Dexamethasone Induces Changes in Osteogenic Differentiation of Human Mesenchymal Stromal Cells via SOX9 and PPARG, but Not RUNX2
Abstract
:1. Introduction
2. Results
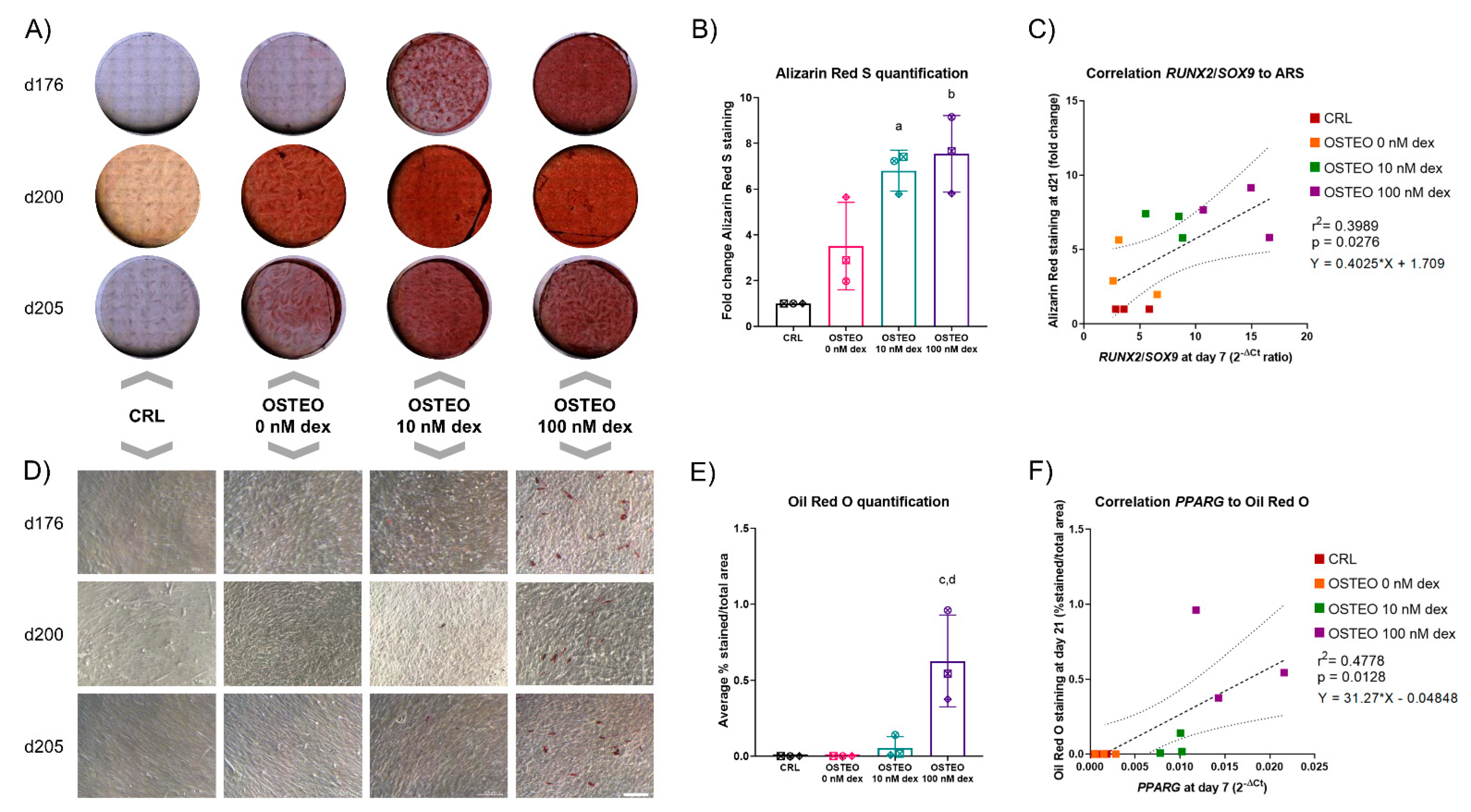
2.1. Dose-Dependent Effect of Dex on Differentiation
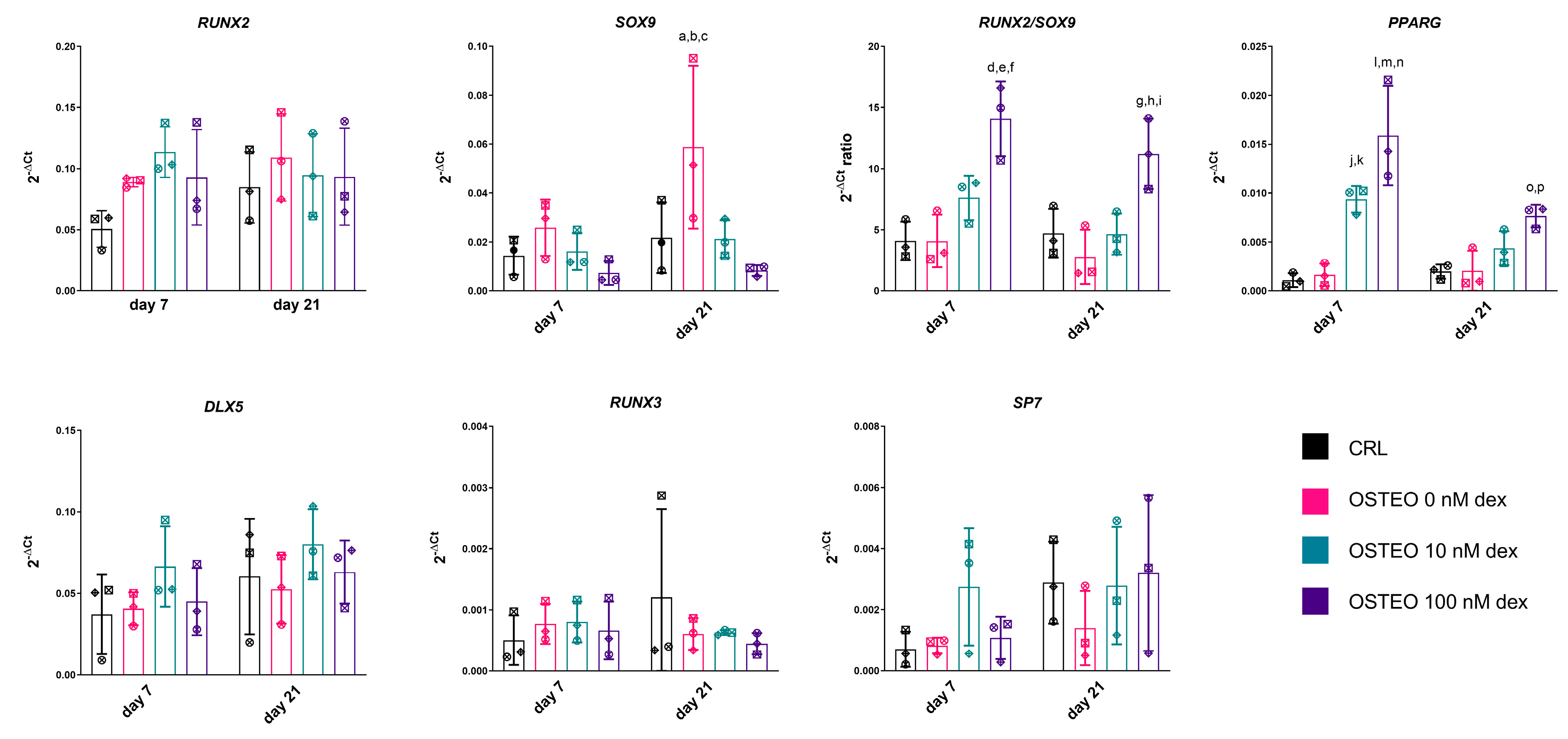
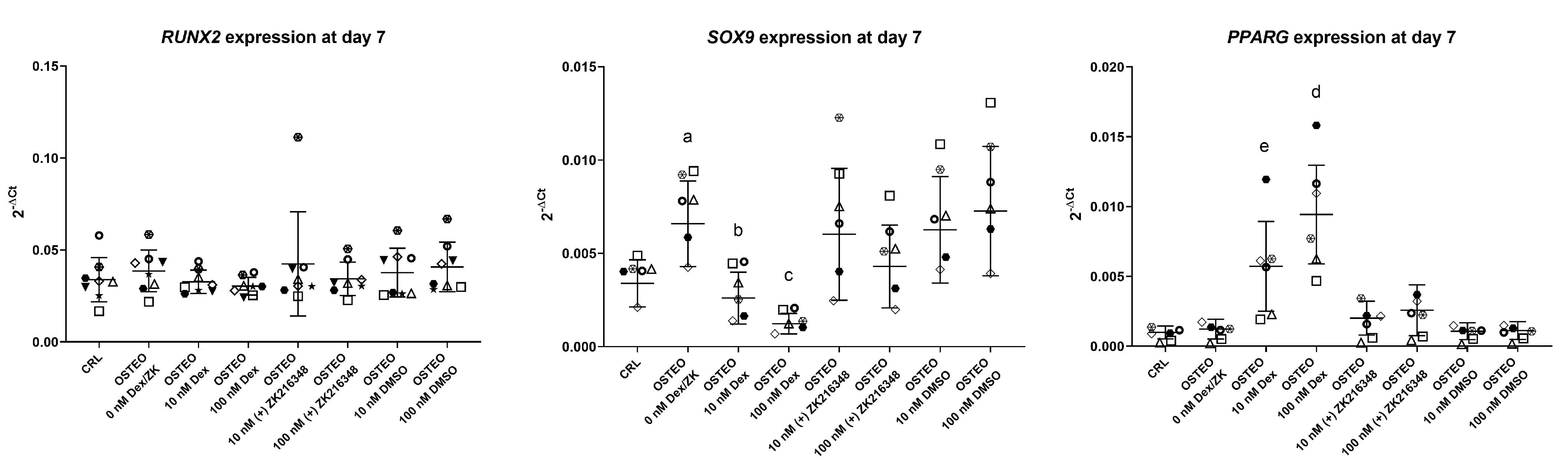
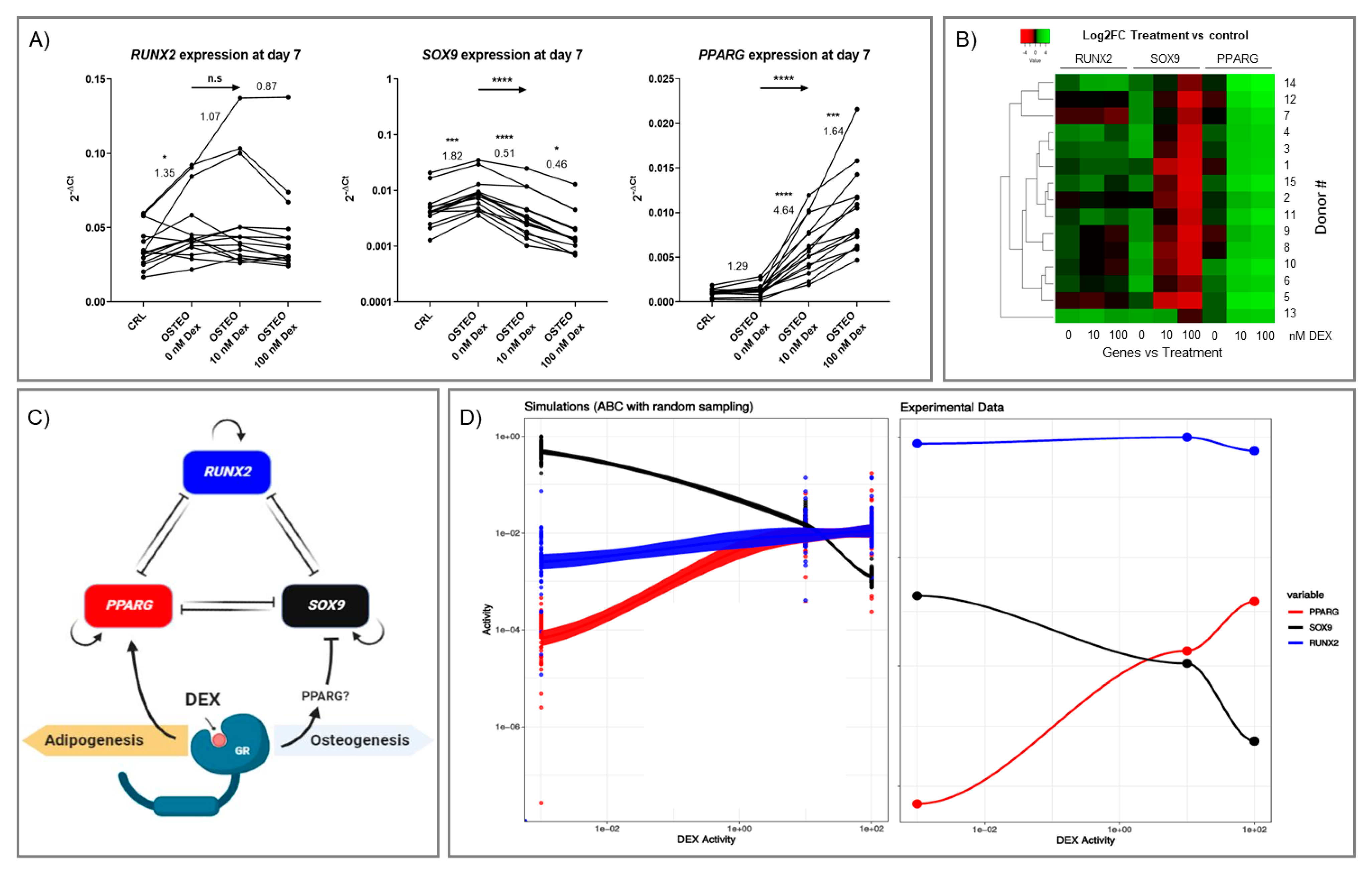
2.2. Effect of Dex on RUNX2, SOX9, and PPARG Expression
2.3. Two-Stage Osteogenic Differentiation
2.4. Expression Profiles of RUNX2, SOX9, and PPARG
3. Discussion
4. Materials and Methods
4.1. Cell Isolation and Induction of Osteogenic Differentiation
4.2. Alizarin Red Staining and Quantification
4.3. Oil Red O Staining and Quantification
4.4. Isolation of RNA
4.5. Gene Expression Analysis
4.6. Approximate Bayesian Computation Simulations
4.7. Heatmap Generation and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Heest, A.; Swiontkowski, M. Bone-graft substitutes. Lancet 1999, 353 (Suppl. 1), SI28–SI29. [Google Scholar] [CrossRef]
- Giannoudis, P.V.; Dinopoulos, H.; Tsiridis, E. Bone substitutes: An update. Injury 2005, 36 (Suppl. 3), S20–S27. [Google Scholar] [CrossRef] [PubMed]
- Hulsart-Billstrom, G.; Dawson, J.I.; Hofmann, S.; Muller, R.; Stoddart, M.J.; Alini, M.; Redl, H.; El Haj, A.; Brown, R.; Salih, V.; et al. A surprisingly poor correlation between in vitro and in vivo testing of biomaterials for bone regeneration: Results of a multicentre analysis. Eur. Cell Mater. 2016, 31, 312–322. [Google Scholar] [CrossRef]
- Barnes, P.J.; Adcock, I.M. Glucocorticoid resistance in inflammatory diseases. Lancet 2009, 373, 1905–1917. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, R.; Gu, Z.; Dong, C.; Guo, G.; Li, L. Effects of glucocorticoids on osteoporosis in rheumatoid arthritis: A systematic review and meta-analysis. Osteoporos. Int. 2020, 31, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Buckley, L.; Humphrey, M.B. Glucocorticoid-Induced Osteoporosis. N. Engl. J. Med. 2018, 379, 2547–2556. [Google Scholar] [CrossRef]
