Differential Subcellular Distribution of Cytokinins: How Does Membrane Transport Fit into the Big Picture?
Abstract
1. Introduction
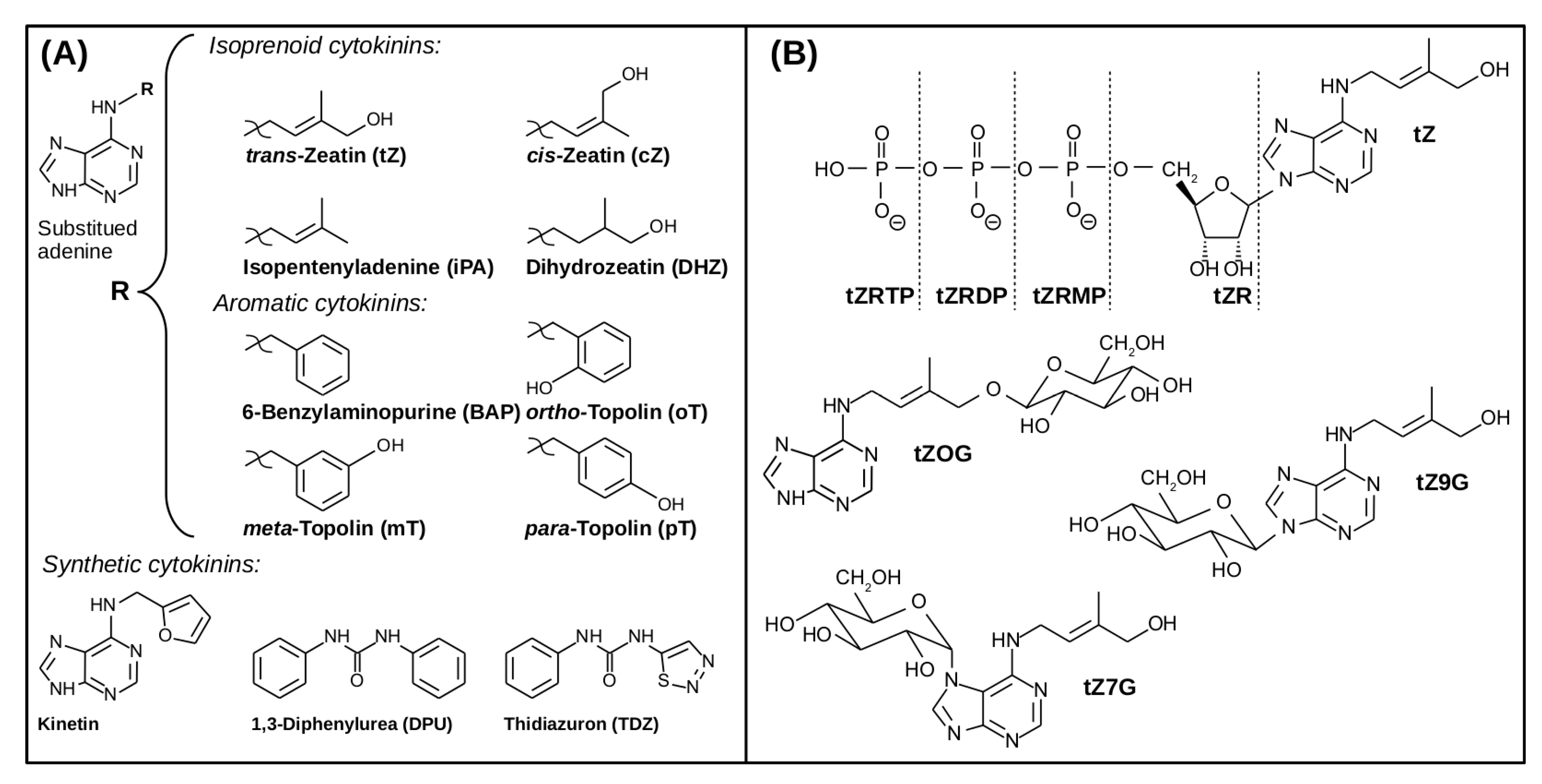
2. The Structural Variety of Naturally Occurring and Synthetic Cytokinins
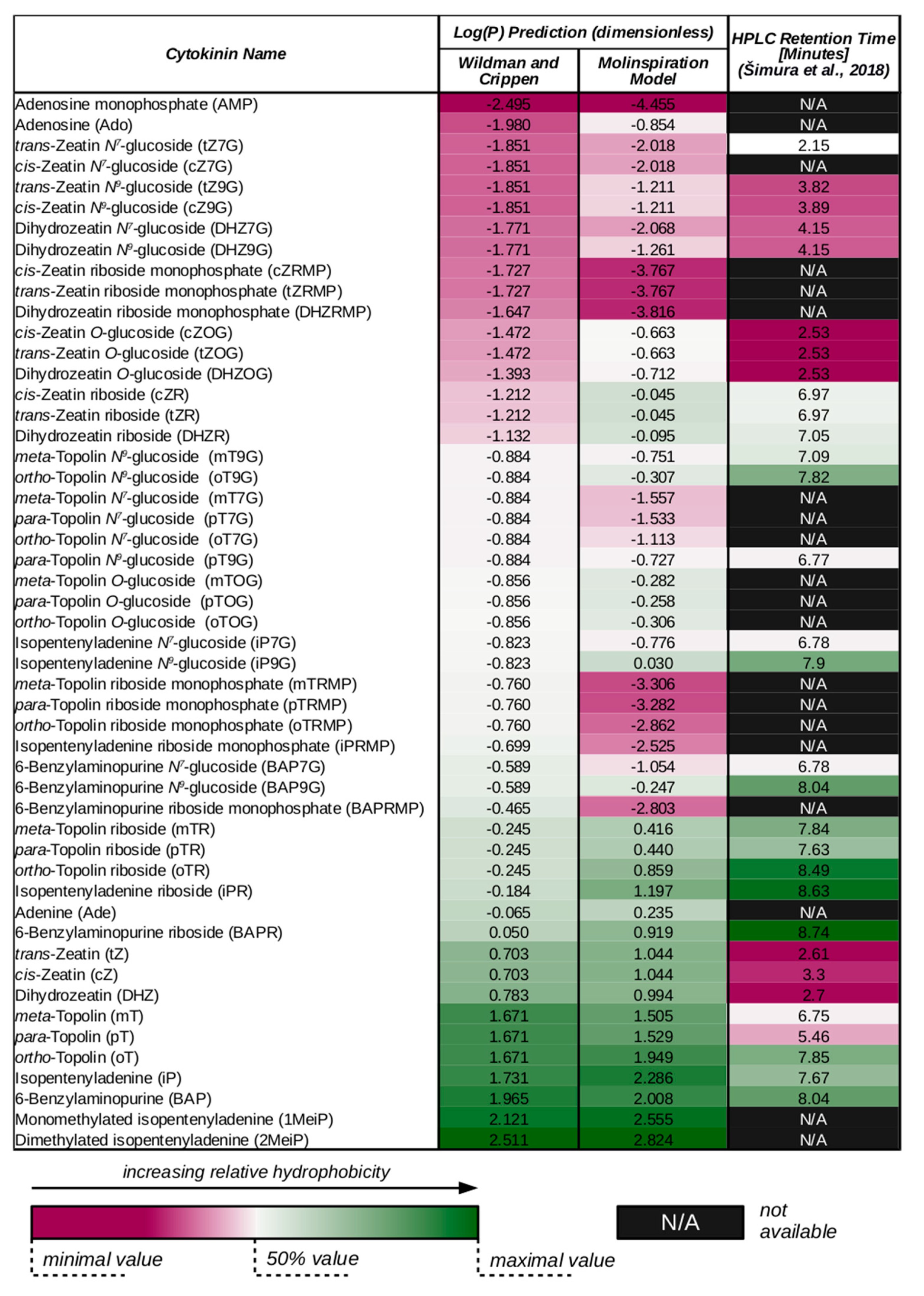
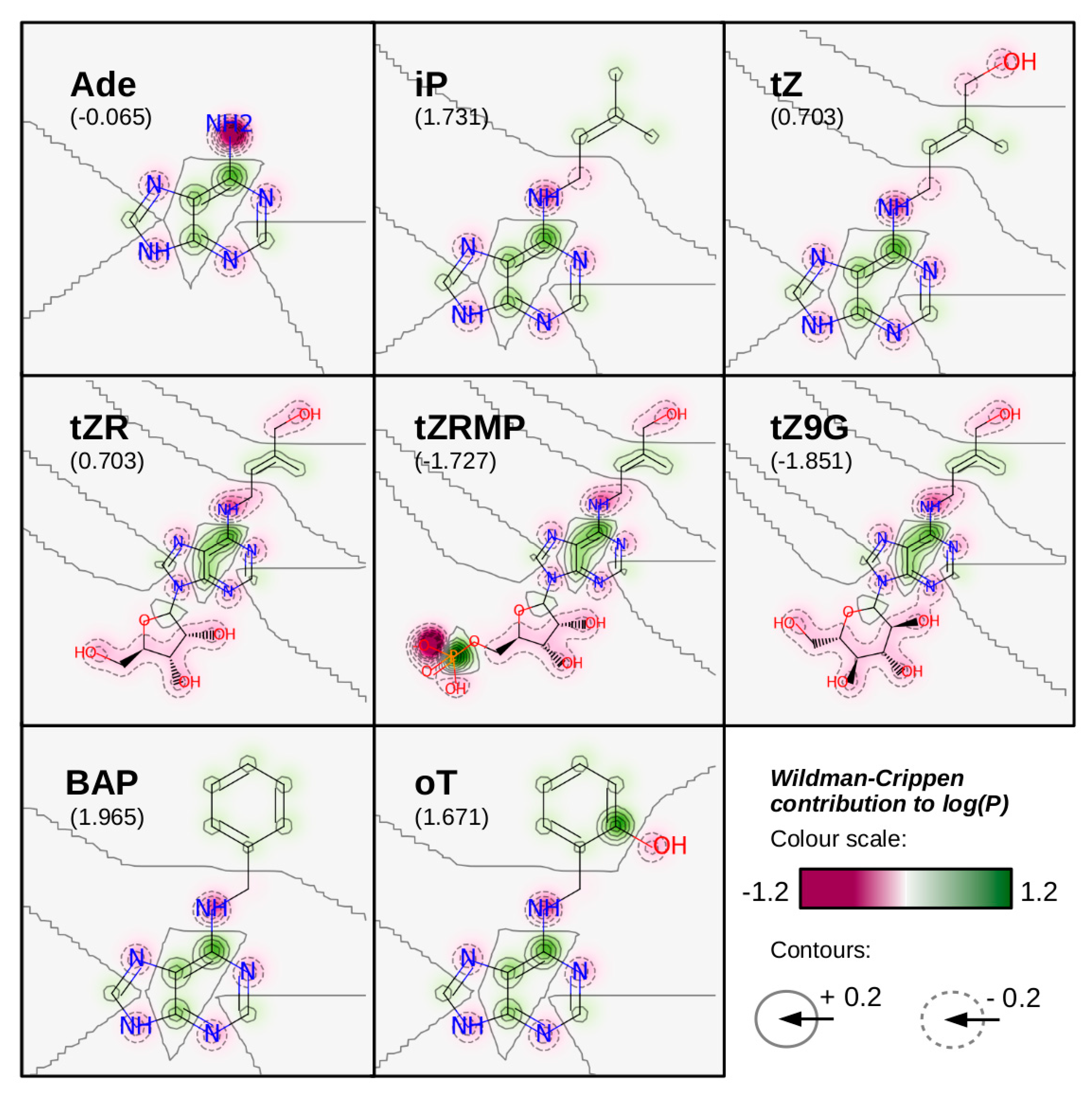
3. Cytokinin Structures Affect Their Physical and Chemical Properties
4. Cytokinin Content Varies in Different Subcellular Compartments
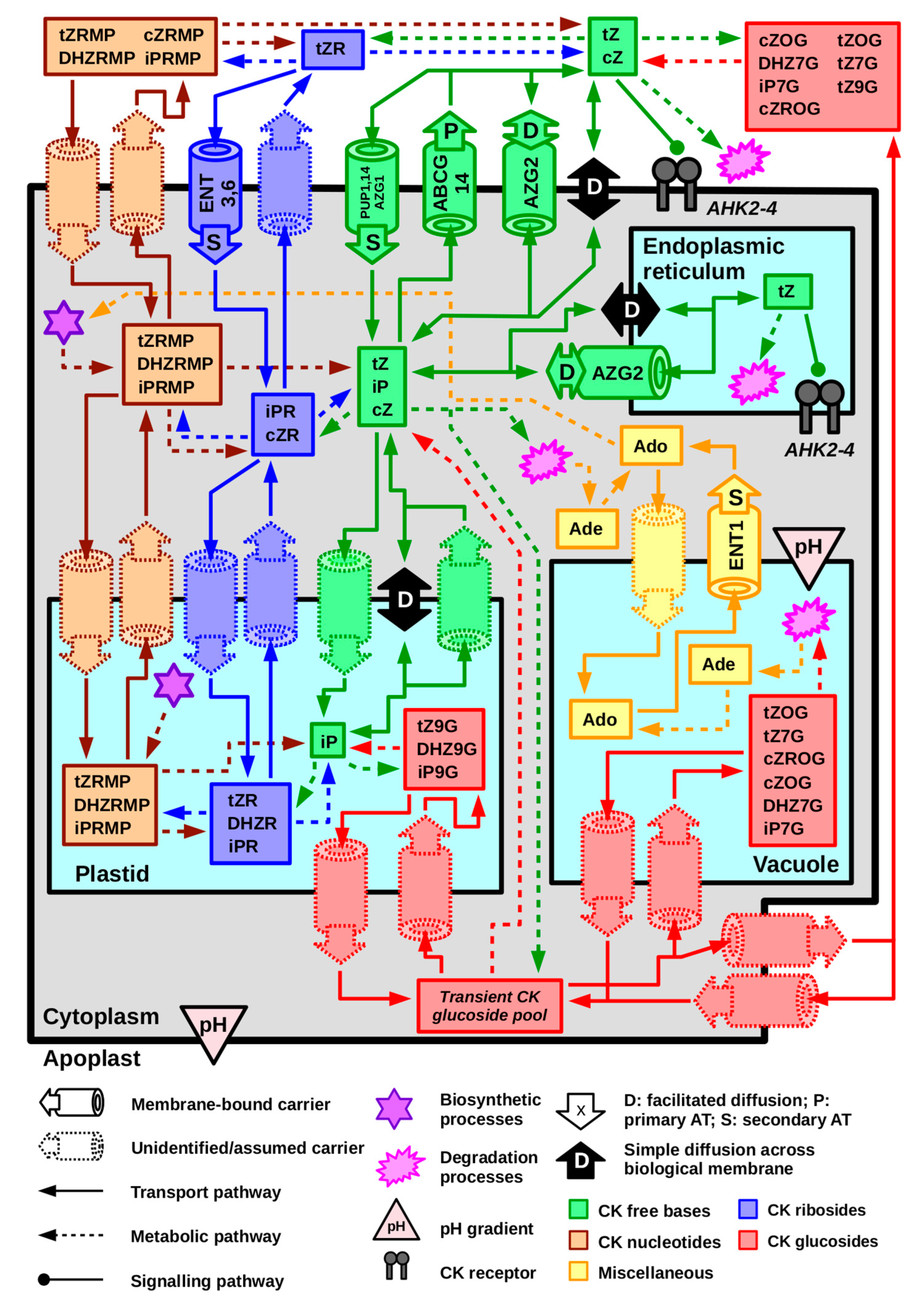
5. Cytokinin Transport at the Cellular Level Occurs via Thermodynamically and Kinetically Diverse Processes
6. Equilibrative Nucleoside Transporters Mediate Proton-Dependent Transport of Cytokinin Ribosides
7. Purine Permeases Are Involved in Proton-Dependent Transport of Free Cytokinin Bases
8. Recent Findings Suggest That Proteins AtPUP14, AtABCG14, AtAZG1, and AtAZG2 Are Cytokinin-Specific Transporters with an Important Role in Cytokinin Signalization
9. Cytokinin Paracrine Movement and Long-Distance Transport Open Up another Layer of Cytokinin Homeostasis Maintenance
10. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The Yin-Yang of Hormones: Cytokinin and Auxin Interactions in Plant Development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin Signaling in Plant Development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef]
- Skalický, V.; Kubeš, M.; Napier, R.; Novák, O. Auxins and Cytokinins—The Role of Subcellular Organization on Homeostasis. Int. J. Mol. Sci. 2018, 19, 3115. [Google Scholar] [CrossRef]
- Akiyoshi, D.E.; Klee, H.; Amasino, R.M.; Nester, E.W.; Gordon, M.P. T-DNA of Agrobacterium Tumefaciens Encodes an Enzyme of Cytokinin Biosynthesis. Proc. Natl. Acad. Sci. USA 1984, 81, 5994–5998. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 Encode Cytokinin Hydroxylases That Catalyze the Biosynthesis of Trans-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Arkhipov, D.V.; Osolodkin, D.I.; Schmülling, T.; Romanov, G.A. Plant Membrane Assays with Cytokinin Receptors Underpin the Unique Role of Free Cytokinin Bases as Biologically Active Ligands. J. Exp. Bot. 2015, 66, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-M.; Kristopeit, S.M. Metabolism of Cytokinin: Dephosphorylation of Cytokinin Ribonucleotide by 5′-Nucleotidases from Wheat Germ Cytosol. Plant Physiol. 1981, 67, 494–498. [Google Scholar] [CrossRef]
- Chen, C.-M.; Kristopeit, S.M. Metabolism of Cytokinin: Deribosylation of Cytokinin Ribonucleoside by Adenosine Nucleosidase from Wheat Germ Cells. Plant Physiol. 1981, 68, 1020–1023. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional Analyses of LONELY GUY Cytokinin-Activating Enzymes Reveal the Importance of the Direct Activation Pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef]
- Tokunaga, H.; Kojima, M.; Kuroha, T.; Ishida, T.; Sugimoto, K.; Kiba, T.; Sakakibara, H. Arabidopsis Lonely Guy (LOG) Multiple Mutants Reveal a Central Role of the LOG-Dependent Pathway in Cytokinin Activation. Plant J. 2012, 69, 355–365. [Google Scholar] [CrossRef]
- Galuszka, P.; Frébort, I.; Šebela, M.; Sauer, P.; Jacobsen, S.; Peč, P. Cytokinin Oxidase or Dehydrogenase? Eur. J. Biochem. 2001, 268, 450–461. [Google Scholar] [CrossRef]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Bartrina y Manns, I. Structure and Function of Cytokinin Oxidase/Dehydrogenase Genes of Maize, Rice, Arabidopsis and Other Species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef]
- Hou, B.; Lim, E.-K.; Higgins, G.S.; Bowles, D.J. N-Glucosylation of Cytokinins by Glycosyltransferases of Arabidopsis Thaliana. J. Biol. Chem. 2004, 279, 47822–47832. [Google Scholar] [CrossRef] [PubMed]
