Exploiting Structural Modelling Tools to Explore Host-Translocated Effector Proteins
Abstract
:1. Introduction
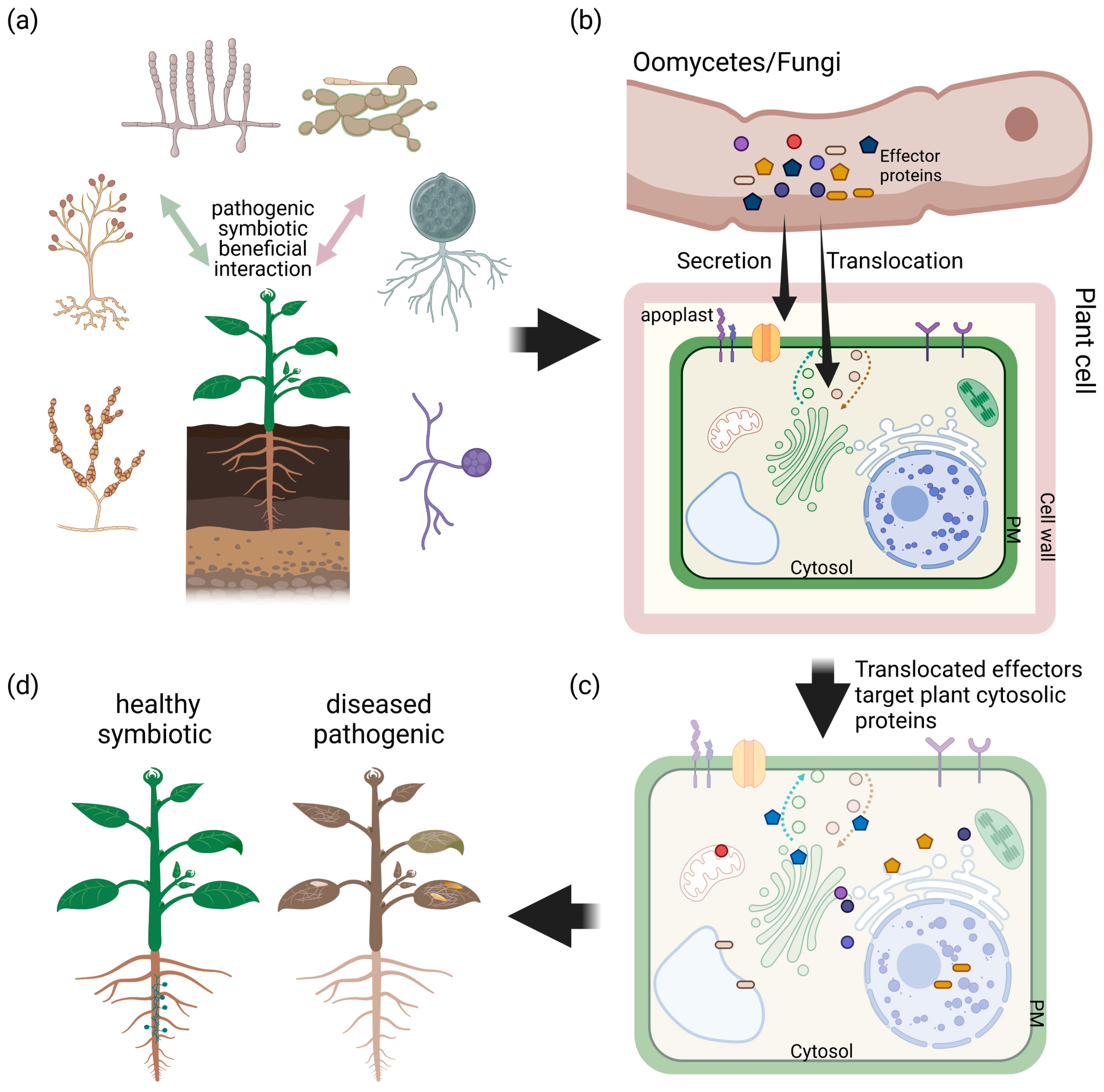
2. Effector Proteins in Plant–Microbe Interaction
2.1. All Lifestyles of Filamentous Microbes Use Effector Proteins to Establish Colonization
2.2. Effector Proteins of Filamentous Microbes
3. Structures and Computational Modelling of Effector Proteins
4. Protein Modelling Approaches for Effector Proteins
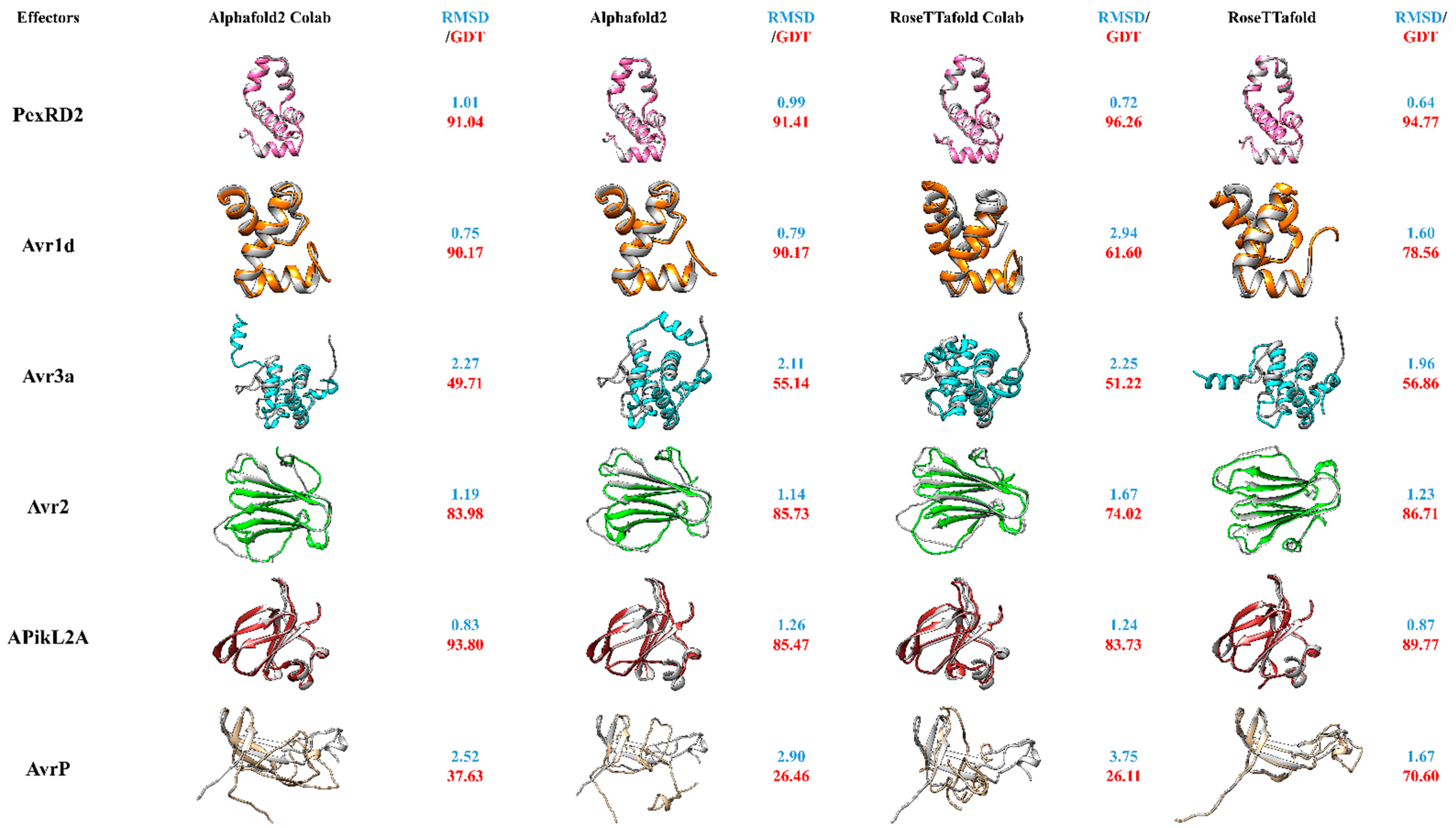
4.1. New Developments: AlphaFold2 and RoseTTafold
4.2. User-Friendly Colabfold Alternatives
4.3. AlphaFold2 and RoseTTafold for Structural Prediction of Effector Proteins
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Nishad, R.; Ahmed, T.; Rahman, V.J.; Kareem, A. Modulation of Plant Defense System in Response to Microbial Interactions. Front. Microbiol. 2020, 11, 1298. [Google Scholar] [CrossRef]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems Biology of Plant-Microbiome Interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Hong, L.; Li, X.-Y.; Yao, Y.; Hu, B.; Li, L. Improved Drought and Salt Tolerance in Transgenic Arabidopsis Overexpressing a NAC Transcriptional Factor fromArachis hypogaea. Biosci. Biotechnol. Biochem. 2011, 75, 443–450. [Google Scholar] [CrossRef] [Green Version]
- Newton, A.C.; Johnson, S.; Gregory, P. Implications of climate change for diseases, crop yields and food security. Euphytica 2011, 179, 3–18. [Google Scholar] [CrossRef]
- Margulis, L.; Fester, R. Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis; Mit Press: Cambridge, MA, USA, 1991. [Google Scholar]
- De Bary, A. Die Erscheinung der Symbiose: Vortrag Gehalten auf der Versammlung Deutscher Naturforscher und Aerzte zu Cassel; Trübner: London, UK, 1879. [Google Scholar]
- Pieterse, C.M.; de Jonge, R.; Berendsen, R. The Soil-Borne Supremacy. Trends Plant Sci. 2016, 21, 171–173. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Pieterse, C.M.J. Modulation of Host Immunity by Beneficial Microbes. Mol. Plant-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Shiu, S.-H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiu, S.-H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, 2001, re22. [Google Scholar] [CrossRef]
- Darvill, A.G.; Albersheim, P. Phytoalexins and their Elicitors-A Defense against Microbial Infection in Plants. Annu. Rev. Plant Physiol. 1984, 35, 243–275. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.-R.; Zhang, R. Exploring Elicitors of the Beneficial Rhizobacterium Bacillus amyloliquefaciens SQR9 to Induce Plant Systemic Resistance and Their Interactions With Plant Signaling Pathways. Mol. Plant-Microbe Interact. 2018, 31, 560–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-Triggered Immunity: From Pathogen Perception to Robust Defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [Green Version]
