Beyond Arabidopsis: BBX Regulators in Crop Plants
Abstract
1. Introduction
2. Structural Characteristics of BBX Genes
The Function of the B-Box Domains and Conserved Motifs Outside
3. Look into the Genomes: BBX Family from Arabidopsis to Crops
3.1. Classification of BBX Genes in Crops
3.1.1. Cereal Crops
3.1.2. Rosaceae Species
3.1.3. Solanaceae Species
3.1.4. Other Crops
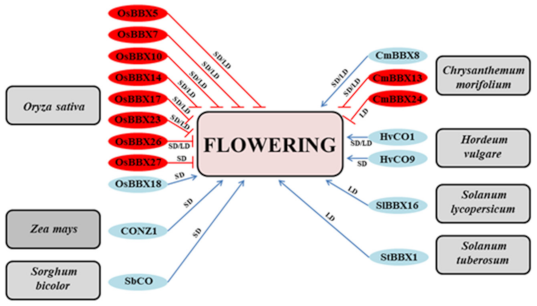
4. Time to Switch from Vegetative to Generative Development
5. Crops BBX Genes in the Anthocyanins Biosynthesis
6. Involvement of the BBX Proteins in Stress Response and Hormonal Pathways
7. Stress Response of Transgenic Plants Overexpressing the BBX Regulators
8. The Interplay between BBX Proteins and the Circadian Clock
9. Summary and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; LI, H.; Ouyang, B. Genomic organization, phylogenetic and expression analysis of the B-box gene family in tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef]
- Talar, U.; Kiełbowicz-Matuk, A.; Czarnecka, J.; Rorat, T. Genome-wide survey of B-box proteins in potato (Solanum tuberosum) – identification, characterization and expression patterns during diurnal cycle, etiolation and de-etiolation. PLoS ONE 2017, 12, e0177471. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the apple (Malus domestica Borkh.) genome. Mol. Genet. Genomics 2017, 293, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Wang, R.; Wang, R.; Yang, S.; Yang, Y. Genome-wide identification, phylogenetic analysis, and expression profiling of the BBX family genes in pear. J. Hort. Sci. Biotechnol. 2017, 93, 1–14. [Google Scholar] [CrossRef]
- Wei, H.; Wang, P.; Chen, J.; Li, C.; Wang, Y.; Yuan, Y.; Fang, J.; Leng, X. Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol. 2020, 20, 72. [Google Scholar] [CrossRef]
- Holm, M.; Hardtke, C.S.; Gaudet, R.; Deng, X.W. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001, 20, 118–127. [Google Scholar] [CrossRef]
- Datta, S.; Hettiarachchi, G.H.; Deng, X.W.; Holm, M. Arabidopsis CONSTANS- LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 2006, 18, 70–84. [Google Scholar] [CrossRef]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007, 19, 3242–3255. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Johansson, H.; Hettiarachchi, C.; Irigoyen, M.L.; Desai, M.; Rubio, V.; Holm, M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1 mediated ubiquitination. Plant Cell 2008, 20, 2324–2338. [Google Scholar] [CrossRef] [PubMed]
- Indorf, M.; Cordero, J.; Neuhaus, G.; Rodríguez-Franco, M. Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 2007, 51, 563–574. [Google Scholar] [CrossRef]
- Chang, C.S.; Li, Y.H.; Chen, L.T.; Chen, W.; Hsieh, W.P.; Shin, J.; Jane, W.N.; Chou, S.J.; Choi, G.; Hu, J.M.; et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008, 54, 205–219. [Google Scholar] [CrossRef]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.; Preuss, A.; et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef]
- Fan, X.Y.; Sun, Y.; Cao, D.M.; Bai, M.Y.; Luo, X.M.; Yang, H.J.; Wei, C.Q.; Zhu, S.W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 2012, 5, 591–600. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Gourrierec, J.L.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef] [PubMed]
- Valverde, F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 2011, 62, 2453–2463. [Google Scholar] [CrossRef]
- Crocco, C.D.; Holm, M.; Yanovsky, M.J.; Botto, J.F. AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 2010, 64, 551–562. [Google Scholar] [CrossRef]
- Crocco, C.D.; Holm, M.; Yanovsky, M.J.; Botto, J.F. Function of B-BOX under shade. Plant Signal. Behav. 2011, 6, 101–104. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-box protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Lippuner, V.; Cyert, M.S.; Gasser, C.S. Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 1996, 271, 12859–12866. [Google Scholar] [CrossRef]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef]