- Brzobohatý, B.; Moore, I.; Kristoffersen, P.; Bako, L.; Campos, N.; Schell, J.; Palme, K. Release of Active Cytokinin by a Beta-Glucosidase Localized to the Maize Root Meristem. Science 1993, 262, 1051–1054. [Google Scholar] [CrossRef]
- Hošek, P.; Hoyerová, K.; Kiran, N.S.; Dobrev, P.I.; Zahajská, L.; Filepová, R.; Motyka, V.; Müller, K.; Kamínek, M. Distinct Metabolism of N-Glucosides of Isopentenyladenine and Trans-Zeatin Determines Cytokinin Metabolic Spectrum in Arabidopsis. New Phytol. 2020, 225, 2423–2438. [Google Scholar] [CrossRef] [PubMed]
- Vylíčilová, H.; Bryksová, M.; Matušková, V.; Doležal, K.; Plíhalová, L.; Strnad, M. Naturally Occurring and Artificial N9-Cytokinin Conjugates: From Synthesis to Biological Activity and Back. Biomolecules 2020, 10, 832. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Lomin, S.N.; Schmülling, T. Cytokinin Signaling: From the ER or from the PM? That is the Question! New Phytol. 2018, 218, 41–53. [Google Scholar] [CrossRef]
- Inoue, T.; Higuchi, M.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Kato, T.; Tabata, S.; Shinozaki, K.; Kakimoto, T. Identification of CRE1 as a Cytokinin Receptor from Arabidopsis. Nature 2001, 409, 1060–1063. [Google Scholar] [CrossRef]
- Ueguchi, C.; Koizumi, H.; Suzuki, T.; Mizuno, T. Novel Family of Sensor Histidine Kinase Genes in Arabidopsis Thaliana. Plant Cell Physiol. 2001, 42, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ryu, H.; Hong, S.H.; Woo, H.R.; Lim, P.O.; Lee, I.C.; Sheen, J.; Nam, H.G.; Hwang, I. Cytokinin-Mediated Control of Leaf Longevity by AHK3 through Phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Caesar, K.; Thamm, A.M.K.; Witthöft, J.; Elgass, K.; Huppenberger, P.; Grefen, C.; Horak, J.; Harter, K. Evidence for the Localization of the Arabidopsis Cytokinin Receptors AHK3 and AHK4 in the Endoplasmic Reticulum. J. Exp. Bot. 2011, 62, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Wulfetange, K.; Lomin, S.N.; Romanov, G.A.; Stolz, A.; Heyl, A.; Schmülling, T. The Cytokinin Receptors of Arabidopsis are Located Mainly to the Endoplasmic Reticulum. Plant Physiol. 2011, 156, 1808–1818. [Google Scholar] [CrossRef]
- Lomin, S.N.; Myakushina, Y.A.; Arkhipov, D.V.; Leonova, O.G.; Popenko, V.I.; Schmülling, T.; Romanov, G.A. Studies of Cytokinin Receptor-Phosphotransmitter Interaction Provide Evidences for the Initiation of Cytokinin Signalling in the Endoplasmic Reticulum. Funct. Plant Biol. 2018, 45, 192–202. [Google Scholar] [CrossRef]
- Antoniadi, I.; Novák, O.; Gelová, Z.; Johnson, A.; Plíhal, O.; Simerský, R.; Mik, V.; Vain, T.; Mateo-Bonmatí, E.; Karady, M.; et al. Cell-Surface Receptors Enable Perception of Extracellular Cytokinins. Nat. Commun. 2020, 11, 4284. [Google Scholar] [CrossRef]
- Kubiasová, K.; Montesinos, J.C.; Šamajová, O.; Nisler, J.; Mik, V.; Semerádová, H.; Plíhalová, L.; Novák, O.; Marhavý, P.; Cavallari, N.; et al. Cytokinin Fluoroprobe Reveals Multiple Sites of Cytokinin Perception at Plasma Membrane and Endoplasmic Reticulum. Nat. Commun. 2020, 11, 4285. [Google Scholar] [CrossRef]
- Zürcher, E.; Liu, J.; di Donato, M.; Geisler, M.; Müller, B. Plant Development Regulated by Cytokinin Sinks. Science 2016, 353, 1027–1030. [Google Scholar] [CrossRef]
- Hluska, T.; Hlusková, L.; Emery, R.J. The Hulks and the Deadpools of the Cytokinin Universe: A Dual Strategy for Cytokinin Production, Translocation, and Signal Transduction. Biomolecules 2021, 11, 209. [Google Scholar] [CrossRef]
- Kieber, J.J. Cytokinins: Regulators of Cell Division. In Plant Physiology; Taiz, L., Zeiger, E., Eds.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2002; pp. 493–517. [Google Scholar]
- Spíchal, L. Cytokinins—Recent News and Views of Evolutionally Old Molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef]