- Osmani, F.; Thakkar, S.; Vigdorchik, J. The utility of conservative treatment modalities in the management of osteonecrosis: A systematic review. Bull. NYU Hosp. Jt. Dis. 2017, 75, 186. [Google Scholar]
- Weinstein, R.S. Glucocorticoid-induced osteonecrosis. Endocrine 2012, 41, 183–190. [Google Scholar] [CrossRef]
- Eijken, M.; Koedam, M.; van Driel, M.; Buurman, C.J.; Pols, H.A.; van Leeuwen, J.P. The essential role of glucocorticoids for proper human osteoblast differentiation and matrix mineralization. Mol. Cell. Endocrinol. 2006, 248, 87–93. [Google Scholar] [CrossRef]
- Li, J.; Zhang, N.; Huang, X.; Xu, J.; Fernandes, J.C.; Dai, K.; Zhang, X. Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by C/EBPalpha promoter methylation. Cell Death Dis. 2013, 4, e832. [Google Scholar] [CrossRef] [Green Version]
- Henneicke, H.; Gasparini, S.J.; Brennan-Speranza, T.C.; Zhou, H.; Seibel, M.J. Glucocorticoids and bone: Local effects and systemic implications. Trends Endocrinol. Metabolism. 2014, 25, 197–211. [Google Scholar] [CrossRef]
- Zhou, H.; Cooper, M.S.; Seibel, M.J. Endogenous glucocorticoids and bone. Bone Res. 2013, 1, 107–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, A.V.; LeBoff, M.S. Osteoanabolic agents for osteoporosis. J. Endocr. Soc. 2018, 2, 922–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, S.L.; Yang, J.W.; Rifas, L.; Zhang, S.F.; Avioli, L.V. Differentiation of human bone marrow osteogenic stromal cells in vitro: Induction of the osteoblast phenotype by dexamethasone. Endocrinology 1994, 134, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Langenbach, F.; Handschel, J. Effects of dexamethasone, ascorbic acid and beta-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res. Ther. 2013, 4, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardner, O.F.W.; Alini, M.; Stoddart, M.J. Mesenchymal Stem Cells Derived from Human Bone Marrow. In Cartilage Tissue Engineering: Methods and Protocols; Doran, P.M., Ed.; Springer: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Hanna, H.; Mir, L.M.; Andre, F.M. In vitro osteoblastic differentiation of mesenchymal stem cells generates cell layers with distinct properties. Stem Cell Res. Ther. 2018, 9, 203. [Google Scholar] [CrossRef]
- Hamidouche, Z.; Hay, E.; Vaudin, P.; Charbord, P.; Schule, R.; Marie, P.J.; Fromigue, O. FHL2 mediates dexamethasone-induced mesenchymal cell differentiation into osteoblasts by activating Wnt/beta-catenin signaling-dependent Runx2 expression. FASEB J. 2008, 22, 3813–3822. [Google Scholar] [CrossRef]
- Bruderer, M.; Richards, R.G.; Alini, M.; Stoddart, M.J. Role and regulation of RUNX2 in osteogenesis. Eur. Cell Mater. 2014, 28, 269–286. [Google Scholar] [CrossRef]
- Felthaus, O.; Gosau, M.; Klein, S.; Prantl, L.; Reichert, T.E.; Schmalz, G.; Morsczeck, C. Dexamethasone-related osteogenic differentiation of dental follicle cells depends on ZBTB16 but not Runx2. Cell Tissue Res. 2014, 357, 695–705. [Google Scholar] [CrossRef]
- Maioli, M.; Basoli, V.; Santaniello, S.; Cruciani, S.; Delitala, A.P.; Pinna, R.; Milia, E.; Grillari-Voglauer, R.; Fontani, V.; Rinaldi, S.; et al. Osteogenesis from Dental Pulp Derived Stem Cells: A Novel Conditioned Medium Including Melatonin within a Mixture of Hyaluronic, Butyric, and Retinoic Acids. Stem Cells Int. 2016, 2016, 2056416. [Google Scholar] [CrossRef] [Green Version]
