Prokaryotic Solute/Sodium Symporters: Versatile Functions and Mechanisms of a Transporter Family †
Abstract
:1. Introduction
2. Functional Properties and Physiological Significance of Prokaryotic Transporters of the SSS Family
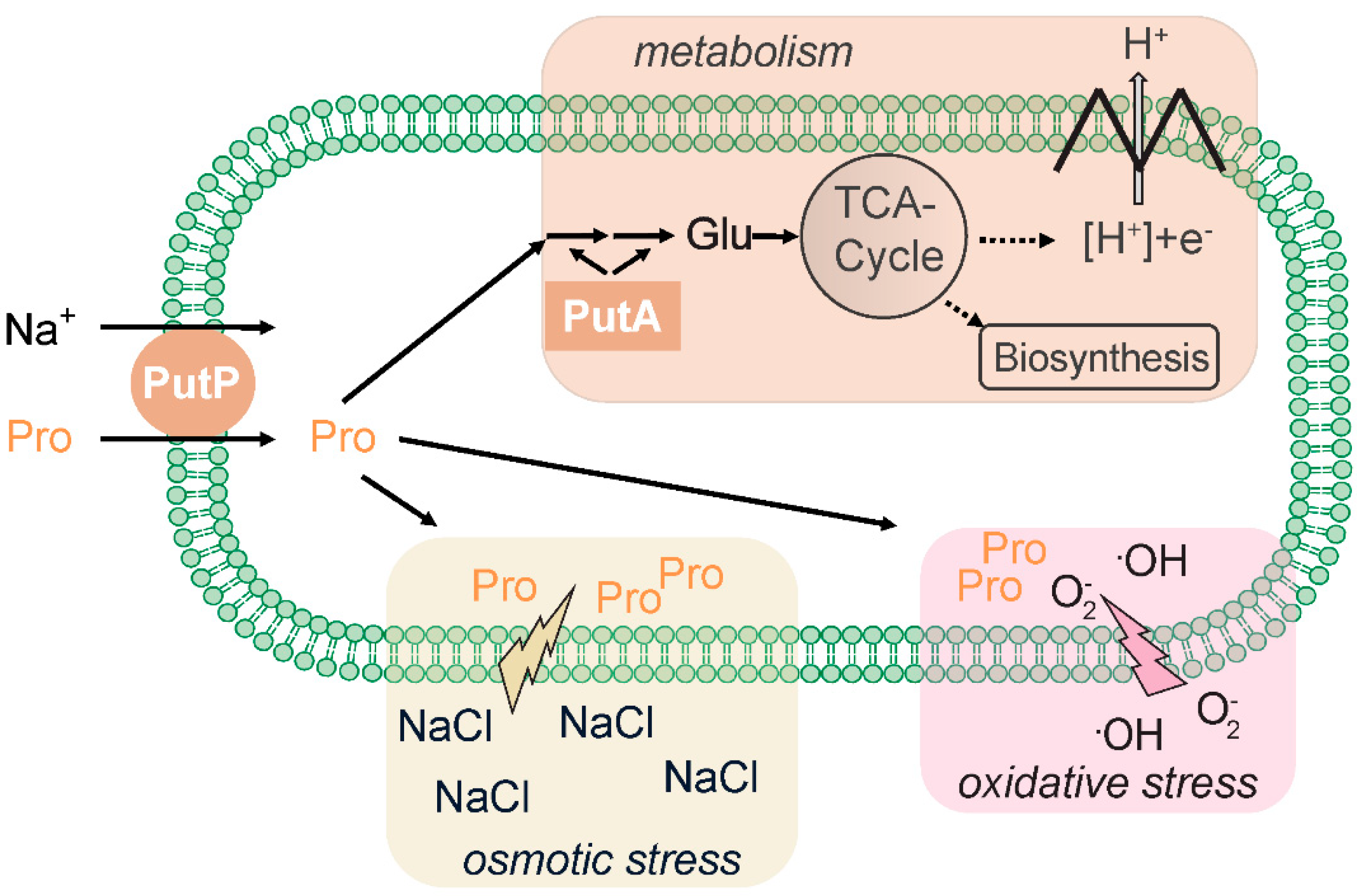
2.1. PutP-Mediated Nutrient Supply and Osmoadaptation
2.2. Sugar Uptake via SSS Family Transporters
2.3. Uptake of Short Chain Organic Acids via SSS Family Transporters
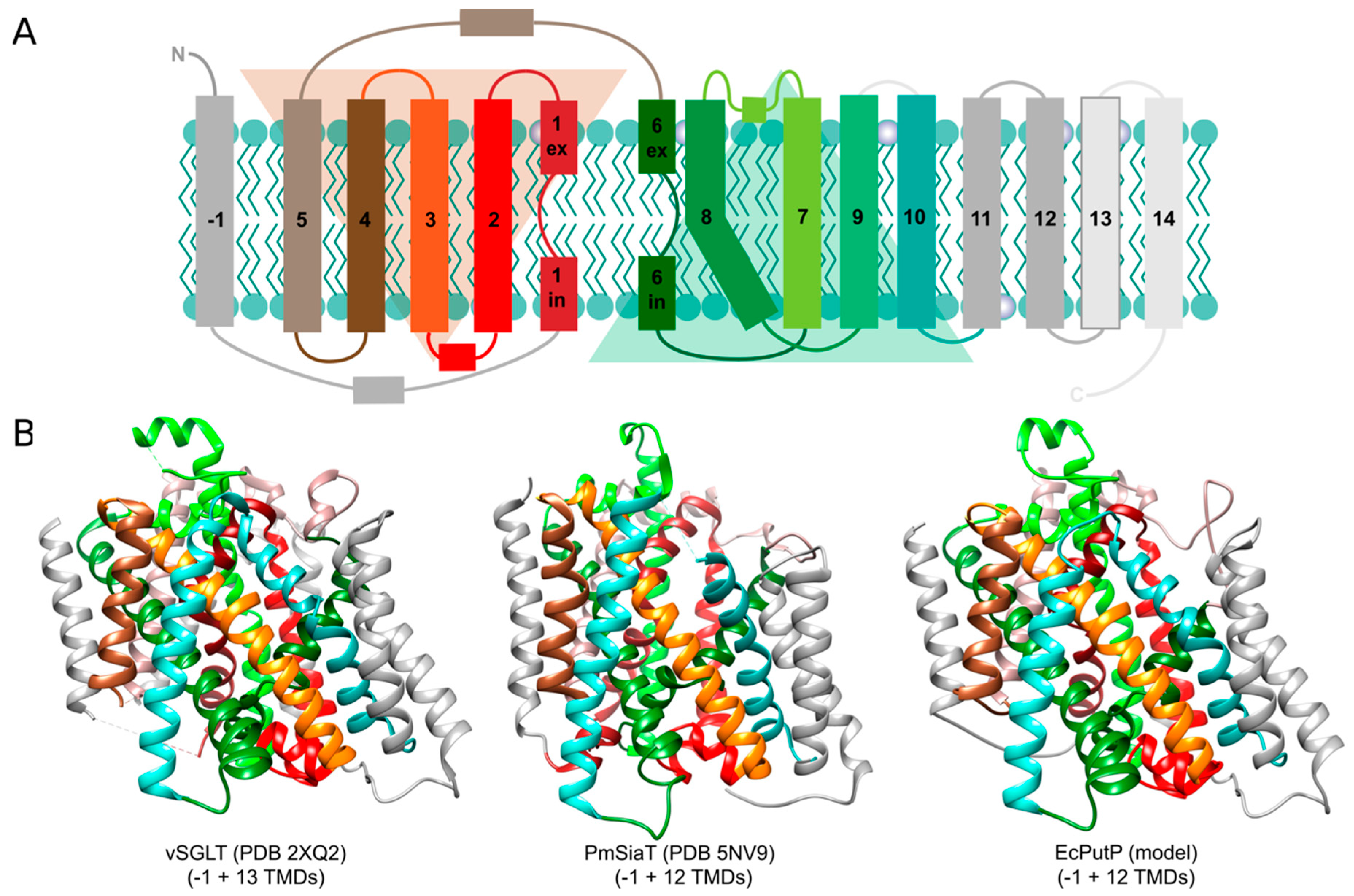
3. Structural Basis of Transport
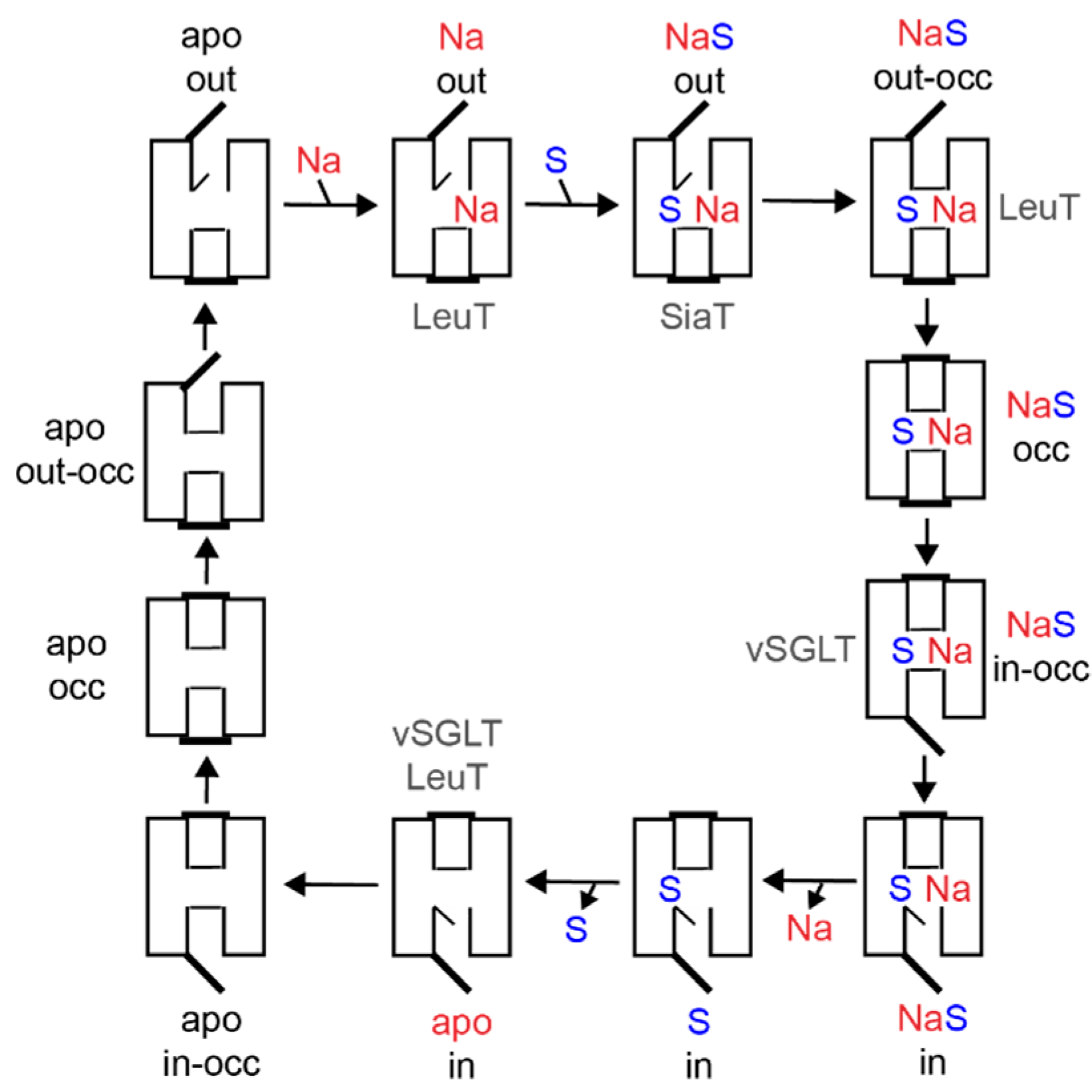
4. Molecular Mechanism of Transport
4.1. Sites of Substrate and Sodium Ion Binding
4.2. Transport by Alternating Access
5. SSS Domain-Dependent Sensor Kinases
5.1. Occurrence, Significance and Targets of SSS Domain-Dependent Signal Transduction Systems
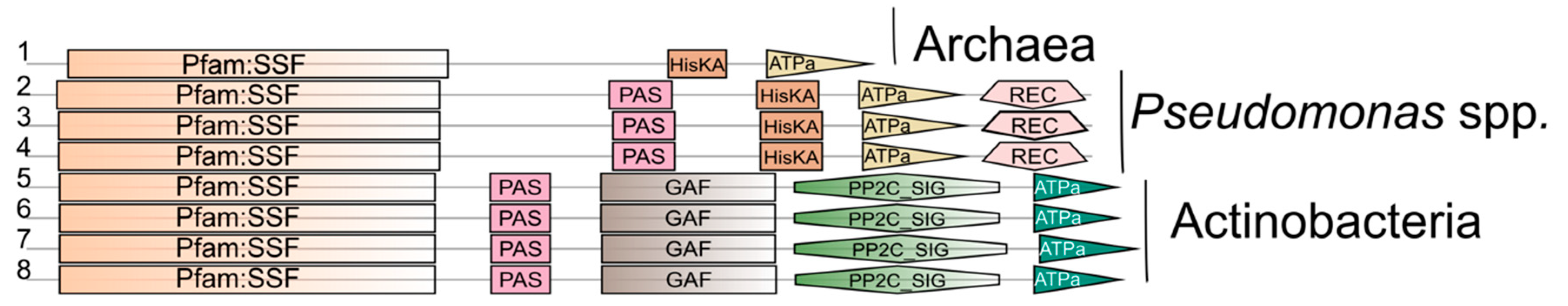
5.1.1. Distribution in Prokaryotes
5.1.2. Biological Significance and Targets of Two-Component Systems Containing SSS Domains
5.2. Structure-Functions Relationsships in SSS Domain-Dependent Sensor Kinases
5.2.1. General Characteristics of SSS-Containing Sensor Kinases
5.2.2. Transport Activity of SSS Family-Containing Sensor Kinase CbrA
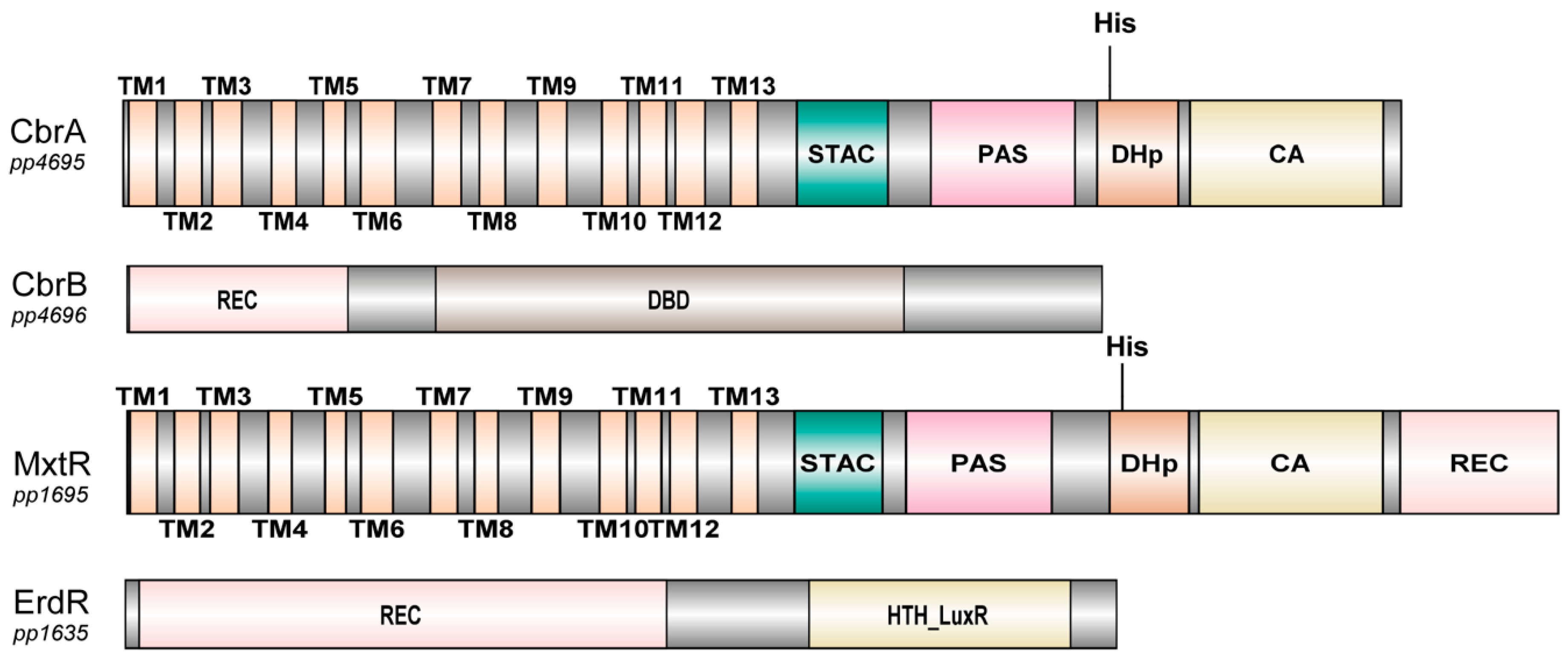
5.2.3. Properties of Domains Associated with SSS Domain-Containing Sensor Kinases
6. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crane, R.; Miller, D.; Bihler, I. The restrictions on possible mechanisms of intestinal transport of sugars. In Membrane Transport and Metabolism, Proceedings of a Symposium held in Prague, August 22–27, 1960; Kleinzeller, A., Kotyk, A., Eds.; ISRC 11795501; Czech Academy of Sciences: Prague, Czech Republic, 1961; pp. 439–449. [Google Scholar]
- Wright, E.M.; Loo, D.D.; Hirayama, B.A. Biology of human sodium glucose transporters. Physiol. Rev. 2011, 91, 733–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinzeller, A.; Kotyk, A. Membrane Transport and Metabolism: Proceedings of a Symposium, Prague, Czechia, 22–27 August 1960; Academic Press: London, UK, 1961. [Google Scholar]
- Mitchell, P. Molecule, group and electron translocation through natural membranes. Biochem. Soc. Symp. 1962, 22, 142–169. [Google Scholar]
- Schmidt, S.; Biegel, E.; Müller, V. The ins and outs of Na+ bioenergetics in Acetobacterium woodii. Biochim. Biophys. Acta (BBA)-Bioenerg. 2009, 1787, 691–696. [Google Scholar] [CrossRef] [Green Version]
- Steffen, W.; Steuber, J. Cation transport by the respiratory NADH:quinone oxidoreductase (complex I): Facts and hypotheses. Biochem Soc. Trans. 2013, 41, 1280–1287. [Google Scholar] [CrossRef] [Green Version]
- Dimroth, P.; von Ballmoos, C. ATP synthesis by decarboxylation phosphorylation. Results Probl. Cell Differ. 2008, 45, 153–184. [Google Scholar] [CrossRef] [PubMed]
- Häse, C.C.; Fedorova, N.D.; Galperin, M.Y.; Dibrov, P.A. Sodium ion cycle in bacterial pathogens: Evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. MMBR 2001, 65, 353–370. [Google Scholar] [CrossRef] [Green Version]
- Padan, E.; Landau, M. Sodium-Proton (Na+/H+) Antiporters: Properties and roles in health and disease. Metal. Ions Life Sci. 2016, 16, 391–458. [Google Scholar]
- Saier, M.H., Jr.; Reddy, V.S.; Tamang, D.G.; Västermark, Å. The Transporter Classification Database. Nucleic Acids Res. 2013, 42, D251–D258. [Google Scholar] [CrossRef]
- Krishnamurthy, H.; Piscitelli, C.L.; Gouaux, E. Unlocking the molecular secrets of sodium-coupled transporters. Nature 2009, 459, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Wright, E.M. Structure and function of Na+-symporters with inverted repeats. Curr. Opin. Struct. Biol. 2009, 19, 425–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalayil, S.; Schulze, S.; Kühlbrandt, W. Arginine oscillation explains Na+ independence in the substrate/product antiporter CaiT. Proc. Natl. Acad. Sci. USA 2013, 110, 17296–17301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y. Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 2013, 42, 51–72. [Google Scholar] [CrossRef]