- Nedvěd, D. Transport and Metabolism of Radio-Labelled Cytokinins in Plant Cells and Tissues. Master’s Thesis, Charles University, Prague, Czech Republic, 2020. [Google Scholar]
- Vickers, C.E.; Bongers, M.; Liu, Q.; Delatte, T.; Bouwmeester, H. Metabolic Engineering of Volatile Isoprenoids in Plants and Microbes. Plant Cell Environ. 2014, 37, 1753–1775. [Google Scholar] [CrossRef]
- Gajdošová, S.; Spíchal, L.; Kamínek, M.; Hoyerová, K.; Novák, O.; Dobrev, P.I.; Galuszka, P.; Klíma, P.; Gaudinová, A.; Žižková, E.; et al. Distribution, Biological Activities, Metabolism, and the Conceivable Function of Cis-Zeatin-Type Cytokinins in Plants. J. Exp. Bot. 2011, 62, 2827–2840. [Google Scholar] [CrossRef] [PubMed]
- Heyl, A.; Riefler, M.; Romanov, G.A.; Schmülling, T. Properties, Functions and Evolution of Cytokinin Receptors. Eur. J. Cell Biol. 2012, 91, 246–256. [Google Scholar] [CrossRef]
- Bassil, N.V.; Mok, D.W.S.; Mok, M.C. Partial Purification of a Cis-Trans-Isomerase of Zeatin from Immature Seed of Phaseolus Vulgaris L. Plant Physiol. 1993, 102, 867–872. [Google Scholar] [CrossRef]
- Hluska, T.; Šebela, M.; Lenobel, R.; Frébort, I.; Galuszka, P. Purification of Maize Nucleotide Pyrophosphatase/Phosphodiesterase Casts Doubt on the Existence of Zeatin Cis–Trans Isomerase in Plants. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP Isopentenyltransferases and TRNA Isopentenyltransferases in Cytokinin Biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef]
- Martin, R.C.; Mok, M.C.; Shaw, G.; Mok, D.W.S. An Enzyme Mediating the Conversion of Zeatin to Dihydrozeatin in Phaseolus Embryos. Plant Physiol. 1989, 90, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Gaudinová, A.; Dobrev, P.I.; Šolcová, B.; Novák, O.; Strnad, M.; Friedecký, D.; Motyka, V. The Involvement of Cytokinin Oxidase/Dehydrogenase and Zeatin Reductase in Regulation of Cytokinin Levels in Pea (Pisum Sativum L.) Leaves. J. Plant Growth Regul. 2005, 24, 188–200. [Google Scholar] [CrossRef]
- Kolachevskaya, O.O.; Sergeeva, L.I.; Floková, K.; Getman, I.A.; Lomin, S.N.; Alekseeva, V.V.; Rukavtsova, E.B.; Buryanov, Y.I.; Romanov, G.A. Auxin Synthesis Gene Tms1 Driven by Tuber-Specific Promoter Alters Hormonal Status of Transgenic Potato Plants and Their Responses to Exogenous Phytohormones. Plant Cell Rep. 2017, 36, 419–435. [Google Scholar] [CrossRef]
- Tarkowská, D.; Doležal, K.; Tarkowski, P.; Åstot, C.; Holub, J.; Fuksová, K.; Schmülling, T.; Sandberg, G.; Strnad, M. Identification of New Aromatic Cytokinins in Arabidopsis Thaliana and Populus × Canadensis Leaves by LC-(+)ESI-MS and Capillary Liquid Chromatography/Frit–Fast Atom Bombardment Mass Spectrometry. Physiol. Plant 2003, 117, 579–590. [Google Scholar] [CrossRef]
- Strnad, M. The Aromatic Cytokinins. Physiol. Plant 1997, 101, 674–688. [Google Scholar] [CrossRef]
- Hluska, T.; Dobrev, P.I.; Tarkowská, D.; Frébortová, J.; Zalabák, D.; Kopečný, D.; Plíhal, O.; Kokáš, F.; Briozzo, P.; Zatloukal, M.; et al. Cytokinin Metabolism in Maize: Novel Evidence of Cytokinin Abundance, Interconversions and Formation of a New Trans-Zeatin Metabolic Product with a Weak Anticytokinin Activity. Plant Sci. 2016, 247, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.L.; Benfield, A.H.; Wollenberg, R.D.; Westphal, K.; Wimmer, R.; Nielsen, M.R.; Nielsen, K.F.; Carere, J.; Covarelli, L.; Beccari, G.; et al. The Cereal Pathogen Fusarium Pseudograminearum Produces a New Class of Active Cytokinins during Infection. Mol. Plant Pathol. 2017, 19, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Haidoune, M.; Mornet, R.; Laloue, M. Synthesis of 6-(3-Methylpyrrol-1-Yl)-9-β-D-Ribofuranosyl Purine, a Novel Metabolite of Zeatin Riboside. Tetrahedron Lett. 1990, 31, 1419–1422. [Google Scholar] [CrossRef]