- Alm, J.J.; Heino, T.J.; Hentunen, T.A.; Vaananen, H.K.; Aro, H.T. Transient 100 nM dexamethasone treatment reduces inter- and intraindividual variations in osteoblastic differentiation of bone marrow-derived human mesenchymal stem cells. Tissue Eng. Part C Methods 2012, 18, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.M.; Rao, L.G.; Ly, H.; Hamilton, L.; Tong, J.; Sturtridge, W.; McBroom, R.; Aubin, J.E.; Murray, T.M. Long-term effects of physiologic concentrations of dexamethasone on human bone-derived cells. J. Bone Miner. Res. 1990, 5, 803–813. [Google Scholar] [CrossRef]
- Rimando, M.G.; Wu, H.H.; Liu, Y.A.; Lee, C.W.; Kuo, S.W.; Lo, Y.P.; Tseng, K.F.; Liu, Y.S.; Lee, O.K. Glucocorticoid receptor and Histone deacetylase 6 mediate the differential effect of dexamethasone during osteogenesis of mesenchymal stromal cells (MSCs). Sci. Rep. 2016, 6, 37371. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, N.; Noh, T.; Khokhar, A.; Smith, E.; Frenkel, B. Glucocorticoids inhibit osteocalcin transcription in osteoblasts by suppressing Egr2/Krox20-binding enhancer. Arthritis Rheum. 2005, 52, 929–939. [Google Scholar] [CrossRef]
- Della Bella, E.; Menzel, U.; Basoli, V.; Tourbier, C.; Alini, M.; Stoddart, M.J. Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells 2020, 9, 398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghali, O.; Broux, O.; Falgayrac, G.; Haren, N.; van Leeuwen, J.P.; Penel, G.; Hardouin, P.; Chauveau, C. Dexamethasone in osteogenic medium strongly induces adipocyte differentiation of mouse bone marrow stromal cells and increases osteoblast differentiation. BMC Cell Biol. 2015, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Yoshiko, Y.; Oizumi, K.; Hasegawa, T.; Minamizaki, T.; Tanne, K.; Maeda, N.; Aubin, J.E. A subset of osteoblasts expressing high endogenous levels of PPARgamma switches fate to adipocytes in the rat calvaria cell culture model. PLoS ONE 2010, 5, e11782. [Google Scholar] [CrossRef] [Green Version]
- Frith, J.; Genever, P. Transcriptional control of mesenchymal stem cell differentiation. Transfus. Med. Hemother. 2008, 35, 216–227. [Google Scholar] [CrossRef]