- Wang, Q.; Tu, X.; Zhang, J.; Chen, X.; Rao, L. Heat stress-induced BBX18 negatively regulates the thermos-tolerance in Arabidopsis. Mol. Biol. Rep. 2013, 40, 2679–2688. [Google Scholar] [CrossRef]
- Kiełbowicz-Matuk, A.; Rey, P.; Rorat, T. Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum Double B-box gene. Ann. Bot. 2014, 113, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, T.; Ito, S.; Nakamichi, N.; Niwa, Y.; Murakami, M.; Yamashino, T.; Mizuno, T. The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2008, 72, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Subramanian, S. Classification of the treble CLEF zinc finger: Noteworthy lessons for structure and function evolution. Sci. Rep. 2016, 6, 32070. [Google Scholar] [CrossRef] [PubMed]
- Kluska, K.; Adamczyk, J.; Krężel, A. Metal binding properties, stability and reactivity of zinc fingers. Coord. Chem. Rev. 2018, 367, 18–64. [Google Scholar] [CrossRef]
- Qi, Q.; Gibson, A.; Fu, X.; Zheng, M.; Kuehn, R.; Wang, Y.; Wang, Y.; Navarro, S.; Morrell, J.A.; Jiang, D.; et al. Involvement of the N-terminal B-box domain of Arabidopsis BBX32 protein in interaction with soybean BBX62 protein. J. Biol. Chem. 2012, 287, 31482–31493. [Google Scholar] [CrossRef]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.; Hepworth, S.R.; Vizir, I.; Pineiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Song, N.; Xu, Z.; Wang, J.; Qin, Q.; Jiang, H.; Si, W.; Li, X. Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment. Gene 2018, 673, 1–11. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; Sun, Q.; Li, W.; Yu, Y.; Zhao, M.; Meng, Z. Expression analysis of genes encoding double B-box zinc finger proteins in maize. Funct. Integr. Genomic. 2017, 17, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Shalmani, A.; Fan, S.; Jia, P.; Li, G.; Muhammad, I.; Li, Y.; Sharif, R.; Dong, F.; Zuo, X.; Li, K.; et al. Genome identification of B-box gene family members in seven Rosaceae species and their expression analysis in response to flower induction in Malus domestica. Molecules 2018, 23, 1763. [Google Scholar] [CrossRef]
- Cai, D.; Liu, H.; Sang, N.; Huang, X. Identification and characterization of CONSTANS-like (COL) gene family in upland cotton (Gossypium hirsutum L.). PLoS ONE 2017, 12, e0179038. [Google Scholar] [CrossRef]
- Jin, H.; Xing, M.; Cai, C.; Li, S. B-box Proteins in Arachis duranensis: Genome-wide characterization and expression profiles analysis. Agronomy 2020, 10, 23. [Google Scholar] [CrossRef]
- Chaurasia, A.K.; Patil, H.B.; Azeez, A.; Subramaniam, V.R.; Krishna, B.; Sane, A.P.; Sane, P.V. Molecular characterization of CONSTANS-Like (COL) genes in banana (Musa acuminata L. AAA Group, cv. Grand Nain). Physiol. Mol. Biol. Plants 2016, 22, 1–15. [Google Scholar] [CrossRef]
- Wu, F.; Price, B.W.; Haider, W.; Seufferheld, G.; Nelson, R.; Hanzawa, Y. Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One 2014, 9, e85754. [Google Scholar] [CrossRef]
- Liu, C.; Tang, Q.; Cheng, C.; Xu, Y.; Yang, Z.; Dai, Z.; Su, J. Identification of putative CONSTANS-like genes from the de novo assembled transcriptome of leek. Biol. Plant. 2018, 62, 269–276. [Google Scholar] [CrossRef]
- Chia, T.Y.; Müller, A.; Jung, C.; Mutasa-Göttgens, E.S. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 2008, 59, 2735–2748. [Google Scholar] [CrossRef]
- Purugganan, M.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Pajoro, A.; Biewers, S.; Dougali, E.; Valentim, F.L.; Mendes, M.A.; Porri, A.; Coupland, G.; van de Peer, Y.; van Dijk, A.D.; Colombo, L.; et al. The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: A two-decade history. J. Exp. Bot. 2014, 65, 4731–4745. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Imaizumi, T. Circadian clock and photoperiodic response in Arabidopsis: From seasonal flowering to redox homeostasis. Biochemistry 2015, 54, 157–170. [Google Scholar] [CrossRef]