- Mok, M.C.; Martin, R.C.; Dobrev, P.I.; Vanková, R.; Ho, P.S.; Yonekura-Sakakibara, K.; Sakakibara, H.; Mok, D.W.S. Topolins and Hydroxylated Thidiazuron Derivatives are Substrates of Cytokinin O-Glucosyltransferase with Position Specificity Related to Receptor Recognition. Plant Physiol. 2005, 137, 1057–1066. [Google Scholar] [CrossRef]
- Galuszka, P.; Popelková, H.; Werner, T.; Frébortová, J.; Pospíšilová, H.; Mik, V.; Köllmer, I.; Schmülling, T.; Frébort, I. Biochemical Characterization of Cytokinin Oxidases/Dehydrogenases from Arabidopsis Thaliana Expressed in Nicotiana Tabacum L. J. Plant Growth Regul. 2007, 26, 255–267. [Google Scholar] [CrossRef]
- Evidente, A.; Fujii, T.; Iacobellis, N.S.; Riva, S.; Sisto, A.; Surico, G. Structure-Activity Relationships of Zeatin Cytokinins Produced by Plant PathogenicPseudomonades. Phytochemistry 1991, 30, 3505–3510. [Google Scholar] [CrossRef]
- Pertry, I.; Václavíková, K.; Depuydt, S.; Galuszka, P.; Spíchal, L.; Temmerman, W.; Stes, E.; Schmülling, T.; Kakimoto, T.; Montagu, M.C.E.V.; et al. Identification of Rhodococcus Fascians Cytokinins and Their Modus Operandi to Reshape the Plant. Proc. Natl. Acad. Sci. USA 2009, 106, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Radhika, V.; Ueda, N.; Tsuboi, Y.; Kojima, M.; Kikuchi, J.; Kudo, T.; Sakakibara, H. Methylated Cytokinins from the Phytopathogen Rhodococcus Fascians Mimic Plant Hormone Activity. Plant Physiol. 2015, 169, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Gibb, M.; Kisiala, A.B.; Morrison, E.N.; Emery, R.J.N. The Origins and Roles of Methylthiolated Cytokinins: Evidence from Among Life Kingdoms. Front. Cell Dev. Biol. 2020, 8, 605672. [Google Scholar] [CrossRef]
- Oshchepkov, M.S.; Kalistratova, A.V.; Savelieva, E.M.; Romanov, G.A.; Bystrova, N.A.; Kochetkov, K.A. Natural and Synthetic Cytokinins and Their Applications in Biotechnology, Agrochemistry and Medicine. Russ. Chem. Rev. 2020, 89, 787. [Google Scholar] [CrossRef]
- Wildman, S.A.; Crippen, G.M. Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant Hormonomics: Multiple Phytohormone Profiling by Targeted Metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Gruissem, W.; Jones, R.L. (Eds.) Biochemistry & Molecular Biology of Plants, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2015; ISBN 978-1-118-50219-8. [Google Scholar]
- Chen, W.; Gai, Y.; Liu, S.; Wang, R.; Jiang, X. Quantitative Analysis of Cytokinins in Plants by High Performance Liquid Chromatography: Electronspray Ionization Ion Trap Mass Spectrometry. J. Integr. Plant Biol. 2010, 52, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, P.I.; Hoyerová, K.; Petrášek, J. Analytical Determination of Auxins and Cytokinins. In Auxins and Cytokinins in Plant Biology; Dandekar, T., Naseem, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1569, pp. 31–39. ISBN 978-1-4939-6829-9. [Google Scholar]
- Hoyerová, K.; Gaudinová, A.; Malbeck, J.; Dobrev, P.I.; Kocábek, T.; Šolcová, B.; Trávníčková, A.; Kamínek, M. Efficiency of Different Methods of Extraction and Purification of Cytokinins. Phytochemistry 2006, 67, 1151–1159. [Google Scholar] [CrossRef]
- Gillissen, B.; Bürkle, L.; André, B.; Kühn, C.; Rentsch, D.; Brandl, B.; Frommer, W.B. A New Family of High-Affinity Transporters for Adenine, Cytosine, and Purine Derivatives in Arabidopsis. Plant Cell 2000, 12, 291–300. [Google Scholar] [CrossRef]
- Bürkle, L.; Cedzich, A.; Döpke, C.; Stransky, H.; Okumoto, S.; Gillissen, B.; Kühn, C.; Frommer, W.B. Transport of Cytokinins Mediated by Purine Transporters of the PUP Family Expressed in Phloem, Hydathodes, and Pollen of Arabidopsis. Plant J. 2003, 34, 13–26. [Google Scholar] [CrossRef]
- Sun, J.; Hirose, N.; Wang, X.; Wen, P.; Xue, L.; Sakakibara, H.; Zuo, J. Arabidopsis SOI33/AtENT8 Gene Encodes a Putative Equilibrative Nucleoside Transporter That is Involved in Cytokinin Transport In Planta. J. Integr. Plant Biol. 2005, 47, 588–603. [Google Scholar] [CrossRef]
