Identification of Gastritis Subtypes by Convolutional Neuronal Networks on Histological Images of Antrum and Corpus Biopsies
Abstract
:1. Introduction
2. Results
2.1. Patient Cohort, Annotation, Image Patches Extraction and Subset Analysis
2.2. CNN Training, Validation, and Model Selection
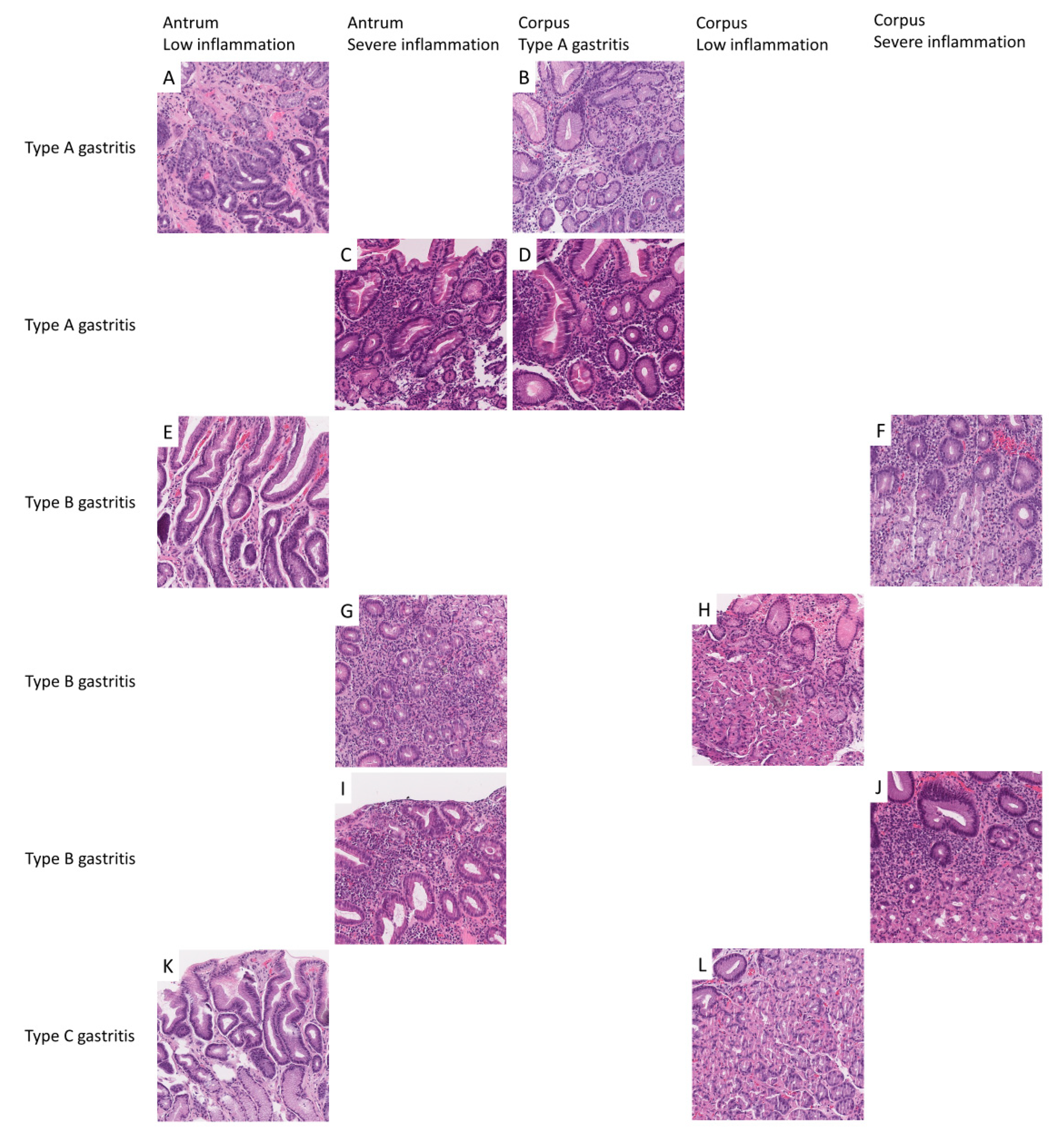
2.2.1. Antrum Classification Model
2.2.2. Corpus Classification Model
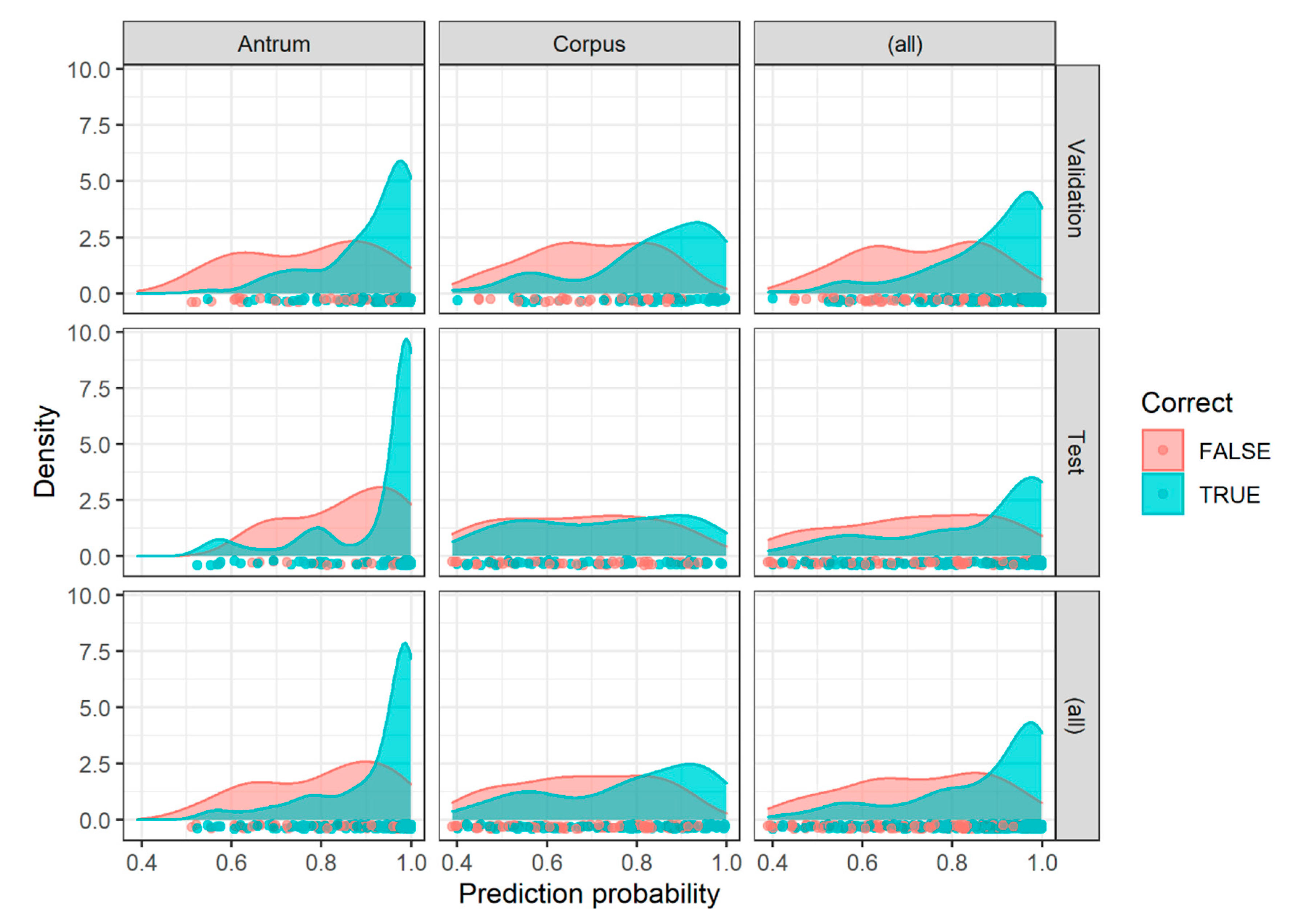
2.3. Image Patch Prediction Results for the Validation and Test Set
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Scanning of Tissue Slides
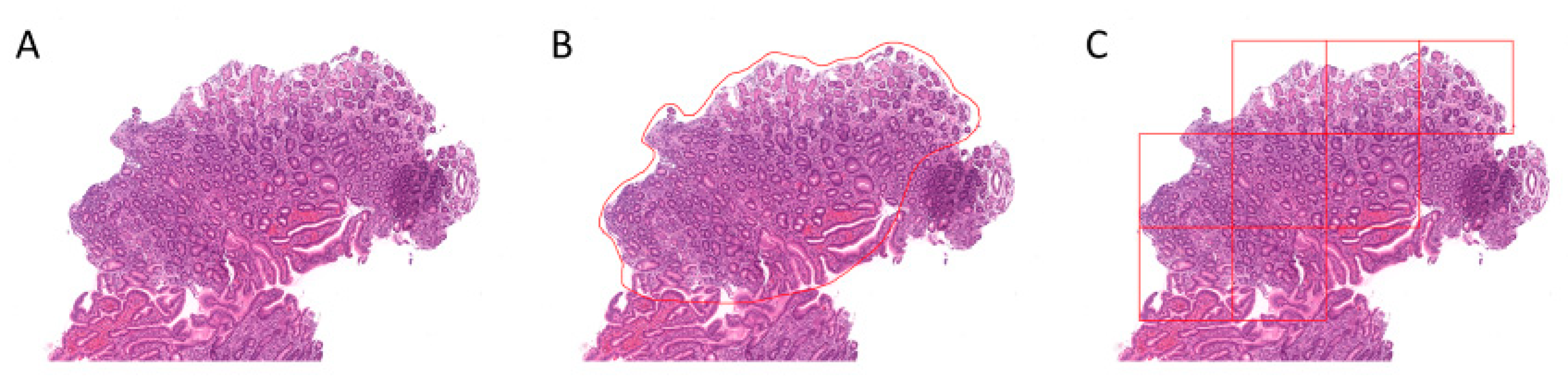
4.2. Region Annotation and Image Patch Extraction
4.3. Nomenclature of Image Patches and Encoding of Diagnosis
4.4. Hardware and Software
4.5. Analytical Subsets
4.6. Convolutional Neuronal Networks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sipponen, P.; Maaroos, H.I. Chronic gastritis. Scand. J. Gastroenterol. 2015, 50, 657–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef] [PubMed]
- Rugge, M.; Genta, R.M. Staging and grading of chronic gastritis. Hum. Pathol. 2005, 36, 228–233. [Google Scholar] [CrossRef]
