Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase
Abstract
1. Introduction
2. Organization and Regulation of the PNPT1 Gene
2.1. PNPT1 Gene and mRNA Features
2.2. PNPT1 Regulation and Expression in Human Tissues
3. Structure of hPNPase: A Catalytic Core Capped by an RNA Binding Pore
3.1. Protomer Domains and Quaternary Structure of hPNPase
3.2. Post-Translational Modifications and Interaction with Small Regulatory Molecules
4. hPNPase Catalytic and RNA Binding Activity
4.1. Catalytic Activity and Active Site Composition
4.2. Both the Enzyme Core and the KH-S1 Domains Contribute to RNA Binding
5. Function of hPNPase in Different Cell Compartments
5.1. hPNPase Controls mtRNA Trafficking
5.2. hPNPase Controls mtRNA Decay and Prevents mtDNA Loss
5.3. Extra-Mitochondrial hPNPase Controls Cell Proliferation and Apoptosis
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leszczyniecka, M.; DeSalle, R.; Kang, D.C.; Fisher, P.B. The origin of polynucleotide phosphorylase domains. Mol. Phylogenet. Evol. 2004, 31, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Briani, F.; Carzaniga, T.; Dehò, G. Regulation and functions of bacterial PNPase. Wiley Interdiscip. Rev. RNA 2016, 7, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Lin-Chao, S.; Chiou, N.T.; Schuster, G. The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. J. Biomed. Sci. 2007, 14, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Grunberg-Manago, M.; Ortiz, P.J.; Ochoa, S. Enzymic synthesis of polynucleotides I. polynucleotide phosphorylase of Azotobacter vinelandii. Biochim. Biophys. Acta 1956, 20, 269–285. [Google Scholar] [CrossRef]
- Hakim, A.A. Synthetic activity of polynucleotide phosphorylase from sperm. Nature 1959, 183, 334. [Google Scholar] [CrossRef]
- Yagi, K.; Ozawa, T.; Konogi, H. Occurrence of Polynucleotide Phosphorylase in Atypical Epithelioma of Rat. Nature 1959, 184, 1939. [Google Scholar] [CrossRef]
- Leszczyniecka, M.; Kang, D.-C.; Sarkar, D.; Su, Z.-Z.; Holmes, M.; Valerie, K.; Fisher, P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. USA 2002, 99, 16636–16641. [Google Scholar] [CrossRef]
- Piwowarski, J.; Grzechnik, P.; Dziembowski, A.; Dmochowska, A.; Minczuk, M.; Stepien, P.P. Human Polynucleotide Phosphorylase, hPNPase, is Localized in Mitochondria. J. Mol. Biol. 2003, 329, 853–857. [Google Scholar] [CrossRef]
- Eaton, A.; Bernier, F.P.; Goedhart, C.; Caluseriu, O.; Lamont, R.E.; Boycott, K.M.; Parboosingh, J.S.; Innes, A.M. Is PNPT1-related hearing loss ever non-syndromic? Whole exome sequencing of adult siblings expands the natural history of PNPT1-related disorders. Am. J. Med. Genet. Part A 2018, 176, 2487–2493. [Google Scholar] [CrossRef]
- Rius, R.; Van Bergen, N.J.; Compton, A.G.; Riley, L.G.; Kava, M.P.; Balasubramaniam, S.; Amor, D.J.; Fanjul-Fernandez, M.; Cowley, M.J.; Fahey, M.C.; et al. Clinical spectrum and functional consequences associated with bi-allelic pathogenic Pnpt1 variants. J. Clin. Med. 2019, 8, 2020. [Google Scholar] [CrossRef]
- Vedrenne, V.; Gowher, A.; De Lonlay, P.; Nitschke, P.; Serre, V.; Boddaert, N.; Altuzarra, C.; Mager-Heckel, A.M.; Chretien, F.; Entelis, N.; et al. Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am. J. Hum. Genet. 2012, 91, 912–918. [Google Scholar] [CrossRef]
- Leszczyniecka, M.; Su, Z.Z.; Kang, D.C.; Sarkar, D.; Fisher, P.B. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPaseold-35, a Type I interferon inducible early response gene. Gene 2003, 316, 143–156. [Google Scholar] [CrossRef]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015, 163, 1515–1526. [Google Scholar] [CrossRef]
- Wang, G.; Chen, H.W.; Oktay, Y.; Zhang, J.; Allen, E.L.; Smith, G.M.; Fan, K.C.; Hong, J.S.; French, S.W.; McCaffery, J.M.; et al. PNPASE regulates RNA import into mitochondria. Cell 2010, 142, 456–467. [Google Scholar] [CrossRef]