- van de Peppel, J.; Strini, T.; Tilburg, J.; Westerhoff, H.; van Wijnen, A.J.; van Leeuwen, J.P. Identification of Three Early Phases of Cell-Fate Determination during Osteogenic and Adipogenic Differentiation by Transcription Factor Dynamics. Stem Cell Rep. 2017, 8, 947–960. [Google Scholar] [CrossRef]
- Loebel, C.; Czekanska, E.M.; Bruderer, M.; Salzmann, G.; Alini, M.; Stoddart, M.J. In vitro osteogenic potential of human mesenchymal stem cells is predicted by Runx2/Sox9 ratio. Tissue Eng. Part A 2015, 21, 115–123. [Google Scholar] [CrossRef] [Green Version]
- Schacke, H.; Schottelius, A.; Docke, W.D.; Strehlke, P.; Jaroch, S.; Schmees, N.; Rehwinkel, H.; Hennekes, H.; Asadullah, K. Dissociation of transactivation from transrepression by a selective glucocorticoid receptor agonist leads to separation of therapeutic effects from side effects. Proc. Natl. Acad. Sci. USA 2004, 101, 227–232. [Google Scholar] [CrossRef] [Green Version]
- MacArthur, B.D.; Please, C.P.; Oreffo, R.O. Stochasticity and the molecular mechanisms of induced pluripotency. PLoS ONE 2008, 3, e3086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buetti-Dinh, A.; Friedman, R. Computer simulations of the signalling network in FLT3 (+)-acute myeloid leukaemia—Indications for an optimal dosage of inhibitors against FLT3 and CDK6. BMC Bioinform. 2018, 19, 155. [Google Scholar] [CrossRef] [Green Version]
- Buetti-Dinh, A.; O’Hare, T.; Friedman, R. Sensitivity Analysis of the NPM-ALK Signalling Network Reveals Important Pathways for Anaplastic Large Cell Lymphoma Combination Therapy. PLoS ONE 2016, 11, e0163011. [Google Scholar] [CrossRef] [PubMed]
- Buetti-Dinh, A.; Pivkin, I.V.; Friedman, R. S100A4 and its role in metastasis–computational integration of data on biological networks. Mol. Biosyst. 2015, 11, 2238–2246. [Google Scholar] [CrossRef] [Green Version]