- Västermark, Å.; Saier, M.H., Jr. Evolutionary relationship between 5+5 and 7+7 inverted repeat folds within the amino acid-polyamine-organocation superfamily. Proteins 2014, 82, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, H.; Gouaux, E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature 2012, 481, 469–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrest, L.R.; Krämer, R.; Ziegler, C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 2011, 1807, 167–188. [Google Scholar] [CrossRef] [Green Version]
- Gotfryd, K.; Boesen, T.; Mortensen, J.S.; Khelashvili, G.; Quick, M.; Terry, D.S.; Missel, J.W.; LeVine, M.V.; Gourdon, P.; Blanchard, S.C.; et al. X-ray structure of LeuT in an inward-facing occluded conformation reveals mechanism of substrate release. Nat. Commun. 2020, 11, 1005. [Google Scholar] [CrossRef] [Green Version]
- McHaourab, H.S.; Steed, P.R.; Kazmier, K. Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy. Structure 2011, 19, 1549–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raba, M.; Dunkel, S.; Hilger, D.; Lipiszko, K.; Polyhach, Y.; Jeschke, G.; Bracher, S.; Klare, J.P.; Quick, M.; Jung, H.; et al. Extracellular loop 4 of the proline transporter PutP controls the periplasmic entrance to ligand binding sites. Structure 2014, 22, 769–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaback, H.R.; Guan, L. It takes two to tango: The dance of the permease. J. Gen. Physiol. 2019, 151, 878–886. [Google Scholar] [CrossRef] [Green Version]
- Hilger, D.; Jung, H. Protein Chemical and Electron Paramagnetic Resonance Spectroscopic Approaches to Monitor Membrane Protein Structure and Dynamics—Methods Chapter; Wiley-Blackwell: Weinheim, Germany, 2010; pp. 247–263. [Google Scholar]
- Hubbell, W.L.; Cafiso, D.S.; Altenbach, C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000, 7, 735–739. [Google Scholar] [CrossRef]
- Lerner, E.; Cordes, T.; Ingargiola, A.; Alhadid, Y.; Chung, S.; Michalet, X.; Weiss, S. Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 2018, 359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Terry, D.; Shi, L.; Weinstein, H.; Blanchard, S.C.; Javitch, J.A. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 2010, 465, 188–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H. The sodium/substrate symporter family: Structural and functional features. FEBS Lett. 2002, 529, 73–77. [Google Scholar] [CrossRef] [Green Version]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, E.M.; Ghezzi, C.; Loo, D.D.F. Novel and unexpected functions of SGLTs. Physiology 2017, 32, 435–443. [Google Scholar] [CrossRef]
- Quick, M.; Shi, L. The sodium/multivitamin transporter: A multipotent system with therapeutic implications. Vitam. Horm. 2015, 98, 63–100. [Google Scholar] [CrossRef] [Green Version]
- Ravera, S.; Reyna-Neyra, A.; Ferrandino, G.; Amzel, L.M.; Carrasco, N. The sodium/iodide symporter (NIS): Molecular physiology and preclinical and clinical applications. Annu. Rev. Physiol. 2017, 79, 261–289. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.; Hilger, D.; Raba, M. The Na+/L-proline transporter PutP. Front. Biosci. 2012, 17, 745–759. [Google Scholar] [CrossRef]
- Jung, H.; Tebbe, S.; Schmid, R.; Jung, K. Unidirectional reconstitution and characterization of purified Na+/proline transporter of Escherichia coli. Biochemistry 1998, 37, 11083–11088. [Google Scholar] [CrossRef]
- Liao, M.K.; Maloy, S. Substrate recognition by proline permease in Salmonella. Amino Acids 2001, 21, 161–174. [Google Scholar] [CrossRef]
- Tanner, J.J. Structural biology of proline catabolic enzymes. Antioxid. Redox Signal. 2019, 30, 650–673. [Google Scholar] [CrossRef] [PubMed]