- Shimada, E.; Ahsan, F.M.; Nili, M.; Huang, D.; Atamdede, S.; TeSlaa, T.; Case, D.; Yu, X.; Gregory, B.D.; Perrin, B.J.; et al. PNPase knockout results in mtDNA loss and an altered metabolic gene expression program. PLoS ONE 2018, 13, e0200925. [Google Scholar] [CrossRef]
- Chen, H.-W.; Rainey, R.N.; Balatoni, C.E.; Dawson, D.W.; Troke, J.J.; Wasiak, S.; Hong, J.S.; McBride, H.M.; Koehler, C.M.; Teitell, M.A.; et al. Mammalian Polynucleotide Phosphorylase Is an Intermembrane Space RNase That Maintains Mitochondrial Homeostasis. Mol. Cell. Biol. 2006, 26, 8475–8487. [Google Scholar] [CrossRef]
- Takahashi, H.; Furukawa, T.; Yano, T.; Sato, N.; Takizawa, J.; Kurasaki, T.; Abe, T.; Narita, M.; Masuko, M.; Koyama, S.; et al. Identification of an overexpressed gene, HSPA4L, the product of which can provoke prevalent humoral immune responses in leukemia patients. Exp. Hematol. 2007, 35, 1091–1099. [Google Scholar] [CrossRef]
- Gewartowski, K.; Tomecki, R.; Muchowski, L.; Dmochowska, A.; Dzwonek, A.; Malecki, M.; Skurzak, H.; Ostrowski, J.; Stepien, P.P. Up-regulation of human PNPase mRNA by β-interferon has no effect on protein level in melanoma cell lines. Acta Biochim. Pol. 2006, 53, 179–187. [Google Scholar] [CrossRef]
- Wang, G.; Shimada, E.; Koehler, C.M.; Teitell, M.A. PNPASE and RNA trafficking into mitochondria. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 998–1007. [Google Scholar] [CrossRef]
- Feng, H.; Zhang, Y.B.; Gui, J.F.; Lemon, S.M.; Yamane, D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 2021, 17, e1009220. [Google Scholar] [CrossRef]
- Michalska, A.; Blaszczyk, K.; Wesoly, J.; Bluyssen, H.A.R. A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses. Front. Immunol. 2018, 9, 1135. [Google Scholar] [CrossRef] [PubMed]
- Pine, R.; Decker, T.; Kessler, D.S.; Levy, D.E.; Darnell, J.E. Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol. 1990, 10, 2448–2457. [Google Scholar] [CrossRef] [PubMed]
- Marchi, P.; Longhi, V.; Zangrossi, S.; Gaetani, E.; Briani, F.; Dehò, G. Autogenous regulation of Escherichia coli polynucleotide phosphorylase during cold acclimation by transcription termination and antitermination. Mol. Genet. Genom. 2007, 278, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Carzaniga, T.; Briani, F.; Zangrossi, S.; Merlino, G.; Marchi, P.; Dehò, G. Autogenous regulation of Escherichia coli polynucleotide phosphorylase expression revisited. J. Bacteriol. 2009, 191, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Jarrige, A.C.; Mathy, N.; Portier, C. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 2001, 20, 6845–6855. [Google Scholar] [CrossRef]
- Shaw, G.; Kamen, R. A conserved AU sequence from the 3’ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 1986, 46, 659–667. [Google Scholar] [CrossRef]
- Chen, C.Y.A.; Shyu, A. Bin Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2011, 2, 167–183. [Google Scholar] [CrossRef]
- Savant-Bhonsale, S.; Cleveland, D.W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 1992, 6, 1927–1939. [Google Scholar] [CrossRef]
- Von Ameln, S.; Wang, G.; Boulouiz, R.; Rutherford, M.A.; Smith, G.M.; Li, Y.; Pogoda, H.M.; Nürnberg, G.; Stiller, B.; Volk, A.E.; et al. A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am. J. Hum. Genet. 2012, 91, 919–927. [Google Scholar] [CrossRef]
- Li, Z.; Deutscher, M.P. Exoribonucleases and Endoribonucleases. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef]
- Bermüdez-Cruz, R.M.; Ramïrez, F.; Kameyama-Kawabe, L.; Montañez, C. Conserved domains in polynucleotide phosphorylase among eubacteria. Biochimie 2005, 87, 737–745. [Google Scholar] [CrossRef]