- Buetti-Dinh, A.; Herold, M.; Christel, S.; El Hajjami, M.; Delogu, F.; Ilie, O.; Bellenberg, S.; Wilmes, P.; Poetsch, A.; Sand, W.; et al. Reverse engineering directed gene regulatory networks from transcriptomics and proteomics data of biomining bacterial communities with approximate Bayesian computation and steady-state signalling simulations. BMC Bioinform. 2020, 21, 23. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, T.; Khademhosseini, A.; Langer, R. Chasing the Paradigm: Clinical Translation of 25 Years of Tissue Engineering. Tissue Eng. Part A 2019, 25, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.S. Glucocorticoids, osteocytes, and skeletal fragility: The role of bone vascularity. Bone 2010, 46, 564–570. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Xu, Y.; Wang, Y.; Jiang, Y. Differential expression profiles of long noncoding RNAs and mRNAs in human bone marrow mesenchymal stem cells after exposure to a high dosage of dexamethasone. Stem Cell Res. Ther. 2021, 12, 9. [Google Scholar] [CrossRef]
- Juszczak, G.R.; Stankiewicz, A.M. Glucocorticoids, genes and brain function. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 82, 136–168. [Google Scholar] [CrossRef]
- Carter, B.S.; Meng, F.; Thompson, R.C. Glucocorticoid treatment of astrocytes results in temporally dynamic transcriptome regulation and astrocyte-enriched mRNA changes in vitro. Physiol. Genom. 2012, 44, 1188–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peffer, M.E.; Chandran, U.R.; Luthra, S.; Volonte, D.; Galbiati, F.; Garabedian, M.J.; Monaghan, A.P.; DeFranco, D.B. Caveolin-1 regulates genomic action of the glucocorticoid receptor in neural stem cells. Mol. Cell. Biol. 2014, 34, 2611–2623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Horikawa, Y.; Iizuka, K.; Sakurai, N.; Tanaka, T.; Shihara, N.; Oshima, A.; Takeda, J.; Mikuni, M. Large-scale analysis of glucocorticoid target genes in rat hypothalamus. J. Neurochem. 2008, 106, 805–814. [Google Scholar] [CrossRef]
