMD3), KIPPR (TMD10-TMD11) residues. | [112,113] |
| AtHMA4 | Mainly expressed in the vascular tissues of roots, stems and leaves | Cell membrane | There are 6 CC pairs, (HX)n, DXXXD residues and two repeats of the GXDSGCCGXKSQQPHQHEXQ sequence at the C-terminus. | [117,118] |
| AtHMA2 | Mainly expressed in the vascular tissues of roots, stems and leaves | Cell membrane | 8 TMDs, an extended C-terminus, potential metal binding residues (13 cysteine pairs and 11 histidine sequences). | [118] |
| OsHMA2 | Root | Cell membrane, Nucleus | 8 TMDs, CC pairs, (HX)n, HXXXD residues are widely distributed at the C-terminus. | [119] |
| The Key Proteins | Tissue Specific Expression | Subcellular Localization | Structural Features | References |
|---|---|---|---|---|
| AtPCR1 | Expressed in all organizations | Plasma membrane | No TMDs, α-membrane protein helix, CCXXXXCPC motif, HXXXD residue | [129] |
| OsPCR1 | Mainly expressed in root of seedling and internode I and II of reproductive stage, but also in spikelet | Plasma membrane | No TMDs, including CCXXXXCPC motif, HXXXD residue | [130,134] |
| OsPCR3 | Root | Plasma membrane | No TMDs, a CCXXXXCPC residue | [130] |
| AtABCG36 | The expression level was highest in root hair and epidermal cells | Plasma membrane | 15 TMDs, including GxxGxGKS/T and DXXXD residues between TMD7-TMD8 | [132] |
| OsABCG36 | It was highly expressed in roots under Cd stress | Plasma membrane | 15 TMDs with DXXXD residues at the N-terminus, GxxGxGKS/T, GNXXE, CXXC residues were found between and TMD7-TMD8, CXXC residues were found on TMD14 | [133] |
| Protein ID | Function Type | Tissue Specific | Summary of Metalloproteins with Research Value |
|---|---|---|---|
| OsNRAMP5 | Influx protein | Cell membrane in roots | The main Cd uptake and transporter in rice; DPGN motis are only found in NRAMP. |
| OsZIP3 | Influx protein | Cell membrane of xylem parenchyma | The appearance of the N-terminal specific signal peptide may induce its expression in xylem parenchyma cells and diversify the functions of members of the ZIP family. |
| AtPDF1.5 | Chelating protein | Cell wall and cytoplasm in nodes and peels | The N-terminal signal peptide may mediate its secretion in the cell wall, chelate Cd in the cell wall, and is rich in cysteine-related motifs. |
| OsMT1e | Chelating protein | Nucleus in roots | Expressed in the nucleus, indicating that members of the MT family seem to be involved in the regulation of gene transcription, and are rich in cysteine-related motifs. |
| AtOPT4 | Long-distance transport protein | Cell membrane in vascular bundle | Participate in the long-distance transport of the GSH-Cd complex, no residues related to cysteine, including NPG and KIPPR motifs unique to this family. |
| OsHMA2 | Long-distance transport protein | Cell membrane and Nucleus in the roots | Participate in the long-distance transportation of Cd, which may be because the C-terminal cysteine-rich motifs can directly bind to Cd. Expressed in the nucleus, they may be involved in gene transcription regulation. |
| AtPCR1 | Efflux protein | Plasma membrane of all tissues | It is a membrane protein but does not contain a TMD. It may excrete Cd ions to the extracellular space through exocytosis, and contains CCXXXXCPC motifs. |
| OsABCG36 | Efflux protein | Plasma membrane of roots | It is highly expressed in roots under Cd stress and contains motifs related to cysteine. Exporting Cd or Cd conjugates. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; He, T.; Saleem, M.; He, G. Metalloprotein-Specific or Critical Amino Acid Residues: Perspectives on Plant-Precise Detoxification and Recognition Mechanisms under Cadmium Stress. Int. J. Mol. Sci. 2022, 23, 1734. https://doi.org/10.3390/ijms23031734
Li D, He T, Saleem M, He G. Metalloprotein-Specific or Critical Amino Acid Residues: Perspectives on Plant-Precise Detoxification and Recognition Mechanisms under Cadmium Stress. International Journal of Molecular Sciences. 2022; 23(3):1734. https://doi.org/10.3390/ijms23031734
Chicago/Turabian StyleLi, Dandan, Tengbing He, Muhammad Saleem, and Guandi He. 2022. "Metalloprotein-Specific or Critical Amino Acid Residues: Perspectives on Plant-Precise Detoxification and Recognition Mechanisms under Cadmium Stress" International Journal of Molecular Sciences 23, no. 3: 1734. https://doi.org/10.3390/ijms23031734
APA StyleLi, D., He, T., Saleem, M., & He, G. (2022). Metalloprotein-Specific or Critical Amino Acid Residues: Perspectives on Plant-Precise Detoxification and Recognition Mechanisms under Cadmium Stress. International Journal of Molecular Sciences, 23(3), 1734. https://doi.org/10.3390/ijms23031734