- Symmons, M.F.; Jones, G.H.; Luisi, B.F. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 2000, 8, 1215–1226. [Google Scholar] [CrossRef]
- Nurmohamed, S.; Vaidialingam, B.; Callaghan, A.J.; Luisi, B.F. Crystal Structure of Escherichia coli Polynucleotide Phosphorylase Core Bound to RNase E, RNA and Manganese: Implications for Catalytic Mechanism and RNA Degradosome Assembly. J. Mol. Biol. 2009, 389, 17–33. [Google Scholar] [CrossRef]
- Hardwick, S.W.; Gubbey, T.; Hug, I.; Jenal, U.; Luisi, B.F. Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open Biol. 2012, 2, 120028. [Google Scholar] [CrossRef]
- Lin, C.L.; Wang, Y.T.; Yang, W.Z.; Hsiao, Y.Y.; Yuan, H.S. Crystal structure of human polynucleotide phosphorylase: Insights into its domain function in RNA binding and degradation. Nucleic Acids Res. 2012, 40, 4146–4157. [Google Scholar] [CrossRef]
- Shi, Z.; Yang, W.Z.; Lin-Chao, S.; Chak, K.F.; Yuan, H.S. Crystal structure of Escherichia coli PNPase: Central channel residues are involved in processive RNA degradation. RNA 2008, 14, 2361–2371. [Google Scholar] [CrossRef]
- Matus-Ortega, M.E.; Regonesi, M.E.; Piña-Escobedo, A.; Tortora, P.; Dehò, G.; García-Mena, J. The KH and S1 domains of Escherichia coli polynucleotide phosphorylase are necessary for autoregulation and growth at low temperature. Biochim. Biophys. Acta 2007, 1769, 194–203. [Google Scholar] [CrossRef]
- García-Mena, J.; Das, A.; Sánchez-Trujillo, A.; Portier, C.; Montanez, C. A novel mutation in the KH domain of polynucleotide phosphorylase affects autoregulation and mRNA decay in Escherichia coli. Mol. Microbiol. 1999, 33, 235–248. [Google Scholar] [CrossRef]
- Jarrige, A.-C.; Bréchemier-Baey, D.; Mathy, N.; Duché, O.; Portier, C. Mutational Analysis of Polynucleotide Phosphorylase from Escherichia coli. J. Mol. Biol. 2002, 321, 397–409. [Google Scholar] [CrossRef]
- Symmons, M.F.; Williams, M.G.; Luisi, B.F.; Jones, G.H.; Carpousis, A.J. Running rings around RNA: A superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002, 27, 11–18. [Google Scholar] [CrossRef]
- Dendooven, T.; Sinha, D.; Roeselová, A.; Cameron, T.A.; De Lay, N.R.; Luisi, B.F.; Bandyra, K.J. A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. Mol. Cell 2021, 81, 2901–2913.e5. [Google Scholar] [CrossRef] [PubMed]
- Carzaniga, T.; Mazzantini, E.; Nardini, M.; Regonesi, M.E.; Greco, C.; Briani, F.; De Gioia, L.; Dehò, G.; Tortora, P. A conserved loop in polynucleotide phosphorylase (PNPase) essential for both RNA and ADP/phosphate binding. Biochimie 2014, 97, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Januszyk, K.; Lima, C.D. Structural components and architectures of RNA exosomes. In RNA Exosomes; Springer: Berlin, Germany, 2010. [Google Scholar] [CrossRef]
- Raijmakers, R.; Vree Egberts, W.; Van Venrooij, W.J.; Pruijn, G.J.M. Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J. Mol. Biol. 2002, 323, 653–663. [Google Scholar] [CrossRef]
- Lorentzen, E.; Walter, P.; Fribourg, S.; Evguenieva-Hackenberg, E.; Klug, G.; Conti, E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005, 12, 575–581. [Google Scholar] [CrossRef]
- Sikorska, N.; Zuber, H.; Gobert, A.; Lange, H.; Gagliardi, D. RNA degradation by the plant RNA exosome involves both phosphorolytic and hydrolytic activities. Nat. Commun. 2017, 8, 2162. [Google Scholar] [CrossRef]
- Dziembowski, A.; Lorentzen, E.; Conti, E.; Séraphin, B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2006, 14, 15–22. [Google Scholar] [CrossRef]
- Liu, Q.; Greimann, J.C.; Lima, C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 2006, 127, 1223–1237. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Yu, Y.L.; Chou, R.H.; Wu, C.H.; Wang, Y.N.; Chang, W.J.; Tseng, Y.J.; Chang, W.C.; Lai, C.C.; Lee, H.J.; Huo, L.; et al. Nuclear EGFR Suppresses Ribonuclease Activity of Polynucleotide Phosphorylase through DNAPK-mediated Phosphorylation at Serine 776. J. Biol. Chem. 2012, 287, 31015. [Google Scholar] [CrossRef]
- Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.A. Cyclic di-GMP Activation of Polynucleotide Phosphorylase Signal-Dependent RNA Processing. J. Mol. Biol. 2011, 407, 633–639. [Google Scholar] [CrossRef]
- Siculella, L.; Damiano, F.; Di Summa, R.; Tredici, S.M.; Alduina, R.; Gnoni, G.V.; Alifano, P. Guanosine 5’-diphosphate 3’-diphosphate (ppGpp) as a negative modulator of polynucleotide phosphorylase activity in a “rare” actinomycete. Mol. Microbiol. 2010, 77, 716–729. [Google Scholar] [CrossRef]
- Gatewood, M.L.; Jones, G.H. (p)ppGpp inhibits polynucleotide phosphorylase from Streptomyces but not from Escherichia coli and increases the stability of bulk mRNA in Streptomyces coelicolor. J. Bacteriol. 2010, 192, 4275–4280. [Google Scholar] [CrossRef]
- Del Favero, M.; Mazzantini, E.; Briani, F.; Zangrossi, S.; Tortora, P.; Dehò, G. Regulation of Escherichia coli polynucleotide phosphorylase by ATP. J. Biol. Chem. 2008, 283, 27355–27359. [Google Scholar] [CrossRef]
- Nurmohamed, S.; Vincent, H.A.; Titman, C.M.; Chandran, V.; Pears, M.R.; Du, D.; Griffin, J.L.; Callaghan, A.J.; Luisi, B.F. Polynucleotide phosphorylase activity may be modulated by metabolites in Escherichia coli. J. Biol. Chem. 2011, 286, 14315–14323. [Google Scholar] [CrossRef]
- Stone, C.M.; Butt, L.E.; Bufton, J.C.; Lourenco, D.C.; Gowers, D.M.; Pickford, A.R.; Cox, P.A.; Vincent, H.A.; Callaghan, A.J. Inhibition of homologous phosphorolytic ribonucleases by citrate may represent an evolutionarily conserved communicative link between RNA degradation and central metabolism. Nucleic Acids Res. 2017, 45, 4655–4666. [Google Scholar] [CrossRef]
- Icard, P.; Poulain, L.; Lincet, H. Understanding the central role of citrate in the metabolism of cancer cells. Biochim. Biophys. Acta 2012, 1825, 111–116. [Google Scholar] [CrossRef]
- Golzarroshan, B.; Lin, C.-L.; Li, C.-L.; Yang, W.-Z.; Chu, L.-Y.; Agrawal, S.; Yuan, H.S. Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities. Nucleic Acids Res. 2018, 46, 8630–8640. [Google Scholar] [CrossRef]
- Portnoy, V.; Palnizky, G.; Yehudai-Resheff, S.; Glaser, F.; Schuster, G. Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts. Rna 2008, 14, 297–309. [Google Scholar] [CrossRef]
- Cheng, Z.F.; Deutscher, M.P. An important role for RNase R in mRNA decay. Mol. Cell 2005, 17, 313–318. [Google Scholar] [CrossRef]
- Spickler, C.; Mackie, G.A. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 2000, 182, 2422–2427. [Google Scholar] [CrossRef]
- Andrade, J.M.; Pobre, V.; Silva, I.J.; Domingues, S.; Arraiano, C.M. The Role of 3′–5′ Exoribonucleases in RNA Degradation. Prog. Mol. Biol. Transl. Sci. 2009, 85, 187–229. [Google Scholar] [CrossRef]
- Viegas, S.C.; Matos, R.G.; Arraiano, C.M. The Bacterial Counterparts of the Eukaryotic Exosome: An Evolutionary Perspective. Methods Mol. Biol. 2020, 2062, 37–46. [Google Scholar] [CrossRef]
- Wang, D.D.H.; Shu, Z.; Lieser, S.A.; Chen, P.L.; Lee, W.H. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J. Biol. Chem. 2009, 284, 20812–20821. [Google Scholar] [CrossRef]
- Liu, X.; Fu, R.; Pan, Y.; Meza-Sosa, K.F.; Zhang, Z.; Lieberman, J. PNPT1 Release from Mitochondria during Apoptosis Triggers Decay of Poly(A) RNAs. Cell 2018, 174, 187–201.e12. [Google Scholar] [CrossRef]
- Soboll, S.; Sholz, R.; Heldt, H.W. Subcellular Metabolite Concentrations: Dependence of Mitochondrial and Cytosolic ATP Systems on the Metabolic State of Perfuse Rat Liver. Eur. J. Biochem. 1978, 87, 377–390. [Google Scholar] [CrossRef]