- Tripathi, P.; Carvallo, M.; Hamilton, E.E.; Preuss, S.; Kay, S.A. Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. USA 2017, 114, 172–177. [Google Scholar] [CrossRef]
- Hassidim, M.; Harir, Y.; Yakir, E.; Kron, I.; Green, M.R. Overexpression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 2009, 230, 481–491. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef]
- Ordonez-Herrera, N.; Trimborn, L.; Menje, M.; Henschel, M.; Robers, L.; Kaufholdt, D.; Hansch, R.; Adrian, J.; Ponnu, J.; Hoecker, U. The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture. Plant Physiol. 2018, 176, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Takase, T.; Kakikubo, Y.; Nakasone, A.; Nishiyama, Y.; Yasuhara, M.; Tokioka-Ono, Y.; Kiyosue, T. Characterization and transgenic study of CONSTANS-LIKE8 (COL8) gene in Arabidopsis thaliana: Expression of 35S:COL8 delays flowering under long-day conditions. Plant Biotechnol. 2011, 28, 439–446. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Jeong, D.H.; Lee, D.Y.; Yi, J.; Ryu, C.H.; Kim, S.L.; Jeong, H.J.; Choi, S.C.; Jin, P.; Yang, J.; et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010, 63, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gu, F.; Dong, S.; Liu, W.; Wang, H.; Chen, Z.; Wang, J. CONSTANS-like 9 (COL9) delays the flowering time in Oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 2016, 479, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Jin, M.; Wang, J.; Wu, F.; Sheng, P.; Cheng, Z.; Wang, J.; Zheng, X.; Chen, L.; Wang, M.; et al. OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant Cell Physiol. 2016, 57, 798–812. [Google Scholar] [CrossRef]
- Sheng, P.; Wu, F.; Tan, J.; Zhang, H.; Ma, W.; Chen, L.; Wang, J.; Wang, J.; Zhu, S.; Guo, X.; et al. A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol. Biol. 2016, 92, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, Y.; Zhang, M.; Zhan, X.; Shen, X.; Yu, P.; Chen, D.; Liu, Q.; Sinumporn, S.; Hussain, K.; et al. The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1. Biochem. Biophys. Res. Commun. 2018, 495, 1349–1355. [Google Scholar] [CrossRef]
- Miller, T.A.; Muslin, E.H.; Dorweiler, J.E. A maize CONSTANS like gene, Conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 2008, 227, 1377–1388. [Google Scholar] [CrossRef]
- Meng, X.; Muszynski, M.G.; Danilevskaya, O.N. The FT-Like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 2011, 23, 942–960. [Google Scholar] [CrossRef]
- Yang, S.; Weers, B.D.; Morishige, D.T.; Mullet, J.E. CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 2014, 14, 148. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z.; et al. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Campoli, C.; Drosse, B.; Searle, I.; Coupland, G.; von Korff, M. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 2012, 69, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Kawahigashi, H.; Oshima, M.; Ando, T.; Handa, H. The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J. Exp. Bot. 2012, 63, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Zhao, J.; Li, Y.; Zhang, F.; Zhou, J.; Chen, F.; Xie, X. OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Sci. 2016, 247, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zheng, X.M.; Chen, D.; Zhang, Y.; Ma, W.; Zhang, H.; Sun, L.; Yang, Z.; Zhao, C.; Zhan, X.; et al. OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 2017, 260, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Yun, C.H.; Lee, J.H.; Jang, Y.H.; Park, H.Y.; Kim, J.K. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 2008, 228, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Ping, Q.; Cheng, P.; Huang, F.; Ren, L.; Cheng, H.; Guan, Z.; Fang, W.; Chen, S.; Jiang, J. The heterologous expression in Arabidopsis thaliana of a chrysanthemum gene encoding the BBX family transcription factor CmBBX13 delays flowering. Plant Physiol. Biochem. 2019, 144. [Google Scholar] [CrossRef]