- Yue, H.; Shan, L.; Bin, L. The significance of OLGA and OLGIM staging systems in the risk assessment of gastric cancer: A systematic review and meta-analysis. Gastric Cancer 2018, 21, 579–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimentel-Nunes, P.; Libanio, D.; Marcos-Pinto, R.; Areia, M.; Leja, M.; Esposito, G.; Garrido, M.; Kikuste, I.; Megraud, F.; Matysiak-Budnik, T.; et al. Management of epithelial precancerous conditions and lesions in the stomach (MAPS II): European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter and Microbiota Study Group (EHMSG), European Society of Pathology (ESP), and Sociedade Portuguesa de Endoscopia Digestiva (SPED) guideline update 2019. Endoscopy 2019, 51, 365–388. [Google Scholar] [CrossRef] [Green Version]
- Toh, B.H.; van Driel, I.R.; Gleeson, P.A. Pernicious anemia. N. Engl. J. Med. 1997, 337, 1441–1448. [Google Scholar] [CrossRef]
- Lenti, M.V.; Rugge, M.; Lahner, E.; Miceli, E.; Toh, B.H.; Genta, R.M.; De Block, C.; Hershko, C.; Di Sabatino, A. Autoimmune gastritis. Nat. Rev. Dis. Primers 2020, 6, 56. [Google Scholar] [CrossRef]
- Toh, B.H. Diagnosis and classification of autoimmune gastritis. Autoimmun. Rev. 2014, 13, 459–462. [Google Scholar] [CrossRef]