- Amin, N.; Peterkofsky, A. A dual mechanism for regulating cAMP levels in Escherichia coli. J. Biol. Chem. 1995, 270, 11803–11805. [Google Scholar] [CrossRef]
- Sarpel, G.; Barp, A.N.; Lubansky, H.J.; Omachi, A. Erythocyte phosphate content in Huntington’s disease. Neurosci. Lett. 1982, 31, 91–96. [Google Scholar] [CrossRef]
- Nesmeyanova, M.A. Polyphosphates and enzymes of polyphosphate metabolism in Escherichia coli. Biochemistry 2000, 65, 309–314. [Google Scholar]
- Unciuleac, M.C.; Ghosh, S.; de la Cruz, M.J.; Goldgur, Y.; Shuman, S. Structure and mechanism of Mycobacterium smegmatis polynucleotide phosphorylase. RNA 2021, 27, 959–969. [Google Scholar] [CrossRef]
- Briani, F.; Del Favero, M.; Capizzuto, R.; Consonni, C.; Zangrossi, S.; Greco, C.; De Gioia, L.; Tortora, P.; Dehò, G. Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie 2007, 89, 145–157. [Google Scholar] [CrossRef]
- Matilainen, S.; Carroll, C.J.; Richter, U.; Euro, L.; Pohjanpelto, M.; Paetau, A.; Isohanni, P.; Suomalainen, A. Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. Hum. Mol. Genet. 2017, 26, 3352–3361. [Google Scholar] [CrossRef] [PubMed]
- Dhir, A.; Dhir, S.; Borowski, L.S.; Jimenez, L.; Teitell, M.; Rötig, A.; Crow, Y.J.; Rice, G.I.; Duffy, D.; Tamby, C.; et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018, 560, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, G.; Littauer, U.Z. Deoxyadenosine diphosphate as substrate for polynucleotide phosphorylase from Escherichia coli. FEBS Lett. 1969, 4, 79–83. [Google Scholar] [CrossRef]
- Gillam, S.; Waterman, K.; Doel, M.; Smith, M. Enzymatic synthesis of deoxyribo-oiigonucleotides of defined sequence. Deoxyribo-oligonucleotide synthesis. Nucleic Acids Res. 1974, 1, 1649–1664. [Google Scholar] [CrossRef]
- Cardenas, P.P.; Carrasco, B.; Sanchez, H.; Deikus, G.; Bechhofer, D.H.; Alonso, J.C. Bacillus subtilis polynucleotide phosphorylase 3’-to-5’ DNase activity is involved in DNA repair. Nucleic Acids Res. 2009, 37, 4157–4169. [Google Scholar] [CrossRef] [PubMed]
- Carzaniga, T.; Sbarufatti, G.; Briani, F.; Dehò, G. Polynucleotide phosphorylase is implicated in homologous recombination and DNA repair in Escherichia coli. BMC Microbiol. 2017, 17, 81. [Google Scholar] [CrossRef]
- Cardenas, P.P.; Carzaniga, T.; Zangrossi, S.; Briani, F.; Garcia-Tirado, E.; Dehò, G.; Alonso, J.C. Polynucleotide phosphorylase exonuclease and polymerase activities on single-stranded DNA ends are modulated by RecN, SsbA and RecA proteins. Nucleic Acids Res. 2011, 39, 9250–9261. [Google Scholar] [CrossRef]
- Becket, E.; Tse, L.; Yung, M.; Cosico, A.; Miller, J.H. Polynucleotide phosphorylase plays an important role in the generation of spontaneous mutations in Escherichia coli. J. Bacteriol. 2012, 194, 5613–5620. [Google Scholar] [CrossRef][Green Version]
- Rath, D.; Mangoli, S.H.; Pagedar, A.R.; Jawali, N. Involvement of pnp in survival of UV radiation in Escherichia coli K-12. Microbiology 2012, 158, 1196–1205. [Google Scholar] [CrossRef]
- Bermúdez-Cruz, R.M.; García-Mena, J.; Montaez, C. Polynucleotide phosphorylase binds to ssRNA with same affinity as to ssDNA. Biochimie 2002, 84, 321–328. [Google Scholar] [CrossRef]
- Lisitsky, I.; Schuster, G. Preferential degradation of polyadenylated and polyuridinylated RNAs by the bacterial exoribonuclease polynucleotide phosphorylase. Eur. J. Biochem. 1999, 261, 468–474. [Google Scholar] [CrossRef]
- Wong, A.G.; McBurney, K.L.; Thompson, K.J.; Stickney, L.M.; Mackie, G.A. S1 and KH domains of polynucleotide phosphorylase determine the efficiency of RNA binding and autoregulation. J. Bacteriol. 2013, 195, 2021–2031. [Google Scholar] [CrossRef]
- Valverde, R.; Edwards, L.; Regan, L. Structure and function of KH domains. FEBS J. 2008, 275, 2712–2726. [Google Scholar] [CrossRef]
