Abstract
Prostaglandins (PGs) play many essential roles in the development, immunity, metabolism, and reproduction of animals. In vertebrates, arachidonic acid (ARA) is generally converted to prostaglandin G2 (PGG2) and H2 (PGH2) by cyclooxygenase (COX); then, various biologically active PGs are produced through different downstream prostaglandin synthases (PGSs), while PGs are inactivated by 15-hydroxyprostaglandin dehydrogenase (PGDH). However, there is very limited knowledge of the PG biochemical pathways in invertebrates, particularly for crustaceans. In this study, nine genes involved in the prostaglandin pathway, including a COX, seven PGSs (PGES, PGES2, PGDS1/2, PGFS, AKR1C3, and TXA2S), and a PGDH were identified based on the Pacific white shrimp (Litopenaeus vannamei) genome, indicating a more complete PG pathway from synthesis to inactivation in crustaceans than in insects and mollusks. The homologous genes are conserved in amino acid sequences and structural domains, similar to those of related species. The expression patterns of these genes were further analyzed in a variety of tissues and developmental processes by RNA sequencing and quantitative real-time PCR. The mRNA expression of PGES was relatively stable in various tissues, while other genes were specifically expressed in distant tissues. During embryo development to post-larvae, COX, PGDS1, GDS2, and AKR1C3 expressions increased significantly, and increasing trends were also observed on PGES, PGDS2, and AKR1C3 at the post-molting stage. During the ovarian maturation, decreasing trends were found on PGES1, PGDS2, and PGDH in the hepatopancreas, but all gene expressions remained relatively stable in ovaries. In conclusion, this study provides basic knowledge for the synthesis and inactivation pathway of PG in crustaceans, which may contribute to the understanding of their regulatory mechanism in ontogenetic development and reproduction.
1. Introduction
Prostaglandins (PGs) are an important class of eicosanoids, which are lipid signals derived from polyunsaturated fatty acids (PUFAs) [1,2,3,4]. In humans, PGs are produced in almost every tissue [5] as autocrine and paracrine regulators and may regulate many physiological processes within the central nervous system [6,7], gastrointestinal system [8,9], and cardiovascular system [10,11,12]. As in terrestrial vertebrates, prostaglandins in marine vertebrates are implicated in reproduction, cardiovascular systems, osmoregulation, and oxygenation regulation [13].
In humans, PGs are produced by the serial metabolic cascades of phospholipids cyclooxygenase (COX) and prostaglandin synthases (PGSs) and finally inactivated by 15-hydroxyprostaglandin dehydrogenase (PGDH). The synthesis and inactivation pathways of PGs are conserved in mammals but partially missing in other animal groups, such as fish [14] and insects [15,16]. However, little is known regarding the PG synthesis and inactivation pathways of crustaceans.
COX, also known as prostaglandin G/H synthase (PGHS) or prostaglandin endoperoxide H synthase (PGHS), is a glycoprotein with both peroxidase and PGS activities [17] that catalyzes the formation of PGs, thromboxanes, and levoglandins, including COX1 and COX2 [18]. In the prostaglandin synthesis pathway, COX can catalyze the formation of arachidonic acid first to PGG2 and subsequently to PGH2 [19,20]. PGH2 serves as a substrate for cell- and tissue-selective prostanoid synthases and isomerases that produce numerous bioactive prostanoids [21]. The exceptions are found in insects, in which ARA converts to PGH2 by the action of peroxidase rather than by COX [15,22].
During the synthesis of various downstream active prostaglandins, prostaglandin E synthases convert PGH2 to PGE2. Three types of PGES have been identified in vertebrates, including membrane-associated cytosolic prostaglandin E synthase 1 (PGES1), membrane-associated prostaglandin E synthase 2 (PGES2), and cytosolic prostaglandin E synthase (cPGES) [23,24]. Likewise, prostaglandin D synthases (PGDS) are classified into two types: lipocalin-type PGDS, which was previously known as the brain-type enzyme or glutathione (GSH)-independent enzyme, and hematopoietic PGDS, the spleen-type enzyme or GSH-requiring enzyme [25]. Prostaglandin F2 (PGF2) is synthesized with three substrates of PGE2, PGD2, or PGH2 through PGE 9-keto reductase, PGD 11-keto reductase (AKR1C3), and PGH 9,11-endoperoxide reductase (PGFS), respectively [26]. In addition, PGH2 is used as a substrate in the synthesis of prostaglandin I2 (PGI2) and thromboxane A2 (TxA2) by PGI synthase (PGIS) and TxA2 synthase (TxA2S), respectively [27,28]. TxA2 is an extremely unstable molecule and may rapidly convert to inactive thromboxane B2 (TxB2) [29].
Furthermore, PGs are primarily decomposed by the initial oxidation of 15 (S)-hydroxyl groups catalyzed by 15-hydroxyprostaglandin dehydrogenases (PGDHs) to 15-ketoprostaglandins (15-keto PGE2s), whose biological activity is significantly lower than that of PGs [30]. PGDH can use a wide range of prostaglandins as substrates, such as PGE1 and PGE2, but exclude PGI2, PGD2, and TXB2 [31].
In crustaceans, the processes of growth, development, and reproduction are delicately regulated by a variety of hormones [32,33,34,35,36,37]. Previous studies have indicated that PGs are involved in reproduction, including vitellogenesis and spawning in the giant river prawn Macrobrachium rosenbergii and the penaeid shrimp Marsupenaeus japonicus [38,39]. During the ovarian development of the penaeid shrimp Penaeus japonicas, particularly in the vitellogenesis stage, the amounts of PGE2 are increased [40]. In the crabs Scylla olivacea, Scylla serrata, and Oziotelphusa senex, PGs have been found to be regulators of molting and ovarian maturation [41,42]. However, studies regarding the whole PG pathway from synthesis to inactivation are still limited in crustaceans.
Pacific white shrimp (Litopenaeus vannamei) is the most economically important crustacean species in global aquaculture [43], and their ovaries are often unable to mature naturally but are induced to mature following artificial unilateral eyestalk ablation and nutritional supplementation [44]. The live polychaetes (Perinereis sp.) that contained PGE2 is one of the most effective nutritional supplementations for the oocyte development of penaeid shrimp, especially during late development and ovulation [45]. In the present study, we first identified nine genes in the PG synthesis and inactivation pathway based on the screening of the L. vannamei genome. These genes were analyzed with bioinformatics methods, and their expression patterns at the transcriptional level were confirmed by transcriptomic RNA sequencing (RNA-seq) and quantitative real-time PCR (qRT–PCR). Based on this study in a penaeid shrimp, we aimed to illustrate the specific synthesis and inactivation models of PG in crustaceans, which may contribute to understanding the potential regulatory mechanism of PGs in the development and reproduction of crustaceans.