- Spíchal, L.; Rakova, N.Y.; Riefler, M.; Mizuno, T.; Romanov, G.A.; Strnad, M.; Schmülling, T. Two Cytokinin Receptors of Arabidopsis Thaliana, CRE1/AHK4 and AHK3, Differ in Their Ligand Specificity in a Bacterial Assay. Plant Cell Physiol. 2004, 45, 1299–1305. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Kojima, M.; Yamaya, T.; Sakakibara, H. Molecular Characterization of Cytokinin-Responsive Histidine Kinases in Maize. Differential Ligand Preferences and Response to Cis-Zeatin. Plant Physiol. 2004, 134, 1654–1661. [Google Scholar] [CrossRef]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Osolodkin, D.I.; Romanov, G.A. Receptor Properties and Features of Cytokinin Signaling. Acta Nat. 2012, 4, 31–45. [Google Scholar] [CrossRef]
- Hirose, N.; Makita, N.; Yamaya, T.; Sakakibara, H. Functional Characterization and Expression Analysis of a Gene, OsENT2, Encoding an Equilibrative Nucleoside Transporter in Rice Suggest a Function in Cytokinin Transport. Plant Physiol. 2005, 138, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Takei, K.; Kuroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of Cytokinin Biosynthesis, Compartmentalization and Translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Xiong, L. Characterization of a Purine Permease Family Gene OsPUP7 Involved in Growth and Development Control in Rice. J. Integr. Plant Biol. 2013, 55, 1119–1135. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, P.I.; Kamínek, M. Fast and Efficient Separation of Cytokinins from Auxin and Abscisic Acid and Their Purification Using Mixed-Mode Solid-Phase Extraction. J. Chromatogr. A 2002, 950, 21–29. [Google Scholar] [CrossRef]
- Jiskrová, E.; Novák, O.; Pospíšilová, H.; Holubová, K.; Karády, M.; Galuszka, P.; Robert, S.; Frébort, I. Extra- and Intracellular Distribution of Cytokinins in the Leaves of Monocots and Dicots. New Biotechnol. 2016, 33, 735–742. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Murfet, I.C.; Kerhoas, L.; Sotta, B.; Miginiac, E.; Rameau, C. The Shoot Controls Zeatin Riboside Export from Pea Roots. Evidence from the Branching Mutant Rms4. Plant J. 1997, 11, 339–345. [Google Scholar] [CrossRef]
- Kuroha, T.; Kato, H.; Asami, T.; Yoshida, S.; Kamada, H.; Satoh, S. A Trans-zeatin Riboside in Root Xylem Sap Negatively Regulates Adventitious Root Formation on Cucumber Hypocotyls. J. Exp. Bot. 2002, 53, 2193–2200. [Google Scholar] [CrossRef]
- Takei, K.; Sakakibara, H.; Taniguchi, M.; Sugiyama, T. Nitrogen-Dependent Accumulation of Cytokinins in Root and TheTranslocation to Leaf: Implication of Cytokinin Species That Induces GeneExpression of Maize ResponseRegulator. Plant Cell Physiol. 2001, 42, 85–93. [Google Scholar] [CrossRef]
- Corbesier, L.; Prinsen, E.; Jacqmard, A.; Lejeune, P.; van Onckelen, H.; Périlleux, C.; Bernier, G. Cytokinin Levels in Leaves, Leaf Exudate and Shoot Apical Meristem of Arabidopsis Thaliana during Floral Transition. J. Exp. Bot. 2003, 54, 2511–2517. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.-H.; Ma, X.-M.; Kojima, M.; Sakakibara, H.; Wang, Y.-W.; Hou, B.-K. Overexpression of Glucosyltransferase UGT85A1 Influences Trans-Zeatin Homeostasis and Trans-Zeatin Responses Likely through O-Glucosylation. Planta 2013, 237, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Šmehilová, M.; Dobrůšková, J.; Novák, O.; Takáč, T.; Galuszka, P. Cytokinin-Specific Glycosyltransferases Possess Different Roles in Cytokinin Homeostasis Maintenance. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Benková, E.; Witters, E.; Dongen, W.V.; Kolar, J.; Motyka, V.; Brzobohatý, B.; Onckelen, H.A.V.; Machácková, I. Cytokinins in Tobacco and Wheat Chloroplasts. Occurrence and Changes Due to Light/Dark Treatment. Plant Physiol. 