- Ye, W.; Wang, Q.; Tripathy, S.; Zhang, M.; Vetukuri, R.R. Editorial: Genomics and Effectomics of Filamentous Plant Pathogens. Front. Genet. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.P.; Srivastava, D.; Jaiswar, A.; Adholeya, A. Effector proteins of Rhizophagus proliferus: Conserved protein domains may play a role in host-specific interaction with different plant species. Braz. J. Microbiol. 2019, 50, 593–601. [Google Scholar] [CrossRef]
- Prasad, P.; Savadi, S.; Bhardwaj, S.C.; Gangwar, O.P.; Kumar, S. Rust pathogen effectors: Perspectives in resistance breeding. Planta 2019, 250, 1–22. [Google Scholar] [CrossRef]
- Zuo, W.; Ökmen, B.; Depotter, J.R.; Ebert, M.K.; Redkar, A.; Misas Villamil, J.; Doehlemann, G. Molecular interactions between smut fungi and their host plants. Annu. Rev. Phytopathol. 2019, 57, 411–430. [Google Scholar] [CrossRef] [PubMed]
- Voß, S.; Betz, R.; Heidt, S.; Corradi, N.; Requena, N. RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Front. Microbiol. 2018, 9, 2068. [Google Scholar] [CrossRef] [PubMed]
- Casarrubia, S.; Daghino, S.; Kohler, A.; Morin, E.; Khouja, H.-R.; Daguerre, Y.; Veneault-Fourrey, C.; Martin, F.M.; Perotto, S.; Martino, E. The Hydrophobin-Like OmSSP1 May Be an Effector in the Ericoid Mycorrhizal Symbiosis. Front. Plant Sci. 2018, 9, 546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Göhre, V.; Robatzek, S. Breaking the Barriers: Microbial Effector Molecules Subvert Plant Immunity. Annu. Rev. Phytopathol. 2008, 46, 189–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macho, A.P.; Zipfel, C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 2015, 23, 14–22. [Google Scholar] [CrossRef]
- Rovenich, H.; Boshoven, J.C.; Thomma, B.P. Filamentous pathogen effector functions: Of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 2014, 20, 96–103. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P. Understanding Plant Immunity as a Surveillance System to Detect Invasion. Annu. Rev. Phytopathol. 2015, 53, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Stotz, H.; Mitrousia, G.K.; de Wit, P.J.; Fitt, B.D. Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 2014, 19, 491–500. [Google Scholar] [CrossRef] [Green Version]
- Benhamou, N.; Rey, P.; Picard, K.; Tirilly, Y. Ultrastructural and Cytochemical Aspects of the Interaction between the Mycoparasite Pythium oligandrum and Soilborne Plant Pathogens. Phytopathology 1999, 89, 506–517. [Google Scholar] [CrossRef] [Green Version]
- Paul, B. Pythium periplocum, an aggressive mycoparasite of Botrytis cinerea causing the gray mould disease of grape-vine. FEMS Microbiol. Lett. 1999, 181, 277–280. [Google Scholar] [CrossRef]
- de Lamo, F.J.; Takken, F.L. Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Front. Plant Sci. 2020, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Plett, J.M.; Martin, F.M. Know your enemy, embrace your friend: Using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 2018, 93, 729–746. [Google Scholar] [CrossRef] [Green Version]