- Mahmud, N.; Stashek, K.; Katona, B.W.; Tondon, R.; Shroff, S.G.; Roses, R.; Furth, E.E.; Metz, D.C. The incidence of neoplasia in patients with autoimmune metaplastic atrophic gastritis: A renewed call for surveillance. Ann. Gastroenterol. 2019, 32, 67–72. [Google Scholar] [CrossRef]
- Warren, J.R.; Marshall, B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983, 1, 1273–1275. [Google Scholar]
- Abbasi, J. Barry Marshall, MD: H pylori 35 Years Later. JAMA 2017, 317, 1400–1402. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miehlke, S.; Hackelsberger, A.; Meining, A.; Hatz, R.; Lehn, N.; Malfertheiner, P.; Stolte, M.; Bayerdorffer, E. Severe expression of corpus gastritis is characteristic in gastric cancer patients infected with Helicobacter pylori. Br. J. Cancer 1998, 78, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Sipponen, P.; Riihela, M.; Hyvarinen, H.; Seppala, K. Chronic nonatropic (‘superficial’) gastritis increases the risk of gastric carcinoma. A case-control study. Scand. J. Gastroenterol. 1994, 29, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Leodolter, A.; Ebert, M.P.; Peitz, U.; Wolle, K.; Kahl, S.; Vieth, M.; Malfertheiner, P. Prevalence of H pylori associated "high risk gastritis" for development of gastric cancer in patients with normal endoscopic findings. World J. Gastroenterol. 2006, 12, 5509–5512. [Google Scholar] [CrossRef]