1999, 121, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, D.; Loureiro, J.; Antoniadi, I.; Bainard, J.; Bureš, P.; Cápal, P.; Castro, M.; Castro, S.; Čertner, M.; Čertnerová, D.; et al. Best Practices in Plant Cytometry. Cytom. Part. A 2021, 1–7. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and Mechanism of ABC Transporters. F1000Prime Rep. 2015, 7. [Google Scholar] [CrossRef]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arab. Book 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Girke, C.; Daumann, M.; Niopek-Witz, S.; Möhlmann, T. Nucleobase and Nucleoside Transport and Integration into Plant Metabolism. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Laurila, T.; Vuorinen, V.; Divinski, S.V. Fick’s Laws of Diffusion. In Thermodynamics, Diffusion and the Kirkendall Effect in Solids; Paul, A., Laurila, T., Vuorinen, V., Divinski, S.V., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 115–139. ISBN 978-3-319-07461-0. [Google Scholar]
- El-Showk, S.; Help-Rinta-Rahko, H.; Blomster, T.; Siligato, R.; Marée, A.F.M.; Mähönen, A.P.; Grieneisen, V.A. Parsimonious Model of Vascular Patterning Links Transverse Hormone Fluxes to Lateral Root Initiation: Auxin Leads the Way, While Cytokinin Levels Out. PLoS Comput. Biol. 2015, 11, e1004450. [Google Scholar] [CrossRef]
- Hošek, P.; Kubeš, M.; Laňková, M.; Dobrev, P.I.; Klíma, P.; Kohoutová, M.; Petrášek, J.; Hoyerová, K.; Jiřina, M.; Zažímalová, E. Auxin Transport at Cellular Level: New Insights Supported by Mathematical Modelling. J. Exp. Bot. 2012, 63, 3815–3827. [Google Scholar] [CrossRef]
- Moore, S.; Zhang, X.; Mudge, A.; Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Spatiotemporal Modelling of Hormonal Crosstalk Explains the Level and Patterning of Hormones and Gene Expression in Arabidopsis Thaliana Wild-Type and Mutant Roots. New Phytol. 2015, 207, 1110–1122. [Google Scholar] [CrossRef]
- Zažímalová, E.; Murphy, A.S.; Yang, H.; Hoyerová, K.; Hošek, P. Auxin Transporters—Why So Many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef]
- Kramer, E.M. How Far Can a Molecule of Weak Acid Travel in the Apoplast or Xylem? Plant Physiol. 2006, 141, 1233–1236. [Google Scholar] [CrossRef]
- Wormit, A.; Traub, M.; Flörchinger, M.; Neuhaus, H.E.; Möhlmann, T. Characterization of Three Novel Members of the Arabidopsis Thaliana Equilibrative Nucleoside Transporter (ENT) Family. Biochem. J. 2004, 383, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, L.; Menten, M.L. Die Kinetik Der Invertinwirkung. Biochemische Zeitschrift 1913, 49, 333–369. [Google Scholar]
- Johnson, K.A.; Goody, R.S. The Original Michaelis Constant: Translation of the 1913 Michaelis-Menten Paper. Biochemistry 2011, 50, 8264–8269. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, D. Cloning and in Vitro Expression of the CDNA Encoding a Putative Nucleoside Transporter from Arabidopsis Thaliana. Plant Sci. 2000, 157, 23–32. [Google Scholar] [CrossRef]
- Möhlmann, T.; Mezher, Z.; Schwerdtfeger, G.; Neuhaus, H.E. Characterisation of a Concentrative Type of Adenosine Transporter from Arabidopsis Thaliana (ENT1,At). FEBS Lett. 2001, 509, 370–374. [Google Scholar] [CrossRef]
- Bernard, C.; Traub, M.; Kunz, H.-H.; Hach, S.; Trentmann, O.; Möhlmann, T. Equilibrative Nucleoside Transporter 1 (ENT1) is Critical for Pollen Germination and Vegetative Growth in Arabidopsis. J. Exp. Bot. 2011, 62, 4627–4637. [Google Scholar] [CrossRef]