- Parniske, M. Intracellular accommodation of microbes by plants: A common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 2000, 3, 320–328. [Google Scholar] [CrossRef]
- Wawra, S.; Fesel, P.; Widmer, H.; Timm, M.; Seibel, J.P.D.; Leson, L.; Kesseler, L.; Nostadt, R.; Hilbert, M.; Langen, G.; et al. The fungal-specific β-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat. Commun. 2016, 7, 13188. [Google Scholar] [CrossRef]
- Kloppholz, S.; Kuhn, H.; Requena, N. A Secreted Fungal Effector of Glomus intraradices Promotes Symbiotic Biotrophy. Curr. Biol. 2011, 21, 1204–1209. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xu, L.; Jia, Q.; Pan, R.; Oelmüller, R.; Zhang, W.; Wu, C. Arms race: Diverse effector proteins with conserved motifs. Plant Signal. Behav. 2019, 14, 1–18. [Google Scholar] [CrossRef]
- Wawra, S.; Belmonte, R.; Löbach, L.; Saraiva, M.; Willems, A.; van West, P. Secretion, delivery and function of oomycete effector proteins. Curr. Opin. Microbiol. 2012, 15, 685–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Contreras, Y.J.; Ramírez-Valdespino, C.A.; Guzmán-Guzmán, P.; Macías-Segoviano, J.I.; Villagómez-Castro, J.C.; Olmedo-Monfil, V. Tal6 From Trichoderma atroviride Is a LysM Effector Involved in Mycoparasitism and Plant Association. Front. Microbiol. 2019, 10, 2231. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Rodriguez-Moreno, L.; Mansurkhodzaev, A.; Wang, P.; van den Berg, W.; Gasciolli, V.; Cottaz, S.; Fort, S.; Thomma, B.P.H.J.; Bono, J.-J.; et al. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2019, 225, 448–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plett, J.M.; Khachane, A.; Ouassou, M.; Sundberg, B.; Kohler, A.; Martin, F. Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between L accaria bicolor and P opulus roots. New Phytol. 2014, 202, 270–286. [Google Scholar] [CrossRef]
- Tsuzuki, S.; Handa, Y.; Takeda, N.; Kawaguchi, M. Strigolactone-Induced Putative Secreted Protein 1 Is Required for the Establishment of Symbiosis by the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis. Mol. Plant-Microbe Interac. 2016, 29, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Jiang, H.; Boeren, S.; Dings, H.; Kulikova, O.; Bisseling, T.; Limpens, E. A nuclear-targeted effector of Rhizophagus irregularis interferes with histone 2B mono-ubiquitination to promote arbuscular mycorrhization. New Phytol. 2021, 230, 1142–1155. [Google Scholar] [CrossRef]
- Akum, F.N.; Steinbrenner, J.; Biedenkopf, D.; Imani, J.; Kogel, K.-H. The Piriformospora indica effector PIIN_08944 promotes the mutualistic Sebacinalean symbiosis. Front. Plant Sci. 2015, 6, 906. [Google Scholar] [CrossRef] [Green Version]