- Moses, S.; Sinner, T.; Zaprasis, A.; Stöveken, N.; Hoffmann, T.; Belitsky, B.R.; Sonenshein, A.L.; Bremer, E. Proline utilization by Bacillus subtilis: Uptake and catabolism. J. Bacteriol. 2012, 194, 745–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köcher, S.; Tausendschön, M.; Thompson, M.; Saum, S.H.; Müller, V. Proline metabolism in the moderately halophilic bacterium Halobacillus halophilus: Differential regulation of isogenes in proline utilization. Environ. Microbiol. Rep. 2011, 3, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Park, N.Y.; Lee, M.H.; Choi, S.H. Characterization of the Vibrio vulnificus putAP operon, encoding proline dehydrogenase and proline permease, and its differential expression in response to osmotic stress. J. Bacteriol. 2003, 185, 3842–3852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, Y.; Nishijyo, T.; Itoh, Y. Divergent structure and regulatory mechanism of proline catabolic systems: Characterization of the putAP proline catabolic operon of Pseudomonas aeruginosa PAO1 and its regulation by PruR, an AraC/XylS family protein. J. Bacteriol. 2002, 184, 5633–5640. [Google Scholar] [CrossRef] [Green Version]
- Vílchez, S.; Molina, L.; Ramos, C.; Ramos, J.L. Proline catabolism by Pseudomonas putida: Cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 2000, 182, 91–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegelhalter, F.; Bremer, E. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: Contributions of the sigma A- and sigma B-dependent stress-responsive promoters. Mol. Microbiol. 1998, 29, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Von Blohn, C.; Kempf, B.; Kappes, R.M.; Bremer, E. Osmostress response in Bacillus subtilis: Characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 1997, 25, 175–187. [Google Scholar] [CrossRef] [Green Version]
- Ozturk, T.N.; Culham, D.E.; Tempelhagen, L.; Wood, J.M.; Lamoureux, G. Salt-dependent interactions between the C-terminal domain of osmoregulatory transporter ProP of Escherichia coli and the lipid membrane. J. Phys. Chem. B 2020, 124, 8209–8220. [Google Scholar] [CrossRef]
- Ziegler, C.; Bremer, E.; Krämer, R. The BCCT family of carriers: From physiology to crystal structure. Mol. Microbiol. 2010, 78, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Racher, K.I.; Voegele, R.T.; Marshall, E.V.; Culham, D.E.; Wood, J.M.; Jung, H.; Bacon, M.; Cairns, M.T.; Ferguson, S.M.; Liang, W.J.; et al. Purification and reconstitution of an osmosensor: Transporter ProP of Escherichia coli senses and responds to osmotic shifts. Biochemistry 1999, 38, 1676–1684. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Doster, A.R.; Duhamel, G.E.; Becker, D.F. Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect. Immun. 2008, 76, 3037. [Google Scholar] [CrossRef] [Green Version]
- Available, N.A.N.; Mohanty, P.; Matysik, J. Effect of proline on the production of singlet oxygen. Amino Acids 2001, 21, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Cumbes, Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Krishnan, N.; Becker, D.F. Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress. J. Bacteriol. 2006, 188, 1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christgen, S.L.; Becker, D.F. Role of proline in pathogen and host interactions. Antioxid. Redox Signal. 2019, 30, 683–709. [Google Scholar] [CrossRef]
- Schwan, W.R.; Wetzel, K.J. Osmolyte transport in Staphylococcus aureus and the role in pathogenesis. World J. Clin. Infect. Dis. 2016, 6, 22–27. [Google Scholar] [CrossRef]
- Schwan, W.R.; Lehmann, L.; McCormick, J. Transcriptional activation of the Staphylococcus aureus putP gene by low-proline-high osmotic conditions and during infection of murine and human tissues. Infect. Immun. 2006, 74, 399–409. [Google Scholar] [CrossRef] [Green Version]
- Schwan, W.R.; Wetzel, K.J.; Gomez, T.S.; Stiles, M.A.; Beitlich, B.D.; Grunwald, S. Low-proline environments impair growth, proline transport and in vivo survival of Staphylococcus aureus strain-specific putP mutants. Microbiology 2004, 150, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.H.; Miller, K.J. Identification of two proline transport systems in Staphylococcus aureus and their possible roles in osmoregulation. Appl. Environ. Microbiol. 1992, 58, 471–475. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.J.; Zelt, S.C.; Bae, J.-H. Glycine betaine and proline are the principal compatible solutes of Staphylococcus aureus. Curr. Microbiol. 1991, 23, 131–137. [Google Scholar] [CrossRef]