- Sarkar, D.; Park, E.S.; Emdad, L.; Randolph, A.; Valerie, K.; Fisher, P.B. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol. Cell. Biol. 2005, 25, 7333–7343. [Google Scholar] [CrossRef]
- Chen, H.W.; Koehler, C.M.; Teitell, M.A. Human polynucleotide phosphorylase: Location matters. Trends Cell Biol. 2007, 17, 600–608. [Google Scholar] [CrossRef]
- Rainey, R.N.; Glavin, J.D.; Chen, H.-W.; French, S.W.; Teitell, M.A.; Koehler, C.M. A New Function in Translocation for the Mitochondrial i-AAA Protease Yme1: Import of Polynucleotide Phosphorylase into the Intermembrane Space. Mol. Cell. Biol. 2006, 26, 8488–8497. [Google Scholar] [CrossRef]
- French, S.W.; Dawson, D.W.; Chen, H.W.; Rainey, R.N.; Sievers, S.A.; Balatoni, C.E.; Wong, L.; Troke, J.J.; Nguyen, M.T.N.; Koehler, C.M.; et al. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer Lett. 2007, 248, 198–210. [Google Scholar] [CrossRef]
- Khaw, S.L.; Min-Wen, C.; Koh, C.G.; Lim, B.; Shyh-Chang, N. Oocyte Factors Suppress Mitochondrial Polynucleotide Phosphorylase to Remodel the Metabolome and Enhance Reprogramming. Cell Rep. 2015, 12, 1080–1088. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Daley, G.Q.; Cantley, L.C. Stem cell metabolism in tissue development and aging. Development 2013, 140, 2535–2547. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Hallas, C.; Pekarsky, Y.; Itoyama, T.; Varnum, J.; Bichi, R.; Rothstein, J.L.; Croce, C.M. Genomic analysis of human and mouse TCL1 loci reveals a complex of tightly clustered genes. Proc. Natl. Acad. Sci. USA 1999, 96, 14418–14423. [Google Scholar] [CrossRef]
- Narducci, M.G.; Fiorenza, M.T.; Kang, S.M.; Bevilacqua, A.; Di Giacomo, M.; Remotti, D.; Picchio, M.C.; Fidanza, V.; Cooper, M.D.; Croce, C.M.; et al. TCL1 participates in early embryonic development and is overexpressed in human seminomas. Proc. Natl. Acad. Sci. USA 2002, 99, 11712–11717. [Google Scholar] [CrossRef]
- Gammage, P.A.; Moraes, C.T.; Minczuk, M. Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR-Ized. Trends Genet. 2018, 34, 101–110. [Google Scholar] [CrossRef]
- Wang, G.; Shimada, E.; Zhang, J.; Hong, J.S.; Smith, G.M.; Teitell, M.A.; Koehler, C.M. Correcting human mitochondrial mutations with targeted RNA import. Proc. Natl. Acad. Sci. USA 2012, 109, 4840–4845. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, P.; Zheng, Q.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Tong, T.; et al. Mitochondrial Trafficking and Processing of Telomerase RNA TERC. Cell Rep. 2018, 24, 2589–2595. [Google Scholar] [CrossRef]
- Jeandard, D.; Smirnova, A.; Tarassov, I.; Barrey, E.; Smirnov, A.; Entelis, N. Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells 2019, 8, 286. [Google Scholar] [CrossRef]
- Smirnov, A.; Comte, C.; Mager-Heckel, A.M.; Addis, V.; Krasheninnikov, I.A.; Martin, R.P.; Entelis, N.; Tarassov, I. Mitochondrial enzyme rhodanese is essential for 5S ribosomal RNA import into human mitochondria. J. Biol. Chem. 2010, 285, 30792–30803. [Google Scholar] [CrossRef]
- Borowski, L.S.; Dziembowski, A.; Hejnowicz, M.S.; Stepien, P.P.; Szczesny, R.J. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013, 41, 1223–1240. [Google Scholar] [CrossRef]
- Chujo, T.; Ohira, T.; Sakaguchi, Y.; Goshima, N.; Nomura, N.; Nagao, A.; Suzuki, T. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012, 40, 8033–8047. [Google Scholar] [CrossRef]
- Rhee, H.W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef]
- Szczesny, R.J.; Borowski, L.S.; Brzezniak, L.K.; Dmochowska, A.; Gewartowski, K.; Bartnik, E.; Stepien, P.P. Human mitochondrial RNA turnover caught in flagranti: Involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2009, 38, 279–298. [Google Scholar] [CrossRef] [PubMed]