- Nostadt, R.; Hilbert, M.; Nizam, S.; Rovenich, H.; Wawra, S.; Martin, J.; Kuepper, H.; Mijovilovich, A.; Ursinus, A.; Langen, G.; et al. A secreted fungal histidine-and alanine-rich protein regulates metal ion homeostasis and oxidative stress. New Phytol. 2020, 227, 1174–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panstruga, R.; Dodds, P.N. Terrific Protein Traffic: The Mystery of Effector Protein Delivery by Filamentous Plant Pathogens. Science 2009, 324, 748–750. [Google Scholar] [CrossRef] [Green Version]
- Whisson, S.C.; Boevink, P.C.; Moleleki, L.; Avrova, A.O.; Morales, J.G.; Gilroy, E.; Armstrong, M.R.; Grouffaud, S.; Van West, P.; Chapman, S.; et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nat. Cell Biol. 2007, 450, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.D.; Gu, B.; Capelluto, D.G.; Dou, D.; Feldman, E.; Rumore, A.; Arredondo, F.D.; Hanlon, R.; Fudal, I.; Rouxel, T.; et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 2010, 142, 284–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wawra, S.; Trusch, F.; Matena, A.; Apostolakis, K.; Linne, U.; Zhukov, I.; Stanek, J.; Koźmiński, W.; Davidson, I.; Secombes, C.J.; et al. The RxLR Motif of the Host Targeting Effector AVR3a of Phytophthora infestans Is Cleaved before Secretion. Plant Cell 2017, 29, 1184–1195. [Google Scholar] [CrossRef] [Green Version]
- Birch, P.R.J.; Armstrong, M.; Bos, J.; Boevink, P.; Gilroy, E.M.; Taylor, R.M.; Wawra, S.; Pritchard, L.; Conti, L.; Ewan, R.; et al. Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. J. Exp. Bot. 2009, 60, 1133–1140. [Google Scholar] [CrossRef] [Green Version]
- Mukhi, N.; Gorenkin, D.; Banfield, M.J. Exploring folds, evolution and host interactions: Understanding effector structure/function in disease and immunity. New Phytol. 2020, 227, 326–333. [Google Scholar] [CrossRef]
- Meisrimler, C.N.; Pelgrom, A.J.; Oud, B.; Out, S.; Van den Ackerveken, G. Multiple downy mildew effectors target the stress-related NAC transcription factor Ls NAC 069 in lettuce. Plant J. 2019, 99, 1098–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Pritchard, L.; Gomez, S.; Morales, J.; Whisson, S.C.; Beynon, J.L.; Birch, P.R.J. An RxLR Effector from Phytophthora infestans Prevents Re-localisation of Two Plant NAC Transcription Factors from the Endoplasmic Reticulum to the Nucleus. PLoS Pathog. 2013, 9, e1003670. [Google Scholar] [CrossRef] [Green Version]
- Stam, R.; Jupe, J.; Howden, A.J.; Morris, J.A.; Boevink, P.C.; Hedley, P.E.; Huitema, E. Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS ONE 2013, 8, e59517. [Google Scholar] [CrossRef]
- Stam, R.; Howden, A.J.M.; Delgado-Cerezo, M.; Amaro, T.M.M.M.; Motion, G.B.; Pham, J.; Huitema, E. Characterization of cell death inducing Phytophthora capsici CRN effectors suggests diverse activities in the host nucleus. Front. Plant Sci. 2013, 4, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Guillen, K.; Ortiz-Vallejo, D.; Gracy, J.; Fournier, E.; Kroj, T.; Padilla, A. Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 2015, 11, e1005228. [Google Scholar] [CrossRef]