2. Results
2.1. Screening of Genes in the Prostaglandin Pathway
As shown in Figure 1A, the gene numbers of COX, PGSs, and PGDH varied among animal species of different taxa. The COX genes were widely present in all species of the Deuterostomia and Lophotrochozoa but lost in some taxa of the Ecdysozoa. In Arthropoda, COX genes can only be found in Crustacea (e.g., Daphnia pulex, Daphnia magna, Tigriopus californicus, and L. vannamei) and Myriapoda (e.g., Strigamia maritima) but not in Insecta (e.g., Bombyx mori and Anopheeles gambiae), Arachnoidea (e.g., Stegodyphus mimosarum and Ixodes scapularis) or Merostomata (e.g., Tachypleus tridentatus). For the three types of PGES, PGES1s were rarely identified in vertebrates, PGES2s were observed in all 20 species, and cPGESs were present in vertebrates and crustaceans. PGDS, PGFS, and AKR1C3 were also found in crustaceans. However, PGIS was present in some vertebrate species. On the other hand, PGIS was limited in some Deuterostomia species. TxA2Ss were widely found in Deuterostomia but lost in Lophotrochozoa, while they were present in some Ecdysozoa arthropod species, including L. vannamei. Furthermore, PGDHs were found in all species we analyzed, while their gene numbers were expanded independently in some species, such as Ptychodera flava, Crassostrea gigas, and B. mori, but retained a single copy in L. vannamei.
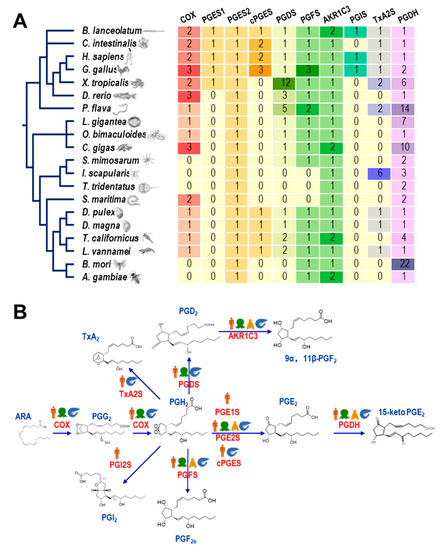
Figure 1.
(A) Gene numbers of COX, PGSs, and PGDH in 20 species from different taxa; (B) presences of genes involved in the prostaglandin pathway from four major taxa.
The synthesis and inactivation pathways of PGs in the four major taxa (mammals, mollusks, insects, and crustaceans) are illustrated in Figure 1B. In total, nine genes involved in the PG pathway were present in L. vannamei (Table 1), as a representative crustacean, while PGIS genes were absent, indicating a relatively complete PG synthesis and inactivation pathway in shrimp compared to mammals.

Table 1.
Genes involved in the PG pathway in L. vannamei.
2.2. Comparative Analysis of COX in L. vannamei and Other Species
Multiple alignments for COX amino acid (a.a.) sequences are shown in Figure 2A. COX proteins from four species (Homo sapiens, C. gigas, D. magna, and L. vannamei) showed considerable consensus sequences, indicating the conservation of these endoperoxidases. Based on the SMART program, the transmembrane domain, epidermal growth factor (EGF) domain, and animal haem peroxidase (An_peroxidase) domain were found in the a.a. sequence of LvCOX. However, for COXs from H. sapiens, M. musculus, and Danio rerio, signal peptides were found instead of the transmembrane domain in LvCOX (Figure 2B), suggesting a possible working model for COX in the shrimp.
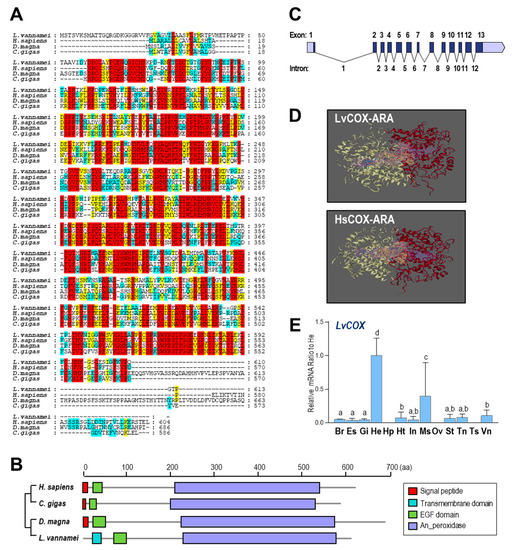
Figure 2.
(A) Alignment of the COX a.a. sequences among four species including L. vannamei (XP027218437.1), H. sapiens (NP000954), D. magna (XP032795448.2), and C. gigas (XP011440168.2). The conserved residues are boxed in red, and similar residues are labeled in yellow; (B) the structural domains of COX proteins predicted by SMART; (C) exon/intron organization of LvCOX gene. The ORFs and UTRs in exons are marked in dark gray and empty boxes, respectively, and the introns are labeled with solid lines. The positions for 13 exons are 231–283, 2444–2557, 2723–2842, 2971–3114, 3307–3450, 3617–3798, 4019–4102, 4493–4633, 4943–5057, 5228–5403, 5561–5672, 5825–6038 and 6196–6444, and for 12 introns are 289–2443, 2558–2722, 2843–2970, 3115–3306, 3451–3616, 3799–4018, 4103–4492, 4634–4942, 5058–5227, 5404–5560, 5673–5824, and 6039–6195; (D) 3D structures and substrate binding regions of COXs in L. vannamei and H. sapiens; (E) relative mRNA ratio of LvCOX transcripts among different tissues including the brain (Br), eyestalk (Es), gills (Gi), hemocyte (He), hepatopancreas (Hp), heart (Ht), intestines (In), muscle (Ms), ovary (Ov), stomach (St), thoracic ganglion (Tn), testis (Ts), ventral ganglion (Vn). The experimental groups denoted by the same letter represent a similar level of transcript expression (p > 0.05, ANOVA, followed by Fisher LSD test).