- Venø, S.T.; Kulikowicz, T.; Pestana, C.; Stepien, P.P.; Stevnsner, T.; Bohr, V.A. The human Suv3 helicase interacts with replication protein A and flap endonuclease 1 in the nucleus. Biochem. J. 2011, 440, 293. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.A.; Vicente, A.M.; Hardwick, S.W.; Alvelos, D.M.; Mazzon, R.R.; Luisi, B.F.; Marques, M.V. Association of the Cold Shock DEAD-Box RNA Helicase RhlE to the RNA Degradosome in Caulobacter crescentus. J. Bacteriol. 2017, 199, e00135-17. [Google Scholar] [CrossRef] [PubMed]
- Regonesi, M.E.; Del Favero, M.; Basilico, F.; Briani, F.; Benazzi, L.; Tortora, P.; Mauri, P.; Dehò, G. Analysis of the Escherichia coli RNA degradosome composition by a proteomic approach. Biochimie 2006, 88, 151–161. [Google Scholar] [CrossRef]
- Khemici, V.; Toesca, I.; Poljak, L.; Vanzo, N.F.; Carpousis, A.J. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: Functional replacement of RhlB by RhlE. Mol. Microbiol. 2004, 54, 1422–1430. [Google Scholar] [CrossRef]
- Prud’homme-Géńreux, A.; Beran, R.K.; Iost, I.; Ramey, C.S.; Mackie, G.A.; Simons, R.W. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: Evidence for a “cold shock degradosome”. Mol. Microbiol. 2004, 54, 1409–1421. [Google Scholar] [CrossRef]
- Tejada-Arranz, A.; de Crécy-Lagard, V.; de Reuse, H. Bacterial RNA degradosomes: Molecular machines under tight control. Trends Biochem. Sci. 2020, 45, 42. [Google Scholar] [CrossRef]
- Py, B.; Higgins, C.F.; Krisch, H.M.; Carpousis, A.J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 1996, 381, 169–172. [Google Scholar] [CrossRef]
- Lin, P.H.; Lin-Chao, S. RhlB helicase rather than enolase is the β-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)–exoribonucleolytic complex. Proc. Natl. Acad. Sci. USA 2005, 102, 16590–16595. [Google Scholar] [CrossRef]
- Dziembowski, A.; Piwowarski, J.; Hoser, R.; Minczuk, M.; Dmochowska, A.; Siep, M.; Van der Spek, H.; Grivell, L.; Stepien, P.P. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 2003, 278, 1603–1611. [Google Scholar] [CrossRef]
- Jedynak-Slyvka, M.; Jabczynska, A.; Szczesny, R.J. Human Mitochondrial RNA Processing and Modifications: Overview. Int. J. Mol. Sci. 2021, 22, 7999. [Google Scholar] [CrossRef]
- Pietras, Z.; Wojcik, M.A.; Borowski, L.S.; Szewczyk, M.; Kulinski, T.M.; Cysewski, D.; Stepien, P.P.; Dziembowski, A.; Szczesny, R.J. Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria. Nat. Commun. 2018, 9, 2558. [Google Scholar] [CrossRef]
- Pietras, Z.; Wojcik, M.A.; Borowski, L.S.; Szewczyk, M.; Kulinski, T.M.; Cysewski, D.; Stepien, P.P.; Dziembowski, A.; Szczesny, R.J. Controlling the mitochondrial antisense–role of the SUV3-PNPase complex and its co-factor GRSF1 in mitochondrial RNA surveillance. Mol. Cell. Oncol. 2018, 5, e1516452. [Google Scholar] [CrossRef]
- Silva, S.; Camino, L.P.; Aguilera, A. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc. Natl. Acad. Sci. USA 2018, 115, 11024–11029. [Google Scholar] [CrossRef]
- Wang, D.D.H.; Guo, X.E.; Modrek, A.S.; Chen, C.F.; Chen, P.L.; Lee, W.H. Helicase SUV3, polynucleotide phosphorylase, and mitochondrial polyadenylation polymerase form a transient complex to modulate mitochondrial mRNA polyadenylated tail lengths in response to energetic changes. J. Biol. Chem. 2014, 289, 16727–16735. [Google Scholar] [CrossRef]
- Mildenhall, K.B.; Wiese, N.; Chung, D.; Maples, V.F.; Mohanty, B.K.; Kushner, S.R. RNase E-based degradosome modulates polyadenylation of mRNAs after Rho-independent transcription terminators in Escherichia coli. Mol. Microbiol. 2016, 101, 645–655. [Google Scholar] [CrossRef]
- Mohanty, B.K.; Maples, V.F.; Kushner, S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004, 54, 905–920. [Google Scholar] [CrossRef]