- Zhang, X.; He, D.; Zhao, Y.; Cheng, X.; Zhao, W.; Taylor, I.A.; Yang, J.; Liu, J.; Peng, Y.-L. A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus. Plant J. 2018, 96, 133–146. [Google Scholar] [CrossRef] [Green Version]
- Pennington, H.G.; Jones, R.; Kwon, S.; Bonciani, G.; Thieron, H.; Chandler, T.; Luong, P.; Morgan, S.N.; Przydacz, M.; Bozkurt, T.; et al. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 2019, 15, e1007620. [Google Scholar] [CrossRef]
- Zhang, X.; Farah, N.; Rolston, L.; Ericsson, D.J.; Catanzariti, A.; Bernoux, M.; Ve, T.; Bendak, K.; Chen, C.; Mackay, J.P.; et al. Crystal structure of the Melampsora lini effector AvrP reveals insights into a possible nuclear function and recognition by the flax disease resistance protein P. Mol. Plant Pathol. 2018, 19, 1196–1209. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franceschetti, M.; Maqbool, A.; Jiménez-Dalmaroni, M.J.; Pennington, H.G.; Kamoun, S.; Banfield, M.J. Effectors of Filamentous Plant Pathogens: Commonalities amid Diversity. Microbiol. Mol. Biol. Rev. 2017, 81, e00066-16. [Google Scholar] [CrossRef] [Green Version]
- Win, J.; Krasileva, K.; Kamoun, S.; Shirasu, K.; Staskawicz, B.J.; Banfield, M.J. Sequence Divergent RXLR Effectors Share a Structural Fold Conserved across Plant Pathogenic Oomycete Species. PLoS Pathog. 2012, 8, e1002400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopakumar, O.D. Bioinformatics: Sequence and Structural Analysis; Alpha Science International, Ltd.: Oxford, UK, 2010. [Google Scholar]
- Zhang, Q.; Veretnik, S.; Bourne, P.E. Overview of Structural Bioinformatics; Springer: Singapore, 2005; pp. 15–44. [Google Scholar]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Anfinsen, C.B. Principles that Govern the Folding of Protein Chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tramontano, A. Protein Structure Prediction: Concepts and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, G.N.M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Mravic, M.; Thomaston, J.L.; Tucker, M.; Solomon, P.E.; Liu, L.; DeGrado, W.F. Packing of apolar side chains enables accurate design of highly stable membrane proteins. Science 2019, 363, 1418–1423. [Google Scholar] [CrossRef]
- Kristianingsih, R.; MacLean, D. Accurate plant pathogen effector protein classification ab initio with deepredeff: An ensemble of convolutional neural networks. BMC Bioinform. 2021, 22, 372. [Google Scholar] [CrossRef]
- Kiran, U.; Abdin, M.Z. Transgenic Technology Based Value Addition in Plant Biotechnology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- RCSB PDB. Available online: https://www.rcsb.org (accessed on 1 July 2020).