- Wang, L.; Sun, J.; Ren, L.; Zhou, M.; Han, X.; Ding, L.; Zhang, F.; Guan, Z.; Fang, W.; Chen, S.; et al. CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum. Plant Biotechnol. J. 2020, 18, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Dusi, V.; Pennisi, F.; Di Sansebastiano, G.P.; Zanzoni, S.; Manara, A.; Furini, A.; Martini, F.; Rotino, G.L.; Pandolfini, T. TCMP-2 affects tomato flowering and interacts with BBX16, a homolog of the Arabidopsis B-box MiP1b. Plant Direct. 2020, 4, e00283. [Google Scholar] [CrossRef] [PubMed]
- Abelenda, J.A.; Cruz-Oró, E.; Franco-Zorrilla, J.M.; Prat, S. Potato St CONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 2016, 26, 872–881. [Google Scholar] [CrossRef]
- Martínez-García, J.F.; Virgós-Soler, A.; Prat, S. Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. Proc. Natl. Acad. Sci. USA 2002, 99, 15211–15216. [Google Scholar] [CrossRef]
- Martin, J.; Storgaard, M.; Andersen, C.H.; Nielsen, K.K. Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Mol. Biol. 2004, 56, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Almada, R.; Cabrera, N.; Casaretto, J.A.; Ruiz-Lara, S.; Villanueva, E.G. VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 2009, 28, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, D.; Barre, P.; Santoni, S.; Julier, B. Association of a CONSTANS-LIKE gene to flowering and height in autotetraploid alfalfa. Theor. Appl. Genet. 2010, 121, 865–876. [Google Scholar] [CrossRef] [PubMed]
- González-Schain, N.D.; Díaz-Mendoza, M.; Zurczak, M.; Suárez-López, P. Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 2012, 70, 678–690. [Google Scholar] [CrossRef]
- Ballerini, E.S.; Kramer, E.M. In the light of evolution: A reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front. Plant Sci. 2011, 2, 81. [Google Scholar] [CrossRef]
- Abelenda, J.A.; Navarro, C.; Prat, S. Flowering and tuberization: A tale of two night shades. Trends Plant Sci. 2014, 19, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Rotino, G.L.; Dusi, V.; Chignola, R.; Sala, T.; Mennella, G.; Francese, G.; Pandolfini, T. Two metallocarboxypeptidase inhibitors are implicated in tomato fruit development and regulated by the inner no outer transcription factor. Plant Sci. 2018, 266, 19–26. [Google Scholar] [CrossRef]
- Chen, X.R.; Wang, Y.; Zhao, H.H.; Zhang, X.Y.; Wang, X.B.; Li, D.W.; Yu, J.L.; Han, C.G. Brassica yellows virus’ movement protein upregulates anthocyanin accumulation, leading to the development of purple leaf symptoms on Arabidopsis thaliana. Sci. Rep. 2018, 8, 16273. [Google Scholar] [CrossRef]
- Li, X.L.; Lv, X.; Wang, X.; Wang, L.; Zhang, M.; Ren, M. Effects of abiotic stress on anthocyanin accumulation and grain weight in purple wheat. Crop Pasture Sci. 2018, 69. [Google Scholar] [CrossRef]
- Zheng, X.T.; Yu, Z.C.; Tang, J.W.; Cai, M.L.; Chen, Y.L.; Yang, C.W.; Chow, W.S.; Peng, C.L. The major photoprotective role of anthocyanins in leaves of Arabidopsis thaliana under long-term high light treatment: Antioxidant or light attenuator? Photosynth. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, B.J.; Henry-Kirk, R.; Friend, A.; Diack, R.; Helbig, S.; Mouhu, K.; Tomes, S.; Dare, A.P.; Espley, R.V.; Putterill, J.; et al. Apple B-box factors regulate light-responsive anthocyanin biosynthesis genes. Sci. Rep. 2019, 9, 17762. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotech. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Espley, R.V.; Lin-Wang, K.