- Suerbaum, S.; Josenhans, C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 2007, 5, 441–452. [Google Scholar] [CrossRef]
- Kavermann, H.; Burns, B.P.; Angermuller, K.; Odenbreit, S.; Fischer, W.; Melchers, K.; Haas, R. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 2003, 197, 813–822. [Google Scholar] [CrossRef]
- Nakajima, K.; Inatsu, S.; Mizote, T.; Nagata, Y.; Aoyama, K.; Fukuda, Y.; Nagata, K. Possible involvement of putA gene in Helicobacter pylori colonization in the stomach and motility. Biomed. Res. 2008, 29, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Rivera-Ordaz, A.; Bracher, S.; Sarrach, S.; Li, Z.; Shi, L.; Quick, M.; Hilger, D.; Haas, R.; Jung, H. The sodium/proline transporter PutP of Helicobacter pylori. PLoS ONE 2013, 8, e83576. [Google Scholar] [CrossRef] [PubMed]
- Park, H.B.; Sampathkumar, P.; Perez, C.E.; Lee, J.H.; Tran, J.; Bonanno, J.B.; Hallem, E.A.; Almo, S.C.; Crawford, J.M. Stilbene epoxidation and detoxification in a Photorhabdus luminescens-nematode symbiosis. J. Biol. Chem. 2017, 292, 6680–6694. [Google Scholar] [CrossRef] [Green Version]
- Kontnik, R.; Crawford, J.M.; Clardy, J. Exploiting a global regulator for small molecule discovery in Photorhabdus luminescens. ACS Chem. Biol. 2010, 5, 659–665. [Google Scholar] [CrossRef] [Green Version]
- Kraemer, P.S.; Mitchell, A.; Pelletier, M.R.; Gallagher, L.A.; Wasnick, M.; Rohmer, L.; Brittnacher, M.J.; Manoil, C.; Skerett, S.J.; Salama, N.R. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect. Immun. 2009, 77, 232–244. [Google Scholar] [CrossRef] [Green Version]
- Wilson, T.H.; Ding, P.Z. Sodium-substrate cotransport in bacteria. Biochim. Biophys. Acta 2001, 1505, 121–130. [Google Scholar] [CrossRef] [Green Version]
- Turk, E.; Kim, O.; le Coutre, J.; Whitelegge, J.P.; Eskandari, S.; Lam, J.T.; Kreman, M.; Zampighi, G.; Faull, K.F.; Wright, E.M. Molecular characterization of Vibrio parahaemolyticus vSGLT: A model for sodium-coupled sugar cotransporters. J. Biol. Chem. 2000, 275, 25711–25716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahlgren, W.Y.; Dunevall, E.; North, R.A.; Paz, A.; Scalise, M.; Bisignano, P.; Bengtsson-Palme, J.; Goyal, P.; Claesson, E.; Caing-Carlsson, R.; et al. Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+ site. Nat. Commun. 2018, 9, 1753. [Google Scholar] [CrossRef] [PubMed]
- Jeckelmann, J.M.; Erni, B. Transporters of glucose and other carbohydrates in bacteria. Pflugers Arch. 2020, 472, 1129–1153. [Google Scholar] [CrossRef]
- Sarker, R.I.; Ogawa, W.; Tsuda, M.; Tanaka, S.; Tsuchiya, T. Characterization of a glucose transport system in Vibrio parahaemolyticus. J. Bacteriol. 1994, 176, 7378–7382. [Google Scholar] [CrossRef] [Green Version]
- Rodionov, D.A.; Yang, C.; Li, X.; Rodionova, I.A.; Wang, Y.; Obraztsova, A.Y.; Zagnitko, O.P.; Overbeek, R.; Romine, M.F.; Reed, S.; et al. Genomic encyclopedia of sugar utilization pathways in the Shewanella genus. BMC Genom. 2010, 11, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiczew, D.; Borowska, A.; Szkaradek, K.; Biegus, T.; Wozniak, K.; Pyclik, M.; Sitarska, M.; Jaszewski, L.; Radosinski, L.; Hanus-Lorenz, B.; et al. Molecular mechanism of vSGLT inhibition by gneyulin reveals antiseptic properties against multidrug-resistant Gram-negative bacteria. J. Mol. Model. 2019, 25, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, G.H. Sialic acid acquisition in bacteria-one substrate, many transporters. Biochem. Soc. Trans. 2016, 44, 760–765. [Google Scholar] [CrossRef]