The gene sequence of LvCOX was obtained and deposited in GenBank under accession no. LOC113810970. The LvCOX gene is 7268 bp in length, containing 13 exons separated by 12 introns. All 12 introns begin with GT and end with AG, conforming to the GT/AG rule of intron splicing (Figure 2C). By using the SWISS-MODEL program, the three-dimensional (3D) structures of COX from H. sapiens and L. vannamei were constructed for comparison. In this case, the binding regions of HsCOX and LvCOX were similar in their 3D structures. Each COX protein was composed of two subunits that formed a binding pocket between them, as shown by the purple region in Figure 2D, which can combine ARA and bind in the yellow region.
The mRNA expression profile of LvCOX among different tissues was analyzed by qRT–PCR. As shown in Figure 2E, LvCOX was highly expressed in the hemocytes followed by the muscle and showed much lower expression in other tissues. However, no detectable expression of LvCOX was found in the hepatopancreas and reproductive organs—namely, the ovary and testis.
2.3. Comparative Analysis of PGSs in L. vannamei and Other Species
In L. vannamei, two PGES genes were identified, corresponding to two types of PGES—namely, LvPGES2 with the GST_N_3 domain and LvcPGES with the CS domain, both of which were conserved with PGESs in other species. In addition, PGFS, AKR1C3, and TxA2S from different species shared high conservation within their functional domains, with an exception of human TxA2S, which does not contain a transmembrane region. Furthermore, the LvPGDS1 was not conserved with its counterparts in other species since it lacked the GST_N domain (Figure 3A).
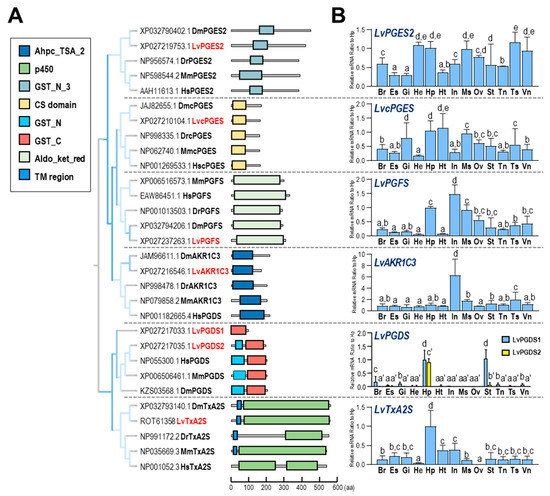
Figure 3.
(A) phylogenetic analysis and the structural domain organization of multiple PGS genes in various species. The selected species include D. magna, L. vannamei, H. sapiens, D. rerio, and Mus musculus; (B) relative mRNA ratio of multiple LvPGS genes (LvPGES2, LvcPGES, LvPGFS, LvAKR1C3, LvPGDS1, LvPGDS2, and LvTxA2S) among different tissues, including brain (Br), eyestalk (Es), gills (Gi), hemocyte (He), hepatopancreas (Hp), heart (Ht), intestines (In), muscle (Ms), ovary (Ov), stomach (St), thoracic ganglion (Tn), testis (Ts), and ventral ganglion (Vn). The experimental groups denoted by the same letter represent a similar level of transcript expression (p > 0.05, ANOVA, followed by Fisher LSD test).
The genes for different types of PGESs were expressed in distant patterns in selected tissues (Figure 3B). In this case, LvPGES2 was primarily expressed in the testis, hemocyte, hepatopancreas, and ventral ganglion, and LvcPGES was predominantly expressed in the heart, hepatopancreas, and gills. In addition, high expression of both LvPGFS and LvAKR1C3 transcripts was observed in the intestines, and LvTxA2S expression was abundant in the hepatopancreas. Moreover, the expression profiles of LvPGDS1 and LvPGDS2 were distinct, in which the former was highly expressed in the hepatopancreas and stomach, and the latter was restrictedly expressed in the hepatopancreas.
2.4. Comparative Analysis of PGDH in L. vannamei and Other Species
As shown in Figure 4A, multiple alignments of a.a. sequences indicated that the PGDH shared considerable consensus sequences between H. sapiens, Xenopus tropicalis, C. gigas, and L. vannamei, indicating a conserved inactivation pathway of PGs among diversified animal species. In addition, LvPGDH transcripts were found to be ubiquitously expressed in all examined tissues, and the highest expression was found in the muscle, followed by the ventral ganglion.
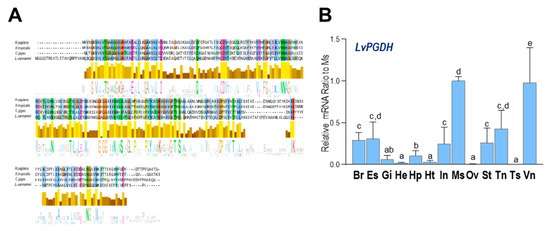
Figure 4.
(A) Alignment of PGDH a.a. sequences among four species (H. sapiens, X. tropicalis, C. gigas, and L. vannamei); (B) relative mRNA ratio of LvPGDH transcripts among different tissues including the brain (Br), eyestalk (Es), gills (Gi), hemocyte (He), hepatopancreas (Hp), heart (Ht), intestines (In), muscle (Ms), ovary (Ov), stomach (St), thoracic ganglion (Tn), testis (Ts), ventral ganglion (Vn). The experimental groups denoted by the same letter represent a similar level of transcript expression (p > 0.05, ANOVA, followed by Fisher LSD test).
2.5. Expression Patterns during Ontogenetic Development, Ovarian Development, and Molting
Based on the RNA-seq data, transcript levels of the COX, PGSs, and PGDH in L. vannamei were analyzed during the developmental processes of ontogenetic stages, ovarian development, and the molting cycle. As shown in Figure 5A, the expression patterns of LvPGES2, LvPEGS, LvPGFS, LvTxA2S, and LvPGDH were almost identical and did not change noticeably at all stages during ontogenesis, except that the expression level of LvPGES2 was relatively higher than other genes. On the other hand, the expression of the remaining genes showed dynamic changes, e.g., LvCOX, LvPGDS2, and LvAKR1C3 showed their highest expression levels at the mysis stage than at other stages, while the highest expression of LvPGDS1 exhibited at the zoea stage. During the molting cycle (Figure 5B), the P1 stage was a transition point for the expression of most genes in the PG-related pathways. In this case, the transcript levels of LvPGES2, LvcPGES, LvPGDS1, LvPGDS2, LvAKR1C3, and LvTxA2S were all reduced at the P1 stage, while LvCOX showed the highest expression level at this stage. In addition, significantly low expression of LvCOX and LvPGDH appeared at the D1 stage, and the expression of LvPGFS did not change markedly at any stage. During ovarian development (Figure 5C), LvCOX, LvPGES2, and LvcPGES were stably expressed in both the hepatopancreas and ovary, while the LvPGDS1, LvPGDS2, LvPGFS, LvTxA2S, and LvPGDH expression was higher in the hepatopancreas than in the ovary.