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple B-Box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2020, 61, 130–143. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Takuhara, Y.; Kobayashi, M.; Suzuki, S. Low-temperature-induced transcription factors in grapevine enhance cold tolerance in transgenic Arabidopsis plants. J. Plant Physiol. 2011, 168, 967–975. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 437–447. [Google Scholar] [CrossRef]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Zhou, M.R.; Wei, X.Y.; Chen, Q.Q.; Li, W.Q.; Liu, W.T.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genomics 2019, 20, 27. [Google Scholar] [CrossRef]
- Liu, J.; Shen, J.; Xu, Y.; Li, X.; Xiao, J.; Xiong, L. Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 2016, 67, 5785–5798. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.Y.; Wang, J.N.; Kuang, J.F.; Shan, W.; Lu, W.J. Molecular characterization and expression profiles of MaCOL1, a CONSTANS-like gene in banana fruit. Gene 2012, 496, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Q.; Zhai, H.; Gao, S.; Yang, L.; Wang, Z.; Xu, Y.; Huo, J.; Ren, Z.; Zhao, N.; et al. IbBBX24 promotes the jasmonic acid pathway and enhances fusarium wilt resistance in sweet potato. Plant Cell 2020, 32, 1102–1123. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, S.; Sun, D.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; Chen, Z. CONSTANS-like 9 (OsCOL9) interacts with receptor for activated C-Kinase 1 (OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS ONE 2016, 11, e0166249. [Google Scholar] [CrossRef] [PubMed]
- Vaishak, K.P.; Yadukrishnan, P.; Bakshi, S.; Kushwaha, A.K.; Ramachandran, H.; Job, N.; Babu, D.; Datta, S. The B-box bridge between light and hormones in plants. J. Photochem. Photobiol. B. 2019, 191, 164–174. [Google Scholar] [CrossRef]
- Hou, H.; Jia, H.; Yan, Q.; Wang, X. Overexpression of a SBP-Box Gene (VpSBP16) from chinese wild vitis species in Arabidopsis improves salinity and drought stress tolerance. Int. J. Mol. Sci. 2018, 19, 940. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef]
- Mbambalala, N.; Panda, S.K.; van der Vyver, C. Overexpression of AtBBX29 improves drought tolerance by maintaining photosynthesis and enhancing the antioxidant and osmolyte capacity of sugarcane plants. Plant Mol. Biol. Rep. 2020. [Google Scholar] [CrossRef]
- Kiełbowicz-Matuk, A.; Czarnecka, J. Interplays of plant circadian clock and abiotic stress response network. Emerging Technologies and Management of Crop Stress Tolerance, Volume 1-Biological Techniques, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 20, pp. 487–506. [Google Scholar]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian clock genes universally control key agricultural traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef] [PubMed]
- Piechulla, B.; Merforth, N.; Rudolph, B. Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol. Biol. 1998, 38, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Kiełbowicz-Matuk, A.; Czarnecka, J.; Banachowicz, E.; Rey, P.; Rorat, T. Solanum tuberosum ZPR1 encodes a light-regulated nuclear DNA-binding protein adjusting the circadian expression of StBBX24 to light cycle. Plant Cell Environ. 2017, 40, 424–440. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Bian, Y.; Liu, J.; Sun, Y.; Xu, D. B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 2020, 62, 1293–1309. [Google Scholar] [CrossRef] [PubMed]

| Plant Species | No. of BBX Genes | No. of Chromosomes Having BBX/No. of Plant Chromosomes | Chromosome with no BBX | References |
|---|---|---|---|---|
| Rice (Oryza sativa) | 30 | 10/12 | 10 and 11 | [2] |
| Maize (Zea mays) | 31 | 9/10 | 8 | [36,37] |
| Apple (Malus domestica) | 64 | 15/17 | 4 and 15 | [6] |
| Pear (Pyrus bretschneideri) | 25 | 12/17 | 1,2,4,7,12 | [5] |