- North, R.A.; Wahlgren, W.Y.; Remus, D.M.; Scalise, M.; Kessans, S.A.; Dunevall, E.; Claesson, E.; Soares da Costa, T.P.; Perugini, M.A.; Ramaswamy, S.; et al. The sodium sialic acid symporter from Staphylococcus aureus has altered substrate specificity. Front. Chem. 2018, 6, 233. [Google Scholar] [CrossRef]
- Ng, K.M.; Ferreyra, J.A.; Higginbottom, S.K.; Lynch, J.B.; Kashyap, P.C.; Gopinath, S.; Naidu, N.; Choudhury, B.; Weimer, B.C.; Monack, D.M.; et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502, 96–99. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- D’Souza, G.; Shitut, S.; Preussger, D.; Yousif, G.; Waschina, S.; Kost, C. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 2018, 35, 455–488. [Google Scholar] [CrossRef] [Green Version]
- Gimenez, R.; Nuñez, M.F.; Badia, J.; Aguilar, J.; Baldoma, L. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 2003, 185, 6448–6455. [Google Scholar] [CrossRef] [Green Version]
- Jolkver, E.; Emer, D.; Ballan, S.; Krämer, R.; Eikmanns, B.J.; Marin, K. Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J. Bacteriol. 2009, 191, 940–948. [Google Scholar] [CrossRef] [Green Version]
- Hosie, A.H.; Allaway, D.; Poole, P.S. A monocarboxylate permease of Rhizobium leguminosarum is the first member of a new subfamily of transporters. J. Bacteriol. 2002, 184, 5436–5448. [Google Scholar] [CrossRef] [Green Version]
- Borghese, R.; Zannoni, D. Acetate permease (ActP) is responsible for tellurite (TeO32−) uptake and resistance in cells of the facultative phototroph Rhodobacter capsulatus. Appl. Environ. Microbiol. 2010, 76, 942–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.; Rübenhagen, R.; Tebbe, S.; Leifker, K.; Tholema, N.; Quick, M.; Schmid, R. Topology of the Na+/proline transporter of Escherichia coli. J. Biol. Chem. 1998, 273, 26400–26407. [Google Scholar] [CrossRef] [Green Version]
- Wegener, C.; Tebbe, S.; Steinhoff, H.-J.; Jung, H. Spin labeling analysis of structure and dynamics of the Na+/proline transporter of Escherichia coli. Biochemistry 2000, 39, 4831–4837. [Google Scholar] [CrossRef] [PubMed]
- Turk, E.; Wright, E.M. Membrane topology motifs in the SGLT cotransporter family. J. Membr. Biol. 1997, 159, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hediger, M.A.; Turk, E.; Wright, E.M. Homology of the human intestinal Na+/glucose and Escherichia coli Na+/proline cotransporters. Proc. Natl. Acad. Sci. USA 1989, 86, 5748. [Google Scholar] [CrossRef] [Green Version]
- Faham, S.; Watanabe, A.; Besserer, G.M.; Cascio, D.; Specht, A.; Hirayama, B.A.; Wright, E.M.; Abramson, J. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 2008, 321, 810–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, A.; Choe, S.; Chaptal, V.; Rosenberg, J.M.; Wright, E.M.; Grabe, M.; Abramson, J. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 2010, 468, 988–991. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, A.; Singh, S.K.; Kawate, T.; Jin, Y.; Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature 2005, 437, 215–223. [Google Scholar] [CrossRef]
- Claxton, D.P.; Quick, M.; Shi, L.; de Carvalho, F.D.; Weinstein, H.; Javitch, J.A.; McHaourab, H.S. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat. Struct. Mol. Biol. 2010, 17, 822–829. [Google Scholar] [CrossRef]
- Kazmier, K.; Claxton, D.P.; McHaourab, H.S. Alternating access mechanisms of LeuT-fold transporters: Trailblazing towards the promised energy landscapes. Curr. Opin. Struct. Biol. 2017, 45, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Bracher, S.; Schmidt, C.C.; Dittmer, S.I.; Jung, H. Core transmembrane domain 6 plays a pivotal role in the transport cycle of the sodium/proline symporter PutP. J. Biol. Chem. 2016, 291, 26208–26215. [Google Scholar] [CrossRef] [Green Version]
- Penmatsa, A.; Gouaux, E. How LeuT shapes our understanding of the mechanisms of sodium-coupled neurotransmitter transporters. J. Physiol. 2014, 592, 863–869. [Google Scholar] [CrossRef]