- Naito, M.; Ohashi, A.; Takahashi, T. Dexamethasone inhibits chondrocyte differentiation by suppression of Wnt/beta-catenin signaling in the chondrogenic cell line ATDC5. Histochem. Cell Biol. 2015, 144, 261–272. [Google Scholar] [CrossRef]
- Akiyama, H.; Chaboissier, M.C.; Martin, J.F.; Schedl, A.; de Crombrugghe, B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002, 16, 2813–2828. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Shou, P.; Zheng, C.; Jiang, M.; Cao, G.; Yang, Q.; Cao, J.; Xie, N.; Velletri, T.; Zhang, X.; et al. Fate decision of mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death Differ. 2016, 23, 1128–1139. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, K.; Koenen, M.; Schauer, S.; Wittig-Blaich, S.; Ahmad, M.; Baschant, U.; Tuckermann, J.P. Molecular Actions of Glucocorticoids in Cartilage and Bone During Health, Disease, and Steroid Therapy. Physiol. Rev. 2016, 96, 409–447. [Google Scholar] [CrossRef] [Green Version]
- Kovermann, N.J.; Basoli, V.; Della Bella, E.; Alini, M.; Lischer, C.; Schmal, H.; Kubosch, E.J.; Stoddart, M.J. BMP2 and TGF-beta Cooperate Differently during Synovial-Derived Stem-Cell Chondrogenesis in a Dexamethasone-Dependent Manner. Cells 2019, 8, 636. [Google Scholar] [CrossRef] [Green Version]
- Ge, C.; Cawthorn, W.P.; Li, Y.; Zhao, G.; Macdougald, O.A.; Franceschi, R.T. Reciprocal Control of Osteogenic and Adipogenic Differentiation by ERK/MAP Kinase Phosphorylation of Runx2 and PPARgamma Transcription Factors. J. Cell. Physiol. 2016, 231, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Della Bella, E.; Pagani, S.; Giavaresi, G.; Capelli, I.; Comai, G.; Donadei, C.; Cappuccilli, M.; La Manna, G.; Fini, M. Uremic Serum Impairs Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stromal Cells. J. Cell. Physiol. 2017, 232, 2201–2209. [Google Scholar] [CrossRef]