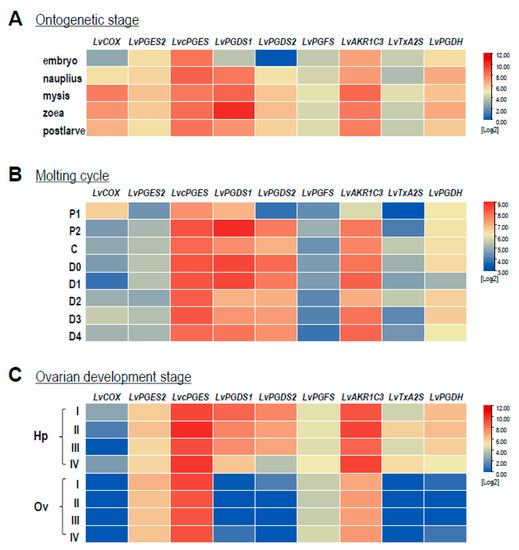
Figure 5.
The expression profiles of genes involved in the PG pathway of L. vannamei during different developmental processes: (A) ontogenetic stages, including embryo, nauplius, mysis, zoea, and post-larvae; (B) molting cycle, including postmolt (P1 and P2 stages), intermolt (C stage) and premolt (D0, D1, D2, D3, and D4 stages); (C) ovarian development, including hepatopancreas (Hp) and ovaries (Ov) in stages I to IV.
3. Discussion
In this study, enzymes related to the prostaglandin pathway were screened based on the L. vannamei genomic data. The identified enzymes can support a complete PG synthesis pathway from ARA to PGG2 (Figure 1B), along with PGH2 to various PGs, followed by the presence of an inactivation pathway. By analysis of functional domains (Figure 2B, Figure 3A, and Figure 4A), most of the enzymes in the PG pathway of L. vannamei show strong conservation with their counterparts in other species, indicating that it is an ancient pathway in metazoans. In addition, the expression of these genes was determined in various tissues and during multiple physiological processes. This evidence will facilitate further studies regarding the specific functions of these enzymes in crustaceans and the PG-mediated applications in shrimp culture.
In most species, COX1 and COX2 are considered constitutive and inducible COXs, respectively [46]. Both enzymes catalyze a cyclooxygenase reaction in which ARA is converted to PGG2 and a peroxidase reaction in which PGG2 is reduced to PGH2. A previous study showed that the major sequence differences between COX isoforms occur in the signal peptide domains [47]. In this study, however, only a single LvCOX gene was identified from the L. vannamei genome, and the N terminus of its protein product is a transmembrane structural domain instead of a signal peptide. Since their main functional domains are highly conserved, LvCOX and HsCOX are speculated to be essentially identical in terms of spatial structure and binding regions. In mammals, COX1 is uniquely expressed in endothelial cells, but COX2 is specifically expressed in some tissues (e.g., epithelial cells and muscle) after stimulation in various ways [48,49,50]. Similarly, LvCOX was highly expressed in the hemocytes and muscle, indicating the functional conservation of these enzymes among distant species. It is very interesting that the expression of LvCOX was absent in the hepatopancreas, ovary, and testis (Figure 2E). However, the effects of PGs on reproduction have been reported in several crustacean species [38,39,40,41,42], and the hepatopancreas and ovary are the major organs for vitellogenesis in L. vannamei [51]. It is logical to speculate that the PGs acting on the hepatopancreas and ovary are conversed by the exogenous intermediate PGH2.
PGES can isomerize PGH2 to PGE2, and molecular identification of multiple PGES enzymes has revealed the existence of two distant pathways for the biosynthesis of PGE2—namely, the constitutive COX1-cPGES pathway and the inducible COX2-mPGES pathway [52,53]. Both mPGES and cPGES were identified in shrimp, and their differences and specificity still need to be investigated. In addition, cPGES is most abundantly expressed in the testes of rats [53], identical to our current study, in which LvcPGES was expressed in the testis at the highest level.
In mammals, two PGDSs can isomerize PGH2 to PGD2—namely, lipocalin-type PGD synthase (L-PGDS) [54] and hematopoietic PGDS (H-PGDS) [55,56]. H-PGDS is a 26 kDa cytosolic protein responsible for the biosynthesis of PGD2 in immune and inflammatory cells, such as mast cells [57]. In L. vannamei, both LvPGDS1 and LvPGDS2 identified were related to human H-PGDS, and they were predominantly expressed in the hepatopancreas, which may respond to pathogenic infection in shrimp. However, L-PGDS has not been found in crustaceans, indicating that it is a specific isoform in vertebrates. PGDH catalyzes the reversible oxidation of the 15-hydroxyl group of prostaglandins to produce a 15-keto metabolite with greatly reduced biological activity [58]. Six a.a. residues (Gly, Gly, Gly, Asp, Tyr, and Lys) in PGDH are strictly conserved in the protein sequences from different species [59]. As shown in Figure 4A, although the LvPGDH a.a. sequences are longer than other species, they are conserved at these key positions for enzyme activities. In addition, the extended sequence in the N terminus of LvPGDH is not a signal peptide.
PGs are widely found in all tissues and body fluids in most metazoan animals. The synthesis and inactivation pathways of PG have been studied extensively in higher-order mammals but are still limited in low-order nonmammalian or invertebrate animals, such as crustaceans. In aquatic animals, studies regarding the function of PGs are mainly focused on ovarian development [60,61]. With its collective findings, this study provides new insights into the synthesis and inactivation pathway of prostaglandins in crustaceans and may contribute to their regulatory mechanism in development and reproduction; nevertheless, further studies need to be conducted to understand the functions and regulatory mechanism of these key genes and PGs in crustaceans in depth.