- Kriegsmann, M.; Haag, C.; Weis, C.A.; Steinbuss, G.; Warth, A.; Zgorzelski, C.; Muley, T.; Winter, H.; Eichhorn, M.E.; Eichhorn, F.; et al. Deep Learning for the Classification of Small-Cell and Non-Small-Cell Lung Cancer. Cancers 2020, 12, 1604. [Google Scholar] [CrossRef]
- Goncalves, W.G.E.; Dos Santos, M.H.P.; Lobato, F.M.F.; Ribeiro-Dos-Santos, A.; de Araujo, G.S. Deep learning in gastric tissue diseases: A systematic review. BMJ Open Gastroenterol. 2020, 7, e000371. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.R.; Hanson, J.A.; Gullapalli, R.R.; Schultz, F.A.; Sethi, A.; Clark, D.P. A Deep Learning Convolutional Neural Network Can Recognize Common Patterns of Injury in Gastric Pathology. Arch. Pathol. Lab. Med. 2020, 144, 370–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, I.J.; Kook, M.C.; Kim, Y.I.; Cho, S.J.; Lee, J.Y.; Kim, C.G.; Park, B.; Nam, B.H. Helicobacter pylori Therapy for the Prevention of Metachronous Gastric Cancer. N. Engl. J. Med. 2018, 378, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Skalski, P. Preventing Deep Neuronal Network from Overfitting. Available online: https://towardsdatascience.com/preventing-deep-neural-network-from-overfitting-953458db800a (accessed on 29 July 2020).