- Buetti-Dinh, A.; Pivkin, I.V.; Friedman, R. S100A4 and its role in metastasis—Simulations of knockout and amplification of epithelial growth factor receptor and matrix metalloproteinases. Mol. Biosyst. 2015, 11, 2247–2254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csilléry, K.; François, O.; Blum, M.G.B. abc: An R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 2012, 3, 475–479. [Google Scholar] [CrossRef] [Green Version]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]

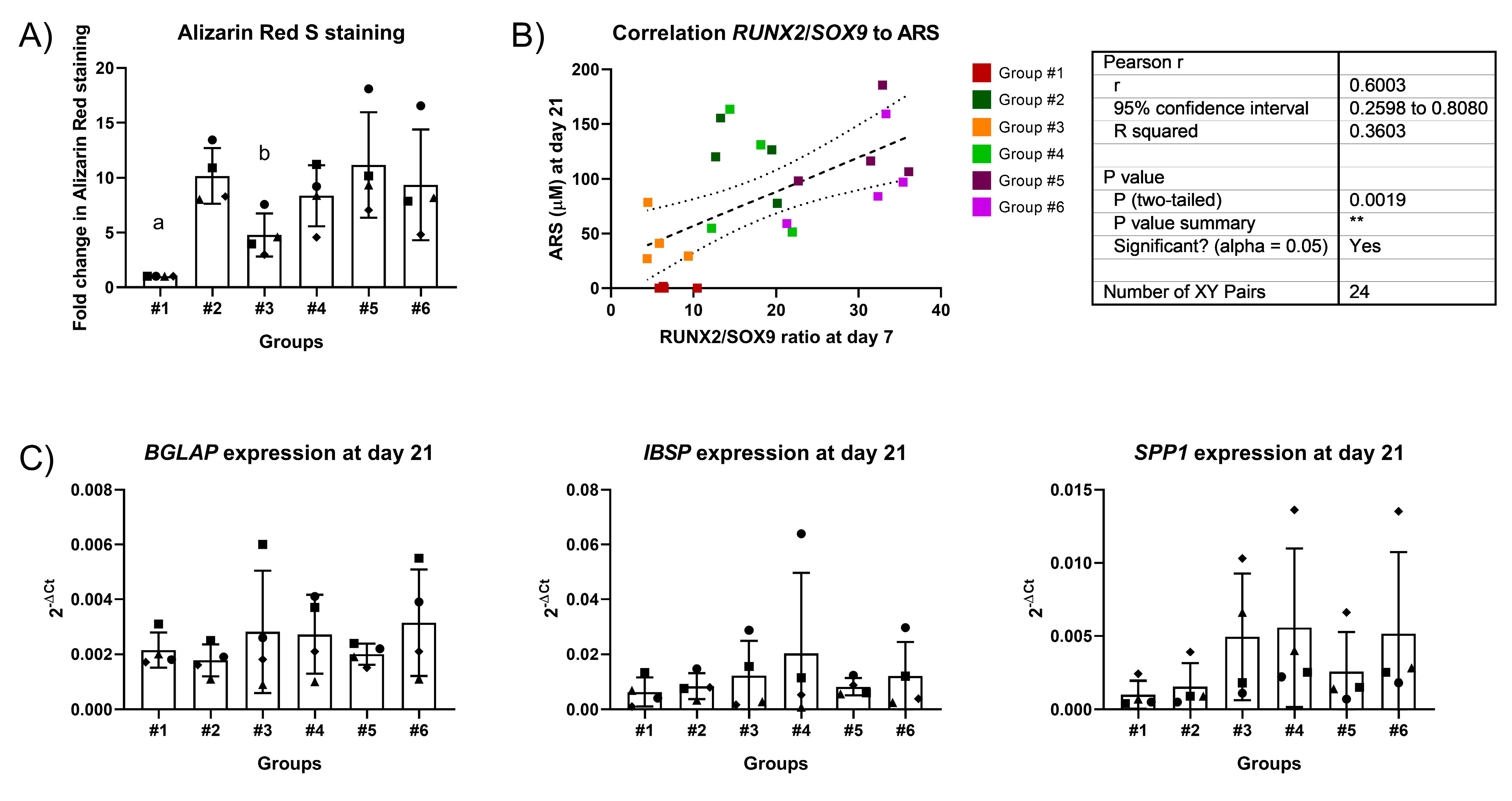
| Group | Week 1 | Week 2 + 3 |
|---|---|---|
| #1 | CRL medium | CRL medium |
| #2 | OSTEO 10 nM dex | OSTEO 10 nM dex |
| #3 | OSTEO 0 nM dex | OSTEO 0 nM dex |
| #4 | OSTEO 10 nM dex | OSTEO 0 nM dex |
| #5 | OSTEO 100 nM dex | OSTEO 10 nM dex |
| #6 | OSTEO 100 nM dex | OSTEO 0 nM dex |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Della Bella, E.; Buetti-Dinh, A.; Licandro, G.; Ahmad, P.; Basoli, V.; Alini, M.; Stoddart, M.J. Dexamethasone Induces Changes in Osteogenic Differentiation of Human Mesenchymal Stromal Cells via SOX9 and PPARG, but Not RUNX2. Int. J. Mol. Sci. 2021, 22, 4785. https://doi.org/10.3390/ijms22094785
Della Bella E, Buetti-Dinh A, Licandro G, Ahmad P, Basoli V, Alini M, Stoddart MJ. Dexamethasone Induces Changes in Osteogenic Differentiation of Human Mesenchymal Stromal Cells via SOX9 and PPARG, but Not RUNX2. International Journal of Molecular Sciences. 2021; 22(9):4785. https://doi.org/10.3390/ijms22094785
Chicago/Turabian StyleDella Bella, Elena, Antoine Buetti-Dinh, Ginevra Licandro, Paras Ahmad, Valentina Basoli, Mauro Alini, and Martin J. Stoddart. 2021. "Dexamethasone Induces Changes in Osteogenic Differentiation of Human Mesenchymal Stromal Cells via SOX9 and PPARG, but Not RUNX2" International Journal of Molecular Sciences 22, no. 9: 4785. https://doi.org/10.3390/ijms22094785