4. Materials and Methods
4.1. Cross-Genomic Analysis of Genes in the Prostaglandin Pathway
For cross-genomic analysis to identify genes in the prostaglandin pathway, the selected species included Branchiostoma lanceolatum (GCA_900088365.1), D. magna (GCA_003990815.1), D. pulex (GCA_911175335.1), T. californicus (GCA_007210705.1), L. vannamei (GCA_003789085.1), A. gambiae (GCA_000005575.1), B. mori (GCA_014905235.2), I. scapularis (GCA_016920785.2), S. mimosarum (GCA_000611955.2), T. tridentatus (GCA_004210375.1), S. maritima (GCA_000239455.1), Octopus bimaculoides (GCA_001194135.1), C. gigas (GCA_902806645.1), Lottia gigantea (GCA_000327385.1), H. sapiens (GCA_000001405.28), Gallus gallus (GCA_016699485.1), X. tropicalis (GCA_000004195.4), D. rerio (GCA_000002035.4), Ciona intestinalis (GCA_000224145.2), and P. flava (GCA_001465055.1). The gene functions were annotated based on the best-matched hits to SwissProt [62] (UniProt < EMBL-EBI, accessed on 21 July 2020) using BLAST 2.9.0+ [63] (-p blastp -e 1e-5) (BLAST: Basic Local Alignment Search Tool (nih.gov), accessed on 21 July 2020). The gene motifs and domains were identified from the InterPro member databases using InterProScanm [64] (v5.27-66.0) (InterPro (Ebi. ac. the UK), accessed on 21 July 2020) with parameters (-appl ProDom, PRINTS, Pfam, SMART, PANTHER, ProSiteProfiles, and ProSitePatterns). Related genes were identified by searching gene and domain description with keywords (such as cyclooxygenase) and manually checked considering phylogeny.
4.2. Phylogenetic Tree and Sequence Analysis
The evolutionary tree of 20 species was constructed through a Lifemap (Lifemap NCBI (univ-lyon1.fr), accessed on 16 August 2021). Alignment of COX, PGSs, and PGDH among various species was performed with the ClustalX program and demonstrated using ESPript 3.0 (ESPript 3.x/ENDscript 2.x (ibcp.fr), accessed on 16 August 2021) and the Genedoc program. The sequences were obtained from GenBank. The phylogenetic tree was constructed based on a.a. (p distance) with the neighbor-joining method (pairwise deletion) with 1000 bootstrap replicates using MEGA X. In this case, different PGS genes were constructed into a phylogenetic tree. The tree was visualized via iTOL (iTOL: Interactive Tree of Life (embl.de), accessed on 16 August 2021) and Evolview (EvolView: login (evolgenius.info), accessed on 16 August 2021).
4.3. Structural Analysis of Proteins
The gene structure for LvCOX was analyzed by mapping with Exon–Intron Graphic Maker (WormWeb.org—Exon–Intron Graphic Maker, accessed on 20 August 2021). The structural domains for COX and PGSs were predicted by using SMART (SMART: Main page (embl-heidelberg.de), accessed on 20 August 2021) and annotated by IBS (IBS—Online (biocuckoo.org), accessed on 20 August 2021). The three-dimensional (3D) models for COX were deduced with the knowledge-based modeling program ProMod II provided by the SWISS-MODEL server and were measured by using SAVES v6.0 (SAVESv6.0—Structure Validation Server (ucla.edu), accessed on 20 August 2021). The 3D chemical structures of ARA and PGs were downloaded from PubChem (PubChem (nih.gov), accessed on 20 August 2021). The ARA binding sites and modes for COXs were predicted by using Proteins Plus (Zentrum für Bioinformatik: Universität Hamburg—Proteins Plus Server, accessed on 20 August 2021).
4.4. Animals and Tissue Samples Collection
Adult Pacific white shrimp were collected from the Jinyang Shrimp Culture Center, Maoming, Guangdong, China, and maintained in artificial seawater (30 parts per thousand ppt and pH 8.2) at 28 °C under a 12 h dark–12 h light photoperiod. Six individuals of shrimp were dissected directly on ice after being collected from the pond. In total, 13 tissues were harvested, including the brain, eyestalk, gills, hemocytes, hepatopancreas, heart, intestines, muscle, ovary, stomach, thoracic ganglion, testis, and ventral ganglion. In this case, ovary and testis samples were harvested from sexually mature shrimp, and other samples were collected from sexually immature shrimp. Samples were immediately frozen in liquid nitrogen and stored at −80 °C for further mRNA experiments. All efforts were made to minimize animal suffering. This article does not contain any studies with human participants.
4.5. Realtime PCR for Tissue Distribution
For tissue distribution, total RNA was isolated using RNAiso Plus (Invitrogen, Carlsbad, CA, USA), digested with gDNA Eraser (TaKaRa, Dalian, China), and reverse transcribed using the PrimeScript RT reagent Kit (TaKaRa). The first-strand cDNA samples obtained were then subjected to qPT–PCR using a RotorGene RG-3000 real-time PCR system (Corbett Research, San Francisco, CA, USA). PCRs were conducted using SYBR Premix Ex Taq II (TaKaRa) with specific primers for their target genes (Table 2). The PCR cycle numbers were fixed to 40 with 30 s at 94 °C for denaturing, 30 s at 60 °C for annealing, 30 s at 72 °C for extension, and 20 s at 82 °C for signal capture. In this case, the transcript expression of the target genes was routinely normalized using β-actin mRNA. The relative expression levels were calculated using the comparative Ct method with the 2−ΔΔCt formula [65]. The raw data for the expression of different genes were transformed as ratios of control groups (e.g., hepatopancreas, hemocyte, or muscle). The data expressed as the mean ± standard error (SE) were analyzed by using one-way ANOVA, followed by Fisher’s least significant difference (LSD) test.

Table 2.
Primers sequences used in this study.
4.6. Transcriptomic Analysis for Gene Expression
Transcript levels of prostaglandin-pathway-involved genes in L. vannamei during different developmental processes were further analyzed with transcriptomes. The transcriptomic data for ontogenetic development stages (PRJNA253518) [66] and molting cycle (PRJNA288849) [67] were obtained from GenBank, and those for ovarian development were previously established by our laboratory (SUB10677615). For the transcriptome of ontogenetic development, the embryo (E) sample was from an equivalent RNA mixture of the zygote, blastula, gastrula, limb bud embryo, and larva in membrane stages; the nauplius (N) sample was from an equivalent RNA mixture of nauplius I, III, and VI stages; the zoea (Z) sample was from an equivalent RNA mixture of zoea I, II, and III stages; the mysis (M) sample was from an equivalent RNA mixture of mysis I, II, and III stages; RNA from post-larvae 1 was considered as the post-larvae (P) sample. For the transcriptome of the molting cycle, the molting stages were classified as postmolt (P1 and P2), inter-molt (C), and pre-molt (D0, D1, D2, D3, and D4). For the transcriptome of ovarian development, the stages were clarified into I–IV, based on the classification of predominant oocytes, as described previously [51]. A matrix of read counts mapped to L. vannamei genomic features (genes) was extracted directly from the files generated by StringTie (run with the -e parameter) with script prepDE.py. The read count per million reads (CPM) was calculated, and cross-sample normalization was performed using edgeR [68] and DESeq2 [69]. The obtained heatmaps were then created and clustered with hierarchical clustering using Tb tools (Guangzhou, China).