- Suh, D.; Lee, J.; Choi, S.; Lee, Y. Recent Applications of Deep Learning Methods on Evolution- and Contact-Based Protein Structure Prediction. Int. J. Mol. Sci. 2021, 22, 6032. [Google Scholar] [CrossRef]
- SWISS-MODEL. Available online: https://swissmodel.expasy.org/ (accessed on 10 September 2021).
- Modeller. Available online: https://salilab.org/modeller/ (accessed on 10 September 2021).
- PHYRE 2. Available online: http://www.sbg.bio.ic.ac.uk/phyre2/ (accessed on 10 September 2021).
- Krieger, E.; Nabuurs, S.B.; Vriend, G. Homology modeling. Methods Biochem. Anal. 2003, 44, 509–524. [Google Scholar]
- Muhammed, M.T.; Aki-Yalcin, E. Homology modeling in drug discovery: Overview, current applications, and future perspectives. Chem. Biol. Drug Des. 2019, 93, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Freddolino, P.L.; Zhang, Y. Ab Initio Protein Structure Prediction. In From Protein Structure to Function with Bioinformatics; Springer: Singapore, 2017; pp. 3–35. [Google Scholar]
- Olechnovič, K.; Monastyrskyy, B.; Kryshtafovych, A.; Venclovas, Č. Comparative analysis of methods for evaluation of protein models against native structures. Bioinformatics 2018, 35, 937–944. [Google Scholar] [CrossRef]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-accuracy protein structure prediction in CASP14. Proteins: Struct. Funct. Bioinform. 2021, 89, 1687–1699. [Google Scholar] [CrossRef]
- Protein Structure Prediction Center CASP14. Available online: https://predictioncenter.org/casp14/ (accessed on 3 October 2021).
- Protein Structure Predcition Center CASP13. Available online: http://predictioncenter.org/casp13/ (accessed on 3 October 2021).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nat. Cell Biol. 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Abagyan, R. Methods of Protein Structure Comparison. In Springer Protocols Handbooks; Springer: Singapore, 2011; Volume 857, pp. 231–257. [Google Scholar]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Mirdita, M.; Ovchinnikov, S.; Steinegger, M. ColabFold—Making protein folding accessible to all. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, X.; Zhang, X.; Song, W.; Li, X.; Feng, R.; Yang, C.; Huang, Z.; Zhu, C. Crystal structure of the RxLR effector PcRxLR12 from Phytophthora capsici. Biochem. Biophys. Res. Commun. 2018, 503, 1830–1835. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Hu, Q.; Zhou, J.; Yin, W.; Yao, D.; Shao, Y.; Zhao, Y.; Guo, B.; Xia, Y.; Chen, Q.; et al. Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Di, X.; Cao, L.; Hughes, R.; Tintor, N.; Banfield, M.J.; Takken, F.L.W. Structure–function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytol. 2017, 216, 897–914. [Google Scholar] [CrossRef] [Green Version]
- Bentham, A.R.; Petit-Houdenot, Y.; Win, J.; Chuma, I.; Terauchi, R.; Banfield, M.J.; Kamoun, S.; Langner, T. A single amino acid polymorphism in a conserved effector of the multihost blast fungus pathogen expands host-target binding spectrum. PLoS Pathog. 2021, 17, e1009957. [Google Scholar] [CrossRef] [PubMed]
- Alphafold2_Advanced Google Colab Notebook. Available online: https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb (accessed on 12 November 2021).
- NeSI (New Zealand eScience Infrastructure). Available online: https://www.nesi.org.nz (accessed on 2 November 2021).