- Steiner, D.F.; MacDonald, R.; Liu, Y.; Truszkowski, P.; Hipp, J.D.; Gammage, C.; Thng, F.; Peng, L.; Stumpe, M.C. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am. J. Surg. Pathol. 2018, 42, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jager, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyo, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; Hulsbergen-van de Kaa, C.; Litjens, G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study. Lancet Oncol. 2020, 21, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Gertych, A.; Swiderska-Chadaj, Z.; Ma, Z.; Ing, N.; Markiewicz, T.; Cierniak, S.; Salemi, H.; Guzman, S.; Walts, A.E.; Knudsen, B.S. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 2019, 9, 1483. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mehra, R. Conventional Machine Learning and Deep Learning Approach for Multi-Classification of Breast Cancer Histopathology Images-a Comparative Insight. J. Digit. Imaging 2020, 33, 632–654. [Google Scholar] [CrossRef]
- Guan, Q.; Wang, Y.; Ping, B.; Li, D.; Du, J.; Qin, Y.; Lu, H.; Wan, X.; Xiang, J. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: A pilot study. J. Cancer 2019, 10, 4876–4882. [Google Scholar] [CrossRef]
- Uchino, E.; Suzuki, K.; Sato, N.; Kojima, R.; Tamada, Y.; Hiragi, S.; Yokoi, H.; Yugami, N.; Minamiguchi, S.; Haga, H.; et al. Classification of glomerular pathological findings using deep learning and nephrologist-AI collective intelligence approach. Int. J. Med. Inform. 2020, 141, 104231. [Google Scholar] [CrossRef]
- Diao, S.; Hou, J.; Yu, H.; Zhao, X.; Sun, Y.; Lambo, R.L.; Xie, Y.; Liu, L.; Qin, W.; Luo, W. Computer-Aided Pathologic Diagnosis of Nasopharyngeal Carcinoma Based on Deep Learning. Am. J. Pathol. 2020, 190, 1691–1700. [Google Scholar] [CrossRef]
- Wang, Y.; Guan, Q.; Lao, I.; Wang, L.; Wu, Y.; Li, D.; Ji, Q.; Wang, Y.; Zhu, Y.; Lu, H.; et al. Using deep convolutional neural networks for multi-classification of thyroid tumor by histopathology: A large-scale pilot study. Ann. Transl. Med. 2019, 7, 468. [Google Scholar] [CrossRef]
- Truong, T.-D.; Nguyen, V.-T.; Tran, M.-T. Lightweight Deep Convolutional Network for Tiny Object Recognition. In Proceedings of the 7th International Conference on Pattern Recognition Applications and Methods, Funchal, Portugal, 16–18 January 2018; Volume 1, pp. 675–682. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Karim, R. Illustrated: 10 CNN Architectures. Available online: https://towardsdatascience.com/illustrated-10-cnn-architectures-95d78ace614d#643c (accessed on 3 April 2020).
- Zhao, X.Y.; Wu, X.; Li, F.F.; Li, Y.; Huang, W.H.; Huang, K.; He, X.Y.; Fan, W.; Wu, Z.; Chen, M.L.; et al. The Application of Deep Learning in the Risk Grading of Skin Tumors for Patients Using Clinical Images. J. Med. Syst. 2019, 43, 283. [Google Scholar] [CrossRef]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhang, J. A machine learning model for detecting invasive ductal carcinoma with Google Cloud AutoML Vision. Comput. Biol. Med. 2020, 122, 103861. [Google Scholar] [CrossRef] [PubMed]
- Motta, D.; Santos, A.A.B.; Machado, B.A.S.; Ribeiro-Filho, O.G.V.; Camargo, L.O.A.; Valdenegro-Toro, M.A.; Kirchner, F.; Badaro, R. Optimization of convolutional neural network hyperparameters for automatic classification of adult mosquitoes. PLoS ONE 2020, 15, e0234959. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Jiang, Q.; Zhou, B.; Chen, D. Lung Nodule Detection using Convolutional Neural Networks with Transfer Learning on CT Images. Comb. Chem. High Throughput Screen. 2020. [Google Scholar] [CrossRef]