- Bracher, S.; Guérin, K.; Polyhach, Y.; Jeschke, G.; Dittmer, S.; Frey, S.; Böhm, M.; Jung, H. Glu-311 in external loop 4 of the sodium/proline transporter PutP is crucial for external gate closure. J. Biol. Chem. 2016, 291, 4998–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Hilger, D.; Böhm, M.; Hackmann, A.; Jung, H. Role of Ser-340 and Thr-341 in transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli in ligand binding and transport. J. Biol. Chem. 2008, 283, 4921–4929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quick, M.; Jung, H. A conserved aspartate residue, Asp187, is important for Na+-dependent proline binding and transport by the Na+/proline transporter of Escherichia coli. Biochemistry 1998, 37, 13800–13806. [Google Scholar] [CrossRef]
- Raba, M.; Baumgartner, T.; Hilger, D.; Klempahn, K.; Härtel, T.; Jung, K.; Jung, H. Function of transmembrane domain IX in the Na+/proline transporter PutP. J. Mol. Biol. 2008, 382, 884–893. [Google Scholar] [CrossRef] [PubMed]
- Quick, M.; Shi, L.; Zehnpfennig, B.; Weinstein, H.; Javitch, J.A. Experimental conditions can obscure the second high-affinity site in LeuT. Nat. Struct. Mol. Biol. 2012, 19, 207–211. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Elferich, J.; Gouaux, E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat. Struct. Mol. Biol. 2012, 19, 212–219. [Google Scholar] [CrossRef]
- Shi, L.; Quick, M.; Zhao, Y.; Weinstein, H.; Javitch, J.A. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 2008, 30, 667–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeVine, M.V.; Cuendet, M.A.; Khelashvili, G.; Weinstein, H. Allosteric mechanisms of molecular machines at the membrane: Transport by sodium-coupled symporters. Chem. Rev. 2016, 116, 6552–6587. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lee, A.S.; Bracher, S.; Jung, H.; Paz, A.; Kumar, J.P.; Abramson, J.; Quick, M.; Shi, L. Identification of a second substrate-binding site in solute-sodium symporters. J. Biol. Chem. 2015, 290, 127–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz, A.; Claxton, D.P.; Kumar, J.P.; Kazmier, K.; Bisignano, P.; Sharma, S.; Nolte, S.A.; Liwag, T.M.; Nayak, V.; Wright, E.M.; et al. Conformational transitions of the sodium-dependent sugar transporter, vSGLT. Proc. Natl. Acad. Sci. USA 2018, 115, E2742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisignano, P.; Ghezzi, C.; Jo, H.; Polizzi, N.F.; Althoff, T.; Kalyanaraman, C.; Friemann, R.; Jacobson, M.P.; Wright, E.M.; Grabe, M. Inhibitor binding mode and allosteric regulation of Na+-glucose symporters. Nat. Commun. 2018, 9, 5245. [Google Scholar] [CrossRef] [PubMed]
- Jardetzky, O. Simple allosteric model for membrane pumps. Nature 1966, 211, 969–970. [Google Scholar] [CrossRef] [PubMed]
- Vidaver, G.A. Inhibition of parallel flux and augmentation of counter flux shown by transport models not involving a mobile carrier. J. Theor. Biol. 1966, 10, 301–306. [Google Scholar] [CrossRef]
- Widdas, W.F. Inability of diffusion to account for placental glucose transfer in the sheep and consideration of the kinetics of a possible carrier transfer. J. Physiol. 1952, 118, 23–39. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Moraes, T.F.; Reithmeier, R.A.F. Structural biology of solute carrier (SLC) membrane transport proteins. Mol. Membr. Biol. 2017, 34, 1–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, A.; Wozniak, A.; Meyer-Lipp, K.; Nietschke, M.; Jung, H.; Fendler, K. Charge translocation during cosubstrate binding in the Na+/proline transporter of E. coli. J. Mol. Biol. 2004, 343, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Pirch, T.; Landmeier, S.; Jung, H. Transmembrane domain II of the Na+/proline transporter PutP of Escherichia coli forms part of a conformationally flexible, cytoplasmic exposed aqueous cavity within the membrane. J. Biol. Chem. 2003, 278, 42942–42949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olkhova, E.; Raba, M.; Bracher, S.; Hilger, D.; Jung, H. Homology model of the Na+/proline transporter PutP of Escherichia coli and its functional implications. J. Mol. Biol. 2011, 406, 59–74. [Google Scholar] [CrossRef]