- RoseTTafold Google Colab Notebook. Available online: https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/RoseTTAFold.ipynb (accessed on 14 November 2021).
- PDBefold. Available online: http://www.ebi.ac.uk/msd-srv/ssm (accessed on 22 November 2021).
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef] [PubMed]
- Zemla, A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003, 31, 3370–3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.W.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Biorender. Available online: https://biorender.com/ (accessed on 26 October 2021).
| Effector Protein | Fungal Species | Host Species | Characterized Biological Function | References |
|---|---|---|---|---|
| SP7 | Glomus intraradices | Medicago truncatula | Interacts with JA/ethylene inducible ERF19 transcription factor and down regulates PTI | [39] |
| Lysm effector Tal6 | Trichoderma atroviride | Arabidopsis thaliana | Binds to chitin of plant’s cell wall and protects the fungi hyphae from plant’s chitinase favoring Trichoderma interaction and increasing mycoparasitic effect | [42] |
| Lysm effector RiSLM | Rhizophagus irregularis | Medicago truncatula | Binds to chitin and chitooligosaccharides of plant’s cell wall and interferes with chitin-triggered immune response protecting hyphae from plant’s chitinase and enabling symbiotic reactions | [43] |
| MiSSP7 | Laccaria bicolor | Populus trichocarpa | Suppresses JA-mediated immune response by preventing JA-dependent degradation of PtJAZ6, a negative regulator of JA-induced genes | [44] |
| RiCRN1 | Rhizophagus irregularis | Medicago truncatula Nicotiana benthamiana | Establishes a functional AM symbiosis and Arbuscules phosphate transporter gene-MtP4-expression | [45] |
| Strigolactone induced secreted protein 1 (SIS1) | Rhizophagus irregularis | Medicago truncatula | Essential for AM symbiosis, gene silencing causes suppression of colonization and production of stunted arbuscules | [25] |
| RP8598 and RP23081 | Rhizophagus proliferus | Medicago truncatula Nicotiana benthamiana Allium schoenoprasum | Interacts with JA/ethylene inducible ERF19 transcription factor and down regulates PTI | [22] |
| Nuclear localizing effector (RiNLE1) | Rhizophagus irregularis | Medicago truncatula | Interferes with mono-ubiquitination of 2B histone and decreases the expression of defense-related genes while enhancing AM colonization process | [46] |
| Hydrophobin-like OmSSP1 | Ericoid mycorrhiza | Vaccinium myrtillu | Mutants are unable to colonize V. myrtillu roots and OmSSP1 may strengthen the attachment of the fungi to the root protecting the hyphae from plant’s immune system | [26] |
| PIIN_08944 | Piriformospora indica | Arabidopsis thaliana | Mutants show delayed colonization and PIIN_08944 expression reveals impairment of SA-defense pathway and reduced expression of flg-22 | [47] |
| Did1 (PIIN_05872) | Piriformospora indica | - | Interferes with iron-mediated defense response which plays an important role in ROS generation | [48] |
| Effector Protein | Organism | Date of Release | Method | PDB Entry | Family |
|---|---|---|---|---|---|
| Fungi | |||||
| Ecp11-1 | Passalora fulva | 4 August 2021 | X-ray | 6ZUS | LARS |
| APikL2A | Magnaporthe oryzae | 24 March 2021 | X-ray | 7NLJ | MAX |
| APikL2F | Magnaporthe oryzae | 24 March 2021 | X-ray | 7NMM | MAX |
| AVR-PikD | Pyricularia oryzae | 17 Februrary 2021 | X-ray | 7BNT | MAX |
| AVR-PikF | Pyricularia oryzae | 3 February 2021 | X-ray | 7B1I | MAX |
| AVR-PikC | Pyricularia oryzae | 3 February 2021 | X-ray | 7A8X | MAX |