- Girke, C.; Arutyunova, E.; Syed, M.; Traub, M.; Möhlmann, T.; Lemieux, M.J. High Yield Expression and Purification of Equilibrative Nucleoside Transporter 7 (ENT7) from Arabidopsis Thaliana. Biochim. Biophys. Acta BBA Gen. Subj. 2015, 1850, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liu, K.; Baldwin, S.A.; Wang, D. Equilibrative Nucleoside Transporters of Arabidopsis Thaliana: CDNA Cloning, Expression Pattern, and Analysis of Transport Activities. J. Biol. Chem. 2003, 278, 35732–35742. [Google Scholar] [CrossRef]
- Traub, M.; Flörchinger, M.; Piecuch, J.; Kunz, H.; Weise-Steinmetz, A.; Deitmer Joachim, W.; Ekkehard Neuhaus, H.; Möhlmann, T. The Fluorouridine Insensitive 1 (Fur1) Mutant is Defective in Equilibrative Nucleoside Transporter 3 (ENT3), and Thus Represents an Important Pyrimidine Nucleoside Uptake System in Arabidopsis Thaliana. Plant J. 2007, 49, 855–864. [Google Scholar] [CrossRef]
- Boswell-Casteel, R.C.; Hays, F.A. Equilibrative Nucleoside Transporters—A Review. Nucleosides Nucleotides Nucleic Acids 2017, 36, 7–30. [Google Scholar] [CrossRef]
- Young, J.D.; Yao, S.Y.M.; Baldwin, J.M.; Cass, C.E.; Baldwin, S.A. The Human Concentrative and Equilibrative Nucleoside Transporter Families, SLC28 and SLC29. Mol. Asp. Med. 2013, 34, 529–547. [Google Scholar] [CrossRef]
- Hildreth, S.B.; Gehman, E.A.; Yang, H.; Lu, R.-H.; Ritech, C.K.; Harich, K.C.; Yu, S.; Lin, J.; Sandoe, J.L.; Okumoto, S.; et al. Tobacco Nicotine Uptake Permease (NUP1) Affects Alkaloid Metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, 18179–18184. [Google Scholar] [CrossRef]
- Cedzich, A.; Stransky, H.; Schulz, B.; Frommer, W.B. Characterization of Cytokinin and Adenine Transport in Arabidopsis Cell Cultures. Plant Physiol. 2008, 148, 1857–1867. [Google Scholar] [CrossRef] [PubMed]
- Durán-Medina, Y.; Díaz-Ramírez, D.; Marsch-Martínez, N. Cytokinins on the Move. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Zhao, Y.; Zhang, K. Cytokinin Transporters: Multisite Players in Cytokinin Homeostasis and Signal Distribution. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Szydlowski, N.; Bürkle, L.; Pourcel, L.; Moulin, M.; Stolz, J.; Fitzpatrick, T.B. Recycling of Pyridoxine (Vitamin B6) by PUP1 in Arabidopsis. Plant J. 2013, 75, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, D.; Zhang, G.; Gao, S.; Liu, L.; Xu, F.; Che, R.; Wang, Y.; Tong, H.; Chu, C. Big Grain3, Encoding a Purine Permease, Regulates Grain Size via Modulating Cytokinin Transport in Rice. J. Integr. Plant Biol. 2019, 61, 581–597. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, J.; Yu, G.; Lu, X.; Mei, W.; Deng, H.; Zhang, G.; Chen, G.; Chu, C.; Tong, H.; et al. Endoplasmic Reticulum-Localized PURINE PERMEASE1 Regulates Plant Height and Grain Weight by Modulating Cytokinin Distribution in Rice. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Zürcher, E.; Tavor-Deslex, D.; Lituiev, D.; Enkerli, K.; Tarr, P.T.; Müller, B. A Robust and Sensitive Synthetic Sensor to Monitor the Transcriptional Output of the Cytokinin Signaling Network in Planta. Plant Physiol. 2013, 161, 1066–1075. [Google Scholar] [CrossRef]
- Ko, D.; Kang, J.; Kiba, T.; Park, J.; Kojima, M.; Do, J.; Kim, K.Y.; Kwon, M.; Endler, A.; Song, W.-Y.; et al. Arabidopsis ABCG14 Is Essential for the Root-to-Shoot Translocation of Cytokinin. Proc. Natl. Acad. Sci. USA 2014, 111, 7150–7155. [Google Scholar] [CrossRef]
- Le Hir, R.; Sorin, C.; Chakraborti, D.; Moritz, T.; Schaller, H.; Tellier, F.; Robert, S.; Morin, H.; Bako, L.; Bellini, C. ABCG9, ABCG11 and ABCG14 ABC Transporters are Required for Vascular Development in Arabidopsis. Plant J. 2013, 76, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Novak, O.; Wei, Z.; Gou, M.; Zhang, X.; Yu, Y.; Yang, H.; Cai, Y.; Strnad, M.; Liu, C.-J. Arabidopsis ABCG14 Protein Controls the Acropetal Translocation of Root-Synthesized Cytokinins. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, N.; Ju, M.; Fan, B.; Zhang, Y.; Zhu, E.; Zhang, M.; Zhang, K. ABC Transporter OsABCG18 Controls the Shootward Transport of Cytokinins and Grain Yield in Rice. J. Exp. Bot. 2019, 70, 6277–6291. [Google Scholar] [CrossRef]