| Pear (Pyrus pyrifolia) | 39 | nd/17 | nd | [38] |
| Rose (Rosa chinensis) | 22 | 6/7 | 1 | [39] |
| Peach (Prunus persica) | 20 | 6/8 | 3 and 6 | [39] |
| Strawberry (Fragaria vesca) | 21 | 6/7 | 7 | [39] |
| Sweet cherry (Prunus avium) | 22 | 6/8 | 2 and 6 | [39] |
| Black raspberry (Rubus occidentalis) | 20 | 6/7 | 7 | [39] |
| Tomato (Solanum lycopersicum) | 29 | 11/12 | 11 | [3] |
| Potato (Solanum tuberosum) | 30 | 10/12 | 11 | [4] |
| Cotton (Gossypium hirsutum) | 42 | 18/18 | - | [40] |
| Grapevine (Vitis vinifera) | 24 | 11/19 | 2,6,8,10,13,15,16,17 | [8] |
| Wild peanut (Arachis duranensis) | 24 | 9/10 | 2 | [41] |
| Banana (Musa acuminata) * | 25 | 7/11 | 4,5,6,8 | [42] |
| Soyabean (Glycine max) * | 26 | nd/10 | nd | [43] |
| Leek (Allium porrum) * | 17 | nd/8 | nd | [44] |
| Sugar beet (Beta vulgaris) * | 10 | nd/9 | nd | [45] |
| Gene | Accession No./ID | Response to Abiotic Stress and Hormones | Species | References | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cold/ Chilling | Drought | Salt | Dehydration | Heat | ABA | GA | SA | MeJA | ||||
| VvZFPL | HQ179976 | + | Vitis vinifera | [91] | ||||||||
| SsBBX24 | ABC25454 | + | + | Solanum sogarandinum | [26] | |||||||
| MdBBX10 | MDP0000733075 | + | Malus domestica | [92] | ||||||||
| OsBBX1 | Os01g0202500 | + | + | + | Oryza sativa | [93] | ||||||
| OsBBX2 | Os02g0176000 | + | + | + | + | + | + | Oryza sativa | [93] | |||
| OsBBX7 | Os02g0724000 | + | + | + | Oryza sativa | [93] | ||||||
| OsBBX8 | Os02g0731700 | + | + | + | Oryza sativa | [93,94] | ||||||
| OsBBX14 | Os05g0204600 | + | Oryza sativa | [93] | ||||||||
| OsBBX17 | Os06g0264200 | + | + | + | Oryza sativa | [93] | ||||||
| OsBBX19 | Os06g0298200 | + | + | + | + | + | + | Oryza sativa | [93] | |||
| OsBBX24 | Os08g0178800 | + | + | + | + | + | + | Oryza sativa | [93] | |||
| SlBBX1 * | Solyc02g089520.1 | + | Solanum lycopersicum | [3] | ||||||||
| SlBBX7 ** | Solyc12g006240.1 | + | Solanum lycopersicum | [3] | ||||||||
| SlBBX16 *** | Solyc12g005750.1 | + | Solanum lycopersicum | [3] | ||||||||
| MaCOL1 | JQ314345 | + | Musa nana | [95] | ||||||||
| Cm-BBX24 | KF385866 | + | + | Chrysanthemum morifolium | [64] | |||||||
| IbBBX24 | MH813941 | + | Ipomoea batatas | [96] | ||||||||
| Gene | Accession No./ID | From | To | Phenotypes | References |
|---|---|---|---|---|---|
| MdBBX10 | MDP0000733075 | Malus domestica | Arabidopsis thaliana | Salt and drought tolerance | [92] |
| VpSBP16 | nd | Vitis pseudoreticulata | Arabidopsis thaliana | Salt and drought tolerance | [99] |
| Cm-BBX24 | KF385866 | Chrysanthemum morifolium | Chrysanthemum morifolium | Tolerance to freezing and drought | [64] |
| VvZFPL | HQ179976 | Vitis vinifera | Arabidopsis thaliana | Cold tolerance | [91] |
| AtBBX29 | At5g54470 | Arabidopsis thaliana | Saccharum | Drought tolerance | [101] |
| MdBBX37 | MDP0000157816 | Malus hupehensis | Malus hupehensis | Cold tolerance | [100] |
| Ghd2(OsBBX8) | Os02g0731700 | Oryza sativa | Oryza sativa | Drought tolerance | [94] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talar, U.; Kiełbowicz-Matuk, A. Beyond Arabidopsis: BBX Regulators in Crop Plants. Int. J. Mol. Sci. 2021, 22, 2906. https://doi.org/10.3390/ijms22062906
Talar U, Kiełbowicz-Matuk A. Beyond Arabidopsis: BBX Regulators in Crop Plants. International Journal of Molecular Sciences. 2021; 22(6):2906. https://doi.org/10.3390/ijms22062906
Chicago/Turabian StyleTalar, Urszula, and Agnieszka Kiełbowicz-Matuk. 2021. "Beyond Arabidopsis: BBX Regulators in Crop Plants" International Journal of Molecular Sciences 22, no. 6: 2906. https://doi.org/10.3390/ijms22062906
APA StyleTalar, U., & Kiełbowicz-Matuk, A. (2021). Beyond Arabidopsis: BBX Regulators in Crop Plants. International Journal of Molecular Sciences, 22(6), 2906. https://doi.org/10.3390/ijms22062906