| SnTox3 | Parastagonospora nodorum | 4 November 2020 | X-ray | 6WES | MAX |
| Zt-KP6-1 | Zymoseptoria tritici | 4 March 2020 | X-ray | 6QPK | LysM |
| MLP124017 | Melampsora larici-populina | 18 December 2019 | Solution NMR | 6SGO | Cys knot, NTF2-like fold |
| Mg1LysM | Zymoseptoria tritici | 16 October 2019 | X-ray | 6Q40 | LysM |
| AVR-Pia | Pyricularia oryzae | 10 July 2019 | X-ray | 6Q76 | MAX |
| AvrPib | Pyricularia oryzae | 5 September 2018 | X-ray | 5Z1V | MAX |
| MlpP4.1 | Melampsora larici-populina | 22 August 2018 | Solution NMR | 6H0I | Cys knot, NTF2-like fold |
| Avr4 | Passalora fulva | 22 August 2018 | X-ray | 6BN0 | Chitin-binding |
| PIIN_05872 | Piriformospora indica | 2 May 2018 | X-ray | 5LOS | DELD |
| BEC1054 | Blumeria hordei | 20 June 2018 | X-ray | 6FMB | RALPH |
| AVR-PikE | Pyricularia oryzae | 13 June 2018 | X-ray | 6G11 | MAX |
| AVR-PikA | Pyricularia oryzae | 3 June 2018 | X-ray | 6FUD | MAX |
| AvrP | Melampsora lini | 30 August 2017 | X-ray | 5VJJ | Zn-binding |
| Avr2 | Fusarium oxysporum | 16 August 2017 | X-ray | 5OD4 | ToxA/TRAF |
| PevD1 | Verticillium dahliae | 5 July 2017 | X-ray | 5XMZ | C2-like |
| Avr4 | Pseudocercospora fuligena | 29 June 2017 | X-ray | 4Z4A | Chitin-binding |
| AVR1-CO39 | Magnaporthe oryzae | 14 October 2015 | Solution NMR | 2MYV | MAX |
| Prp5 | Saccharomyces cerevisiae | 11 December 2013 | X-ray | 4LK2 | DEAD-box |
| AvrLm4-7 | Leptosphaeria maculans | 11 December 2013 | X-ray | 2OPC | LARS |
| AvrM | Melampsora lini | 16 October 2013 | X-ray | 4BJM | RXLR-like |
| AvrM-A | Melampsora lini | 16 October 2013 | X-ray | 4BJN | RXLR-like |
| Ecp6 | Passalora fulva | 17 July 2013 | X-ray | 4B8V | LARS |
| AvrPiz-t | Pyricularia oryzae | 12 September 2012 | Solution NMR | 2LW6 | MAX |
| AvrL567-D | Melampsora lini | 30 October 2007 | X-ray | 2QVT | RXLR-like |
| AvrL567-A | Melampsora lini | 6 March 2007 | X-ray | 2OPC | RXLR-like |
| Oomycetes | |||||
| Avr1d | Phytophthora sojae | 17 March 2021 | X-ray | 7C96 | RXLR |
| PsAvh240 | Phytophthora sojae | 6 February 2019 | X-ray | 6J8L | RXLR/WY |
| SFI3 | Phytophthora infestans | 5 December 2018 | X-ray | 6GU1 | RXLR/WY |
| PcRXLR12 | Phytophthora capsici | 15 August 2018 | X-ray | 5ZC3 | RXLR/WY |
| PSR2 | Phytophthora sojae | 16 August 2017 | X-ray | 5GNC | RXLR/WY |
| Avr3a | Phytophthora infestans | 11 January 2017 | Solution NMR | 2NAR | RXLR/WY |
| PexRD54 | Phytophthora infestans | 3 August 2016 | X-ray | 5L7S | RXLR/WY |
| ATR13 | Hyaloperonospora parasitica | 18 january 2012 | Solution NMR | 2LAI | RXLR |
| AVR3a4 | Phytophthora capsici | 3 August 2011 | Solution NMR | 2LC2 | RXLR |
| PexRD2 | Phytophthora infestans | 3 August 2011 | X-ray | 3ZRG | RXLR/WY |
| Avr3a11 | Phytophthora capsici | 3 August 2011 | X-ray | 3ZR8 | RXLR/WY |
| ATR1 | Hyaloperonospora parasitica | 20 July 2011 | X-ray | 3RMR | RXLR/WY |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amoozadeh, S.; Johnston, J.; Meisrimler, C.-N. Exploiting Structural Modelling Tools to Explore Host-Translocated Effector Proteins. Int. J. Mol. Sci. 2021, 22, 12962. https://doi.org/10.3390/ijms222312962
Amoozadeh S, Johnston J, Meisrimler C-N. Exploiting Structural Modelling Tools to Explore Host-Translocated Effector Proteins. International Journal of Molecular Sciences. 2021; 22(23):12962. https://doi.org/10.3390/ijms222312962
Chicago/Turabian StyleAmoozadeh, Sahel, Jodie Johnston, and Claudia-Nicole Meisrimler. 2021. "Exploiting Structural Modelling Tools to Explore Host-Translocated Effector Proteins" International Journal of Molecular Sciences 22, no. 23: 12962. https://doi.org/10.3390/ijms222312962