- Mansfield, T.A.; Schultes, N.P.; Mourad, G.S. AtAzg1 and AtAzg2 Comprise a Novel Family of Purine Transporters in Arabidopsis. FEBS Lett. 2009, 583, 481–486. [Google Scholar] [CrossRef]
- Tessi, T.M.; Brumm, S.; Winklbauer, E.; Schumacher, B.; Pettinari, G.; Lescano, I.; González, C.A.; Wanke, D.; Maurino, V.G.; Harter, K.; et al. Arabidopsis AZG2 Transports Cytokinins in vivo and Regulates Lateral Root Emergence. New Phytol. 2020, 229, 979–993. [Google Scholar] [CrossRef]
- Tessi, T.M.; Shahriari, M.; Maurino, V.G.; Meissner, E.; Novak, O.; Pasternak, T.; Schumacher, B.S.; Flubacher, N.S.; Nautscher, M.; Williams, A.; et al. The Auxin Transporter PIN1 and the Cytokinin Transporter AZG1 Interact to Regulate the Root Stress Response. bioRxiv 2020. [Google Scholar] [CrossRef]
- Faiss, M.; Zalubilová, J.; Strnad, M.; Schmülling, T. Conditional Transgenic Expression of the Ipt Gene Indicates a Function for Cytokinins in Paracrine Signaling in Whole Tobacco Plants. Plant J. 1997, 12, 401–415. [Google Scholar] [CrossRef]
- De Rybel, B.; Adibi, M.; Breda, A.S.; Wendrich, J.R.; Smit, M.E.; Novák, O.; Yamaguchi, N.; Yoshida, S.; Isterdael, G.V.; Palovaara, J.; et al. Integration of Growth and Patterning during Vascular Tissue Formation in Arabidopsis. Science 2014, 345. [Google Scholar] [CrossRef]
- Badenoch-Jones, J.; Letham, D.S.; Parker, C.W.; Rolfe, B.G. Quantitation of Cytokinins in Biological Samples Using Antibodies Against Zeatin Riboside. Plant Physiol. 1984, 75, 1117–1125. [Google Scholar] [CrossRef]
- Abo-Hamed, S.; Collin, H.A.; Hardwick, K. Biochemical and Physiological Aspects of Leaf Development in Cocoa (Theobroma Cacao L.). New Phytol. 1984, 97, 219–225. [Google Scholar] [CrossRef]
- Kudoyarova, G.R.; Korobova, A.V.; Akhiyarova, G.R.; Arkhipova, T.N.; Zaytsev, D.Y.; Prinsen, E.; Egutkin, N.L.; Medvedev, S.S.; Veselov, S.Y. Accumulation of Cytokinins in Roots and Their Export to the Shoots of Durum Wheat Plants Treated with the Protonophore Carbonyl Cyanide M-Chlorophenylhydrazone (CCCP). J. Exp. Bot. 2014, 65, 2287–2294. [Google Scholar] [CrossRef]
- Osugi, A.; Kojima, M.; Takebayashi, Y.; Ueda, N.; Kiba, T.; Sakakibara, H. Systemic Transport of Trans-Zeatin and Its Precursor Have Differing Roles in Arabidopsis Shoots. Nat. Plants 2017, 3, 17112. [Google Scholar] [CrossRef]
- Matsumoto-Kitano, M.; Kusumoto, T.; Tarkowski, P.; Kinoshita-Tsujimura, K.; Václavíková, K.; Miyawaki, K.; Kakimoto, T. Cytokinins are Central Regulators of Cambial Activity. Proc. Natl. Acad. Sci. USA 2008, 105, 20027–20031. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nedvěd, D.; Hošek, P.; Klíma, P.; Hoyerová, K. Differential Subcellular Distribution of Cytokinins: How Does Membrane Transport Fit into the Big Picture? Int. J. Mol. Sci. 2021, 22, 3428. https://doi.org/10.3390/ijms22073428
Nedvěd D, Hošek P, Klíma P, Hoyerová K. Differential Subcellular Distribution of Cytokinins: How Does Membrane Transport Fit into the Big Picture? International Journal of Molecular Sciences. 2021; 22(7):3428. https://doi.org/10.3390/ijms22073428
Chicago/Turabian StyleNedvěd, Daniel, Petr Hošek, Petr Klíma, and Klára Hoyerová. 2021. "Differential Subcellular Distribution of Cytokinins: How Does Membrane Transport Fit into the Big Picture?" International Journal of Molecular Sciences 22, no. 7: 3428. https://doi.org/10.3390/ijms22073428
APA StyleNedvěd, D., Hošek, P., Klíma, P., & Hoyerová, K. (2021). Differential Subcellular Distribution of Cytokinins: How Does Membrane Transport Fit into the Big Picture? International Journal of Molecular Sciences, 22(7), 3428. https://doi.org/10.3390/ijms22073428