Author Contributions
X.W., T.C. and J.Z. conceived and designed the experiments; H.Y., X.C. and Z.L. performed the experiments; H.Y., X.C., Z.L., M.Z., X.Z., Y.L. and Q.G. analyzed the data; Y.S., C.Z. and T.C. contributed reagents/materials/analysis tools; H.Y., X.W., T.C. and J.Z. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams [2019KJ149], the Science and Technology Project of Guangzhou [201904020009], the Key R&D Program from the Ministry of science and technology of China [2018YFD0900100], and the National Natural Science Foundation of China [41876196, 31872613 and 32172954], and the APC was funded by [41876196].
Institutional Review Board Statement
The animal study protocol was approved by Ethics Committees of the South China Sea Institute of Oceanology, Chinese Academy of Sciences.
Informed Consent Statement
This research not involved humans.
Data Availability Statement
The data presented in this study are available in the article.
Acknowledgments
This study was supported by a Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (2019KJ149), Science and Technology Project of Guangzhou (201904020009), a Key R&D Program (2018YFD0900100) from the Ministry of science and technology of China, and National Natural Science Foundation of China (Grant Nos. 41876196, 31872613 and 32172954).
Conflicts of Interest
Q.G. from EasyATGC Limited Liability Company download the genomic data from Genbank and annotated the genes for the study. EasyATGC Limited Liability Company had no role in the topic selected, experimental design, execution, interpretation, or writing of the study. No fund of the study was supported by EasyATGC Limited Liability Company, and the study did not refer to any product from EasyATGC Limited Liability Company.
Abbreviations
| COX | cyclooxygenase |
| PGES | prostaglandin E synthase |
| PGDS | prostaglandin D synthase |
| PGFS | prostaglandin F synthase/PGH 9,11-endoperoxide reductase |
| PGIS | prostaglandin I synthase |
| AKR1C3 | PGD 11-keto reductase |
| TxA2S | thromboxane A2 synthase |
| PGDH | 15-hydroxyprostaglandin dehydrogenases |
References
- Cha, Y.I.; Solnica-Krezel, L.; DuBois, R.N. Fishing for prostanoids: Deciphering the developmental functions of cyclooxygenase-derived prostaglandins. Dev. Biol. 2006, 289, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Narumiya, S.; Ichikawa, A. Distribution and function of prostanoid receptors: Studies from knockout mice. Prog. Lipid Res. 2000, 39, 289–314. [Google Scholar] [CrossRef]
- Funk, C.D. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science 2001, 294, 1871–1875. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.N.; Breyer, R.M. Pharmacology and signaling of prostaglandin receptors: Multiple roles in inflammation and immune modulation. Pharmacol. Ther. 2004, 103, 147–166. [Google Scholar] [CrossRef] [PubMed]
- Christ, E.J.; van Dorp, D.A. Comparative aspects of prostaglandin biosynthesis in animal tissues. Biochim. Biophys. Acta 1972, 270, 534–545. [Google Scholar] [CrossRef]
- Leslie, J.B.; Watkins, W.D. Eicosanoids in the central nervous system. J. Neurosurg. 1985, 63, 659–668. [Google Scholar] [CrossRef]
- Yagami, T.; Koma, H.; Yamamoto, Y. Pathophysiological Roles of Cyclooxygenases and Prostaglandins in the Central Nervous System. Mol. Neurobiol. 2016, 53, 4754–4771. [Google Scholar] [CrossRef]
- Heeney, A.; Rogers, A.C.; Mohan, H.; Mc Dermott, F.; Baird, A.W.; Winter, D.C. Prostaglandin E2 receptors and their role in gastrointestinal motility—Potential therapeutic targets. Prostaglandins Other Lipid Mediat. 2021, 152, 106499. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, R.N. Role of prostanoids in gastrointestinal cancer. J. Clin. Investig. 2018, 128, 2732–2742. [Google Scholar] [CrossRef]
- Karim, S.M.; Somers, K.; Hillier, K. Cardiovascular actions of prostaglandin Fa-alpha infusion in man. Eur. J. Pharmacol. 1969, 5, 117–120. [Google Scholar] [CrossRef]
- Sasaguri, T.; Miwa, Y. Prostaglandin J2 family and the cardiovascular system. Curr. Vasc. Pharmacol. 2004, 2, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Fan, X.; Xu, F.; Wang, J.; Chen, Y. Prostaglandin E1 attenuates post-cardiac arrest myocardial dysfunction through inhibition of mitochondria-mediated cardiomyocyte apoptosis. Mol. Med. Rep. 2021, 23, 110. [Google Scholar] [CrossRef]
- Brown, J.A.; Bucknall, R.M. Antidiuretic and cardiovascular actions of prostaglandin E2 in the rainbow trout Salmo gairdneri. Gen. Comp. Endocrinol. 1986, 61, 330–337. [Google Scholar] [CrossRef]
- Goetz, F.W. Compartmentalization of prostaglandin synthesis within the fish ovary. Am. J. Physiol. 1991, 260 Pt 2, R862–R865. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Kim, Y. Prostaglandins and Other Eicosanoids in Insects: Biosynthesis and Biological Actions. Front. Physiol. 2018, 9, 1927. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Kim, Y. Prostaglandin catabolism in Spodoptera exigua, a lepidopteran insect. J. Exp. Biol. 2020, 223, jeb233221. [Google Scholar] [CrossRef]
- Van der Ouderaa, F.J.; Buytenhek, M.; Nugteren, D.H.; Van Dorp, D.A. Purification and characterisation of prostaglandin endoperoxide synthetase from sheep vesicular glands. Biochim. Biophys. Acta 1977, 487, 315–331. [Google Scholar] [CrossRef]