- Quick, M.; Jung, H. Aspartate 55 in the Na+/proline permease of Escherichia coli is essential for Na+-coupled proline uptake. Biochemistry 1997, 36, 4631–4636. [Google Scholar] [CrossRef]
- Nishijyo, T.; Haas, D.; Itoh, Y. The CbrA–CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 2001, 40, 917–931. [Google Scholar] [CrossRef]
- Jung, H. Towards the molecular mechanism of Na+/solute symport in prokaryotes. Biochim. Biophys. Acta (BBA)-Bioenerg. 2001, 1505, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galperin, M. Sensory transduction in bacteria. Encycl. Microbiol. 2009, 447–463. [Google Scholar] [CrossRef]
- Tiwari, S.; Jamal, S.B.; Hassan, S.S.; Carvalho, P.V.S.D.; Almeida, S.; Barh, D.; Ghosh, P.; Silva, A.; Castro, T.L.P.; Azevedo, V. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: An Overview. Front. Microbiol. 2017, 8, 1878. [Google Scholar] [CrossRef]
- Valentini, M.; García-Mauriño, S.M.; Pérez-Martínez, I.; Santero, E.; Canosa, I.; Lapouge, K. Hierarchical management of carbon sources is regulated similarly by the CbrA/B systems in Pseudomonas aeruginosa and Pseudomonas putida. Microbiology 2014, 160, 2243–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, R.; Fonseca, P.; Rojo, F. Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol. MicroBiol. 2012, 83, 24–40. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Rainey, P.B. Genetic analysis of the histidine utilization (hut) genes in Pseudomonas fluorescens SBW25. Genetics 2007, 176, 2165–2176. [Google Scholar] [CrossRef] [Green Version]
- Monteagudo-Cascales, E.; García-Mauriño, S.M.; Santero, E.; Canosa, I. Unraveling the role of the CbrA histidine kinase in the signal transduction of the CbrAB two-component system in Pseudomonas putida. Sci. Rep. 2019, 9, 9110. [Google Scholar] [CrossRef] [Green Version]
- Wirtz, L.; Eder, M.; Schipper, K.; Rohrer, S.; Jung, H. Transport and kinase activities of CbrA of Pseudomonas putida KT2440. Sci. Rep. 2020, 10, 5400. [Google Scholar] [CrossRef]
- Yeung, A.T.Y.; Janot, L.; Pena, O.M.; Neidig, A.; Kukavica-Ibrulj, I.; Hilchie, A.; Levesque, R.C.; Overhage, J.; Hancock, R.E.W. Requirement of the Pseudomonas aeruginosa CbrA sensor kinase for full virulence in a murine acute lung infection model. Infect. Immun. 2014, 82, 1256–1267. [Google Scholar] [CrossRef] [Green Version]
- Yeung, A.T.Y.; Bains, M.; Hancock, R.E.W. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J. Bacteriol. 2011, 193, 918–931. [Google Scholar] [CrossRef] [Green Version]
- Quiroz-Rocha, E.; Bonilla-Badía, F.; García-Aguilar, V.; López-Pliego, L.; Serrano-Román, J.; Cocotl-Yañez, M.; Guzmán, J.; Ahumada-Manuel, C.L.; Muriel-Millán, L.F.; Castañeda, M.; et al. Two-component system CbrA/CbrB controls alginate production in Azotobacter vinelandii. Microbiology 2017, 163, 1105–1115. [Google Scholar] [CrossRef]
- Quiroz-Rocha, E.; Moreno, R.; Hernández-Ortíz, A.; Fragoso-Jiménez, J.C.; Muriel-Millán, L.F.; Guzmán, J.; Espín, G.; Rojo, F.; Núñez, C. Glucose uptake in Azotobacter vinelandii occurs through a GluP transporter that is under the control of the CbrA/CbrB and Hfq-Crc systems. Sci. Rep. 2017, 7, 858. [Google Scholar] [CrossRef]
- Hang, S.; Purdy, A.E.; Robins, W.P.; Wang, Z.; Mandal, M.; Chang, S.; Mekalanos, J.J.; Watnick, P.I. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 2014, 16, 592–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, K.; Rasmussen, A.; Tyler, P.; Servos, M.M.; Sylla, M.; Prado, C.; Daniele, E.; Sharp, J.S.; Purdy, A.E. Regulation of acetyl-CoA synthetase transcription by the CrbS/R two-component system is conserved in genetically diverse environmental pathogens. PLoS ONE 2017, 12, e0177825. [Google Scholar] [CrossRef] [PubMed]