- Yagi, M.; Uchiumi, T.; Takazaki, S.; Okuno, B.; Nomura, M.; Yoshida, S.I.; Kanki, T.; Kang, D. p32/gC1qR is indispensable for fetal development and mitochondrial translation: Importance of its RNA-binding ability. Nucleic Acids Res. 2012, 40, 9717–9737. [Google Scholar] [CrossRef]
- Dominguez, C.; Fisette, J.F.; Chabot, B.; Allain, F.H.T. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat. Struct. Mol. Biol. 2010, 17, 853–861. [Google Scholar] [CrossRef]
- Mustafi, S.B.; Aznar, N.; Dwivedi, S.K.D.; Chakraborty, P.K.; Basak, R.; Mukherjee, P.; Ghosh, P.; Bhattacharya, R. Mitochondrial BMI1 maintains bioenergetic homeostasis in cells. FASEB J. 2016, 30, 4042–4055. [Google Scholar] [CrossRef]
- Sarkar, D.; Leszczyniecka, M.; Kang, D.C.; Lebedeva, I.V.; Valerie, K.; Dhar, S.; Pandita, T.K.; Fisher, P.B. Down-regulation of Myc as a Potential Target for Growth Arrest Induced by Human Polynucleotide Phosphorylase (hPNPaseold-35) in Human Melanoma Cells. J. Biol. Chem. 2003, 278, 24542–24551. [Google Scholar] [CrossRef]
- Sarkar, D.; Park, E.S.; Fisher, P.B. Defining the mechanism by which IFN-β dowregulates c-myc expression in human melanoma cells: Pivotal role for human polynucleotide phosphorylase (hPNPaseold-35). Cell Death Differ. 2006, 13, 1541–1553. [Google Scholar] [CrossRef]
- Das, S.K.; Bhutia, S.K.; Sokhi, U.K.; Dash, R.; Azab, B.; Sarkar, D.; Fisher, P.B. Human polynucleotide phosphorylase (hPNPase old-35): An evolutionary conserved gene with an expanding repertoire of RNA degradation functions. Oncogene 2011, 30, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Park, E.S.; Barber, G.N.; Fisher, P.B. Activation of double-stranded RNA-dependent protein kinase, a new pathway by which human polynucleotide phosphorylase (hPNPaseold-35) induces apoptosis. Cancer Res. 2007, 67, 7333–7343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghosh, S.; Guimaraes, J.C.; Lanzafame, M.; Schmidt, A.; Syed, A.P.; Dimitriades, B.; Börsch, A.; Ghosh, S.; Mittal, N.; Montavon, T.; et al. Prevention of dsRNA-induced interferon signaling by AGO1x is linked to breast cancer cell proliferation. EMBO J. 2020, 39, e103922. [Google Scholar] [CrossRef]
- Cailleau, R.; Olivé, M.; Cruciger, Q.V.J. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro 1978, 14, 911–915. [Google Scholar] [CrossRef]
- Hayakawa, H.; Sekiguchi, M. Human Polynucleotide Phosphorylase Protein in Response to Oxidative Stress. Biochemistry 2006, 45, 6749–6755. [Google Scholar] [CrossRef]
- Hayakawa, H.; Kuwano, M.; Sekiguchi, M. Specific Binding of 8-Oxoguanine-Containing RNA to Polynucleotide Phosphorylase Protein. Biochemistry 2001, 40, 9977–9982. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Z.; Liu, M.; Gong, X.; Wu, S.; Burns, C.M.; Li, Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry 2009, 48, 2012–2020. [Google Scholar] [CrossRef]
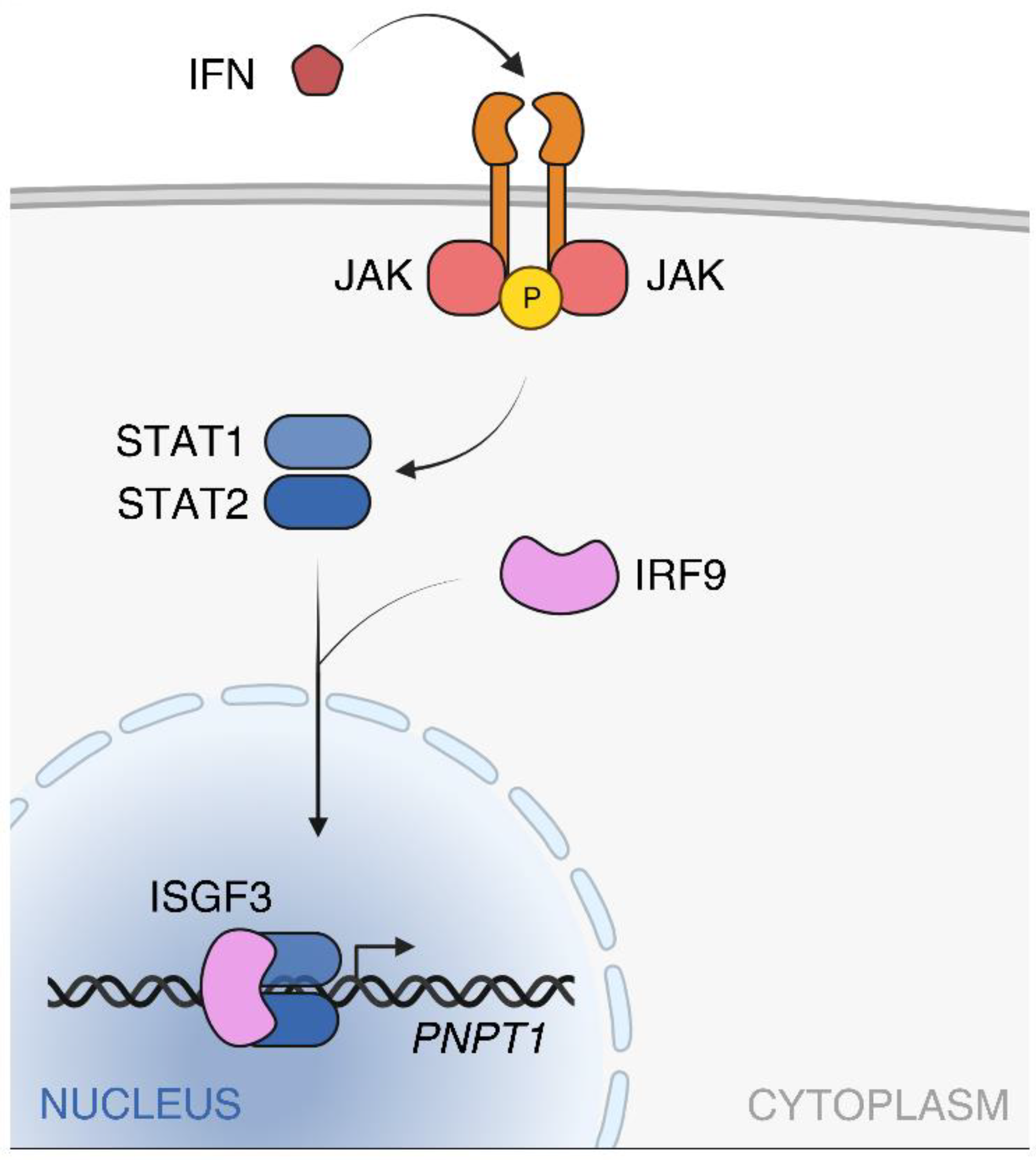
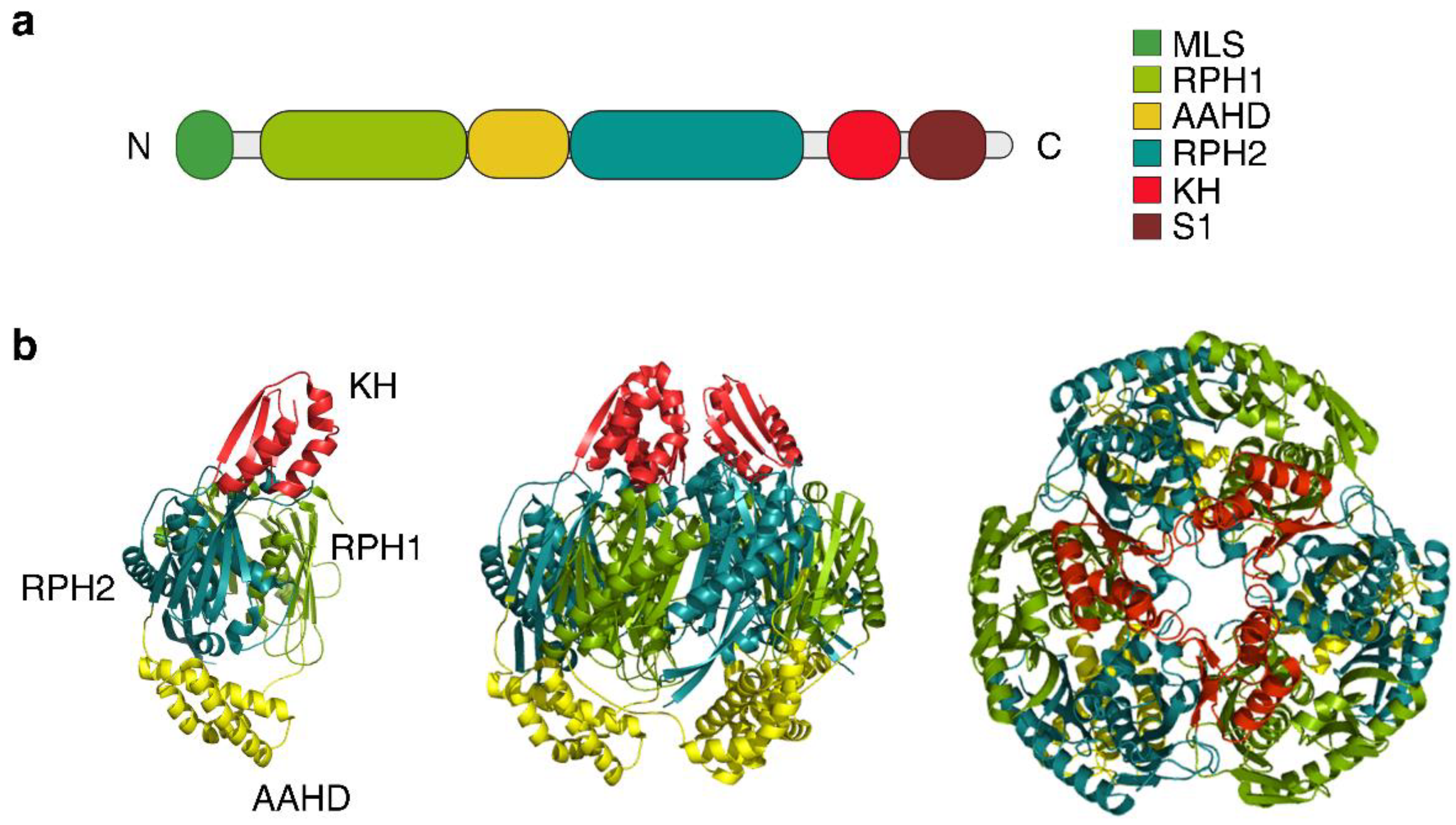

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falchi, F.A.; Pizzoccheri, R.; Briani, F. Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase. Int. J. Mol. Sci. 2022, 23, 1652. https://doi.org/10.3390/ijms23031652
Falchi FA, Pizzoccheri R, Briani F. Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase. International Journal of Molecular Sciences. 2022; 23(3):1652. https://doi.org/10.3390/ijms23031652
Chicago/Turabian StyleFalchi, Federica A., Roberto Pizzoccheri, and Federica Briani. 2022. "Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase" International Journal of Molecular Sciences 23, no. 3: 1652. https://doi.org/10.3390/ijms23031652
APA StyleFalchi, F. A., Pizzoccheri, R., & Briani, F. (2022). Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase. International Journal of Molecular Sciences, 23(3), 1652. https://doi.org/10.3390/ijms23031652