- Urade, Y.; Watanabe, K.; Hayaishi, O. Prostaglandin d, e., and f synthases. J. Lipid Mediat. Cell Signal. 1995, 12, 257–273. [Google Scholar] [CrossRef]
- Smith, W.L.; DeWitt, D.L.; Garavito, R.M. Cyclooxygenases: Structural, cellular, and molecular biology. Annu. Rev. Biochem. 2000, 69, 145–182. [Google Scholar] [CrossRef]
- Mendes, R.T.; Stanczyk, C.P.; Sordi, R.; Otuki, M.F.; dos Santos, F.A.; Fernandes, D. Selective inhibition of cyclooxygenase-2: Risks and benefits. Rev. Bras. Reumatol. 2012, 52, 767–782. [Google Scholar]
- Pelley, J.W. 11—Metabolism of Steroids and Other Lipids. In Elsevier’s Integrated Review Biochemistry, 2nd ed.; Pelley, J.W., Ed.; W.B. Saunders: Philadelphia, PA, USA, 2012; pp. 89–98. [Google Scholar]
- Tootle, T.L.; Williams, D.; Hubb, A.; Frederick, R.; Spradling, A. Drosophila eggshell production: Identification of new genes and coordination by Pxt. PLoS ONE 2011, 6, e19943. [Google Scholar] [CrossRef] [PubMed]
- Hara, S.; Kamei, D.; Sasaki, Y.; Tanemoto, A.; Nakatani, Y.; Murakami, M. Prostaglandin E synthases: Understanding their pathophysiological roles through mouse genetic models. Biochimie 2010, 92, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Tanikawa, N.; Ohmiya, Y.; Ohkubo, H.; Hashimoto, K.; Kangawa, K.; Kojima, M.; Ito, S.; Watanabe, K. Identification and characterization of a novel type of membrane-associated prostaglandin E synthase. Biochem. Biophys. Res. Commun. 2002, 291, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Urade, Y.; Hayaishi, O. Prostaglandin D synthase: Structure and function. Vitam. Horm. 2000, 58, 89–120. [Google Scholar] [PubMed]
- Watanabe, K. Prostaglandin F synthase. Prostaglandins Other Lipid Mediat. 2002, 68, 401–407. [Google Scholar] [CrossRef]
- Dai, D.; Chen, B.; Feng, Y.; Wang, W.; Jiang, Y.; Huang, H.; Liu, J. Prognostic value of prostaglandin I2 synthase and its correlation with tumor-infiltrating immune cells in lung cancer, ovarian cancer, and gastric cancer. Aging 2020, 12, 9658–9685. [Google Scholar] [CrossRef]
- Jones, D.A.; Fitzpatrick, F.A. Thromboxane A2 synthase. Modification during “suicide” inactivation. J. Biol. Chem. 1991, 266, 23510–23514. [Google Scholar] [CrossRef]
- Catella, F.; Healy, D.; Lawson, J.A.; FitzGerald, G.A. 11-Dehydrothromboxane B2: A quantitative index of thromboxane A2 formation in the human circulation. Proc. Natl. Acad. Sci. USA 1986, 83, 5861–5865. [Google Scholar] [CrossRef] [PubMed]
- Hsin-Hsiung, T.; Charles, M.E.; Min, T. Prostaglandin catabolizing enzymes. Prostaglandins Other Lipid Mediat. 2002, 68, 483–493. [Google Scholar]
- Ensor, C.M.; Tai, H.H. 15-Hydroxyprostaglandin dehydrogenase. J. Lipid Mediat Cell Signal. 1995, 12, 313–319. [Google Scholar] [CrossRef]
- Luo, X.; Chen, T.; Zhong, M.; Jiang, X.; Zhang, L.; Ren, C.; Hu, C. Differential regulation of hepatopancreatic vitellogenin (VTG) gene expression by two putative molt-inhibiting hormones (MIH1/2) in Pacific white shrimp (Litopenaeus vannamei). Peptides 2015, 68, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Ren, C.; Wang, Y.; Gao, Y.; Wong, N.-K.; Zhang, L.; Hu, C. Crustacean cardioactive peptide (CCAP) of the Pacific white shrimp (Litopenaeus vannamei): Molecular characterization and its potential roles in osmoregulation and freshwater tolerance. Aquaculture 2016, 451, 405–412. [Google Scholar] [CrossRef]
- Chen, T.; Ren, C.; Jiang, X.; Zhang, L.; Li, H.; Huang, W.; Hu, C. Mechanisms for type-II vitellogenesis-inhibiting hormone suppression of vitellogenin transcription in shrimp hepatopancreas: Crosstalk of GC/cGMP pathway with different MAPK-dependent cascades. PLoS ONE 2018, 13, e0194459. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Shi, W.; Lin, D.; Ye, H. A Possible Role of Allatostatin C in Inhibiting Ecdysone Biosynthesis Revealed in the Mud Crab Scylla Paramamosain. Front. Mar. Sci. 2021, 8, 1293. [Google Scholar] [CrossRef]
- Liu, J.; Liu, A.; Liu, F.; Huang, H.; Ye, H. Role of neuroparsin 1 in vitellogenesis in the mud crab, Scylla paramamosain. Gen. Comp. Endocrinol. 2020, 285, 113248. [Google Scholar] [CrossRef]
- Pan, J.; Liu, M.; Chen, T.; Cheng, Y.; Wu, X. Immunolocalization and changes of 17beta-estradiol during ovarian development of Chinese mitten crab Eriocheir sinensis. Cell Tissue Res. 2018, 373, 509–520. [Google Scholar] [CrossRef]
- Tahara, D.; Yano, I. Maturation-related variations in prostaglandin and fatty acid content of ovary in the kuruma prawn (Marsupenaeus japonicus). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2004, 137, 631–637. [Google Scholar] [CrossRef]
- Sumpownon, C.; Engsusophon, A.; Siangcham, T.; Sugiyama, E.; Soonklang, N.; Meeratana, P.; Wanichanon, C.; Hanna, P.J.; Setou, M.; Sobhon, P. Variation of prostaglandin E2 concentrations in ovaries and its effects on ovarian maturation and oocyte proliferation in the giant fresh water prawn, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 2015, 223, 129–138. [Google Scholar] [CrossRef]
- Tahara, D.; Yano, I. Development of hemolymph prostaglandins assay systems and their concentration variations during ovarian maturation in the kuruma prawn, Penaeus japonicus. Aquaculture 2003, 220, 791–800. [Google Scholar] [CrossRef]