- Muzhingi, I.; Prado, C.; Sylla, M.; Diehl, F.F.; Nguyen, D.K.; Servos, M.M.; Flores Ramos, S.; Purdy, A.E. Modulation of CrbS-dependent activation of the acetate switch in Vibrio cholerae. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaoui, C.; Overhage, J.; Löns, D.; Zimmermann, A.; Müsken, M.; Bielecki, P.; Pustelny, C.; Becker, T.; Nimtz, M.; Häussler, S. An orphan sensor kinase controls quinolone signal production via MexT in Pseudomonas aeruginosa. Mol. Microbiol. 2012, 83, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, E.; Lupas, A.N. Characterization of the CrbS/R two-component system in Pseudomonas fluorescens reveals a new set of genes under its control and a DNA motif required for CrbR-mediated transcriptional activation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Moya, J.; Argandoña, M.; Reina-Bueno, M.; Nieto, J.J.; Iglesias-Guerra, F.; Jebbar, M.; Vargas, C. Involvement of EupR, a response regulator of the NarL/FixJ family, in the control of the uptake of the compatible solutes ectoines by the halophilic bacterium Chromohalobacter salexigens. BMC Microbiol. 2010, 10, 256. [Google Scholar] [CrossRef] [Green Version]
- Silva-Rocha, R.; Martinez-Garcia, E.; Calles, B.; Chavarria, M.; Arce-Rodriguez, A.; de Las Heras, A.; Paez-Espino, A.D.; Durante-Rodriguez, G.; Kim, J.; Nikel, P.I.; et al. The Standard European Vector Architecture (SEVA): A coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. [Google Scholar] [CrossRef]
- Mirabella, A.; Villanueva, R.-M.Y.; Delrue, R.-M.; Uzureau, S.; Zygmunt, M.S.; Cloeckaert, A.; De Bolle, X.; Letesson, J.-J. The two-component system PrlS/PrlR of Brucella melitensis is required for persistence in mice and appears to respond to ionic strength. Microbiology 2012, 158, 2642–2651. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-X.; Gauntlett, J.C.; Oldenburg, D.G.; Cook, G.M.; Rainey, P.B. Role of the transporter-like sensor kinase CbrA in histidine uptake and signal transduction. J. Bacteriol. 2015, 197, 2867–2878. [Google Scholar] [CrossRef] [Green Version]
- Korycinski, M.; Albrecht, R.; Ursinus, A.; Hartmann, M.D.; Coles, M.; Martin, J.; Dunin-Horkawicz, S.; Lupas, A.N. STAC—A new domain associated with transmembrane solute transport and two-component signal transduction systems. J. Mol. Biol. 2015, 427, 3327–3339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, A.H.; Stock, A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001, 26, 369–376. [Google Scholar] [CrossRef]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Wolanin, P.M.; Thomason, P.A.; Stock, J.B. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 2002, 3, REVIEWS3013. [Google Scholar] [CrossRef] [Green Version]
- Möglich, A.; Ayers, R.A.; Moffat, K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 2009, 17, 1282–1294. [Google Scholar] [CrossRef] [Green Version]
- Henry, J.T.; Crosson, S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 2011, 65, 261–286. [Google Scholar] [CrossRef] [Green Version]
- Nambu, J.R.; Lewis, J.O.; Wharton, K.A.; Crews, S.T. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 1991, 67, 1157–1167. [Google Scholar] [CrossRef]
- Bisignano, P.; Lee, M.A.; George, A.; Zuckerman, D.M.; Grabe, M.; Rosenberg, J.M. A kinetic mechanism for enhanced selectivity of membrane transport. PLoS Comput. Biol. 2020, 16, e1007789. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henriquez, T.; Wirtz, L.; Su, D.; Jung, H. Prokaryotic Solute/Sodium Symporters: Versatile Functions and Mechanisms of a Transporter Family. Int. J. Mol. Sci. 2021, 22, 1880. https://doi.org/10.3390/ijms22041880
Henriquez T, Wirtz L, Su D, Jung H. Prokaryotic Solute/Sodium Symporters: Versatile Functions and Mechanisms of a Transporter Family. International Journal of Molecular Sciences. 2021; 22(4):1880. https://doi.org/10.3390/ijms22041880
Chicago/Turabian StyleHenriquez, Tania, Larissa Wirtz, Dan Su, and Heinrich Jung. 2021. "Prokaryotic Solute/Sodium Symporters: Versatile Functions and Mechanisms of a Transporter Family" International Journal of Molecular Sciences 22, no. 4: 1880. https://doi.org/10.3390/ijms22041880