- Nishio, M.; Sugiyama, O.; Yakami, M.; Ueno, S.; Kubo, T.; Kuroda, T.; Togashi, K. Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS ONE 2018, 13, e0200721. [Google Scholar] [CrossRef] [Green Version]
- Ritter, C.; Wollmann, T.; Bernhard, P.; Gunkel, M.; Braun, D.M.; Lee, J.Y.; Meiners, J.; Simon, R.; Sauter, G.; Erfle, H.; et al. Hyperparameter optimization for image analysis: Application to prostate tissue images and live cell data of virus-infected cells. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1847–1857. [Google Scholar] [CrossRef]
- Liu, Y.; Kohlberger, T.; Norouzi, M.; Dahl, G.E.; Smith, J.L.; Mohtashamian, A.; Olson, N.; Peng, L.H.; Hipp, J.D.; Stumpe, M.C. Artificial Intelligence-Based Breast Cancer Nodal Metastasis Detection: Insights Into the Black Box for Pathologists. Arch. Pathol. Lab. Med. 2019, 143, 859–868. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Liu, H.; Ni, H.; Wang, X.; Su, M.; Guo, W.; Wang, K.; Jiang, T.; Qian, Y. A Fast and Refined Cancer Regions Segmentation Framework in Whole-slide Breast Pathological Images. Sci. Rep. 2019, 9, 882. [Google Scholar] [CrossRef] [Green Version]
- Bejnordi, B.E.; Veta, M.; van Diest, P.J.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Bandi, P.; Geessink, O.; Manson, Q.; Van Dijk, M.; Balkenhol, M.; Hermsen, M.; Ehteshami Bejnordi, B.; Lee, B.; Paeng, K.; Zhong, A.; et al. From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge. IEEE Trans. Med. Imaging 2019, 38, 550–560. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Li, F.; Yan, H.; Wang, K.; Ma, Y.; Shen, L.; Xu, M.; Alzheimer’s Disease Neuroimaging Initiative. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. Neuroimage 2020, 208, 116459. [Google Scholar] [CrossRef] [PubMed]
- Metter, D.M.; Colgan, T.J.; Leung, S.T.; Timmons, C.F.; Park, J.Y. Trends in the US and Canadian Pathologist Workforces From 2007 to 2017. JAMA Netw. Open 2019, 2, e194337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markl, B.; Fuzesi, L.; Huss, R.; Bauer, S.; Schaller, T. Number of pathologists in Germany: Comparison with European countries, USA, and Canada. Virchows Arch 2020. [Google Scholar] [CrossRef]
- Provenzano, E.; Driskell, O.J.; O’Connor, D.J.; Rodriguez-Justo, M.; McDermott, J.; Wong, N.; Kendall, T.; Zhang, Y.Z.; Robinson, M.; Kurian, K.M.; et al. The important role of the histopathologist in clinical trials: Challenges and approaches to tackle them. Histopathology 2020, 76, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Baudin, K.; Gustafsson, C.; Frennert, S. Views of Swedish Elder Care Personnel on Ongoing Digital Transformation: Cross-Sectional Study. J. Med. Internet Res. 2020, 22, e15450. [Google Scholar] [CrossRef]
- Hanna, M.G.; Reuter, V.E.; Hameed, M.R.; Tan, L.K.; Chiang, S.; Sigel, C.; Hollmann, T.; Giri, D.; Samboy, J.; Moradel, C.; et al. Whole slide imaging equivalency and efficiency study: Experience at a large academic center. Mod. Pathol. 2019, 32, 916–928. [Google Scholar] [CrossRef]
- Bankhead, P.; Fernandez, J.A.; McArt, D.G.; Boyle, D.P.; Li, G.; Loughrey, M.B.; Irwin, G.W.; Harkin, D.P.; James, J.A.; McQuaid, S.; et al. Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer. Lab. Investig. 2018, 98, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 July 2015; pp. 2818–2826. [Google Scholar]