- Duangprom, S.; Ampansri, W.; Suwansa-Ard, S.; Chotwiwatthanakun, C.; Sobhon, P.; Kornthong, N. Identification and expression of prostaglandin E synthase (PGES) gene in the central nervous system and ovary during ovarian maturation of the female mud crab, Scylla olivacea. Anim. Reprod. Sci. 2018, 198, 220–232. [Google Scholar] [CrossRef]
- Swetha, C.H.; Girish, B.P.; Hemalatha, M.; Reddy, P.S. Induction of vitellogenesis, methyl farnesoate synthesis and ecdysteroidogenesis in two edible crabs by arachidonic acid and prostaglandins. J. Exp. Biol. 2020, 223 Pt 3, jeb212381. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Chen, T.; Luo, P.; Huang, W.; Huo, D.; Yun, L.; Hu, C.; Cheng, C. A fibrinogen-related protein, LvFREP2, from Litopenaeus vannamei facilitates the clearance of Vibrio harveyi. Fish Shellfish Immunol. 2018, 78, 364–371. [Google Scholar] [CrossRef]
- Chen, T.; Lin, T.; Li, H.; Lu, T.; Li, J.; Huang, W.; Sun, H.; Jiang, X.; Zhang, J.; Yan, A.; et al. Heat Shock Protein 40 (HSP40) in Pacific White Shrimp (Litopenaeus vannamei): Molecular Cloning, Tissue Distribution and Ontogeny, Response to Temperature, Acidity/Alkalinity and Salinity Stresses, and Potential Role in Ovarian Development. Front. Physiol. 2018, 9, 1784. [Google Scholar] [CrossRef] [PubMed]
- Meunpol, O.; Duangjai, E.; Yoonpun, R.; Piyatiratitivorakul, S. Detection of prostaglandin E2 in polychaete Perinereis sp. and its effect on Penaeus monodon oocyte development in vitro. Fish. Sci. 2010, 76, 281–286. [Google Scholar] [CrossRef]
- Vane, J.R.; Bakhle, Y.S.; Botting, R.M. Cyclooxygenases 1 and 2. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 97–120. [Google Scholar] [CrossRef] [PubMed]
- Otto, J.C.; Smith, W.L. Prostaglandin endoperoxide synthases-1 and -2. J. Lipid Mediat. Cell Signal. 1995, 12, 139–156. [Google Scholar] [CrossRef]
- DeWitt, D.L.; Day, J.S.; Sonnenburg, W.K.; Smith, W.L. Concentrations of prostaglandin endoperoxide synthase and prostaglandin I2 synthase in the endothelium and smooth muscle of bovine aorta. J. Clin. Investig. 1983, 72, 1882–1888. [Google Scholar] [CrossRef]
- Hatae, T.; Hara, S.; Yokoyama, C.; Yabuki, T.; Tanabe, T.I.J.F.L. Site-directed mutagenesis of human prostacyclin synthase: Alteration of Cys441 of the Cys-pocket, and Glu347 and Arg350 of the EXXR motif. FEBS Lett. 1996, 389, 268–272. [Google Scholar] [CrossRef]
- Pritchard, K.J.J.J.B.C. Induction of cyclooxygenase-2 in rat vascular smooth muscle cells in vitro and in vivo. J. Biol. Chem. 1994, 269, 8504–8509. [Google Scholar] [CrossRef]
- Ruan, Y.; Wong, N.K.; Zhang, X.; Zhu, C.; Wu, X.; Ren, C.; Luo, P.; Jiang, X.; Ji, J.; Wu, X.; et al. Vitellogenin Receptor (VgR) Mediates Oocyte Maturation and Ovarian Development in the Pacific White Shrimp (Litopenaeus vannamei). Front. Physiol. 2020, 11, 485. [Google Scholar] [CrossRef]
- Murakami, M.; Naraba, H.; Tanioka, T.; Semmyo, N.; Nakatani, Y.; Kojima, F.; Ikeda, T.; Fueki, M.; Ueno, A.; Oh, S.; et al. Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. J. Biol. Chem. 2000, 275, 32783–32792. [Google Scholar] [CrossRef] [PubMed]
- Tanioka, T.; Nakatani, Y.; Semmyo, N.; Murakami, M.; Kudo, I. Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with cyclooxygenase-1 in immediate prostaglandin E2 biosynthesis. J. Biol. Chem. 2000, 275, 32775–32782. [Google Scholar] [CrossRef] [PubMed]
- Urade, Y.; Eguchi, N. Lipocalin-type and hematopoietic prostaglandin D synthases as a novel example of functional convergence. Prostaglandins Other Lipid Mediat. 2002, 68–69, 375–382. [Google Scholar] [CrossRef]
- Christ-Hazelhof, E.J.B.B.A. Purification and characterisation of prostaglandin endoperoxide D-isomerase, a cytoplasmic, glutathione-requiring enzyme. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1979, 572, 43–51. [Google Scholar] [CrossRef]
- Urade, Y.; Fujimoto, N.; Ujihara, M.; Hayaishi, O. Biochemical and immunological characterization of rat spleen prostaglandin D synthetase. J. Biol. Chem. 1987, 262, 3820–3825. [Google Scholar] [CrossRef]
- Urade, Y.; Ujihara, M.; Horiguchi, Y.; Igarashi, M.; Nagata, A.; Ikai, K.; Hayaishi, O. Mast cells contain spleen-type prostaglandin D synthetase. J. Biol. Chem. 1990, 265, 371–375. [Google Scholar] [CrossRef]
- Hansen, H.S.J.P. 15-Hydroxyprostaglandin dehydrogenase. A review. Prostaglandins 1976, 12, 647–679. [Google Scholar] [CrossRef]
- Persson, B.; Krook, M.; Jornvall, H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 1991, 200, 537–543. [Google Scholar] [CrossRef]
- Lister, A.L.; Van Der Kraak, G. An investigation into the role of prostaglandins in zebrafish oocyte maturation and ovulation. Gen. Comp. Endocrinol. 2008, 159, 46–57. [Google Scholar] [CrossRef]
- Ruggeri, B.; Thoroughgood, C.A. Prostaglandins in aquatic fauna: A comprehensive review. Mar. Ecol. Prog. Ser. 1985, 23, 301–306. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, X.; Yu, Y.; Huang, H.; Li, F.; Xiang, J. Comparative transcriptomic characterization of the early development in Pacific white shrimp Litopenaeus vannamei. PLoS ONE 2014, 9, e106201. [Google Scholar]
- Gao, Y.; Zhang, X.; Wei, J.; Sun, X.; Yuan, J.; Li, F.; Xiang, J. Whole Transcriptome Analysis Provides Insights into Molecular Mechanisms for Molting in Litopenaeus vannamei. PLoS ONE 2015, 10, e0144350. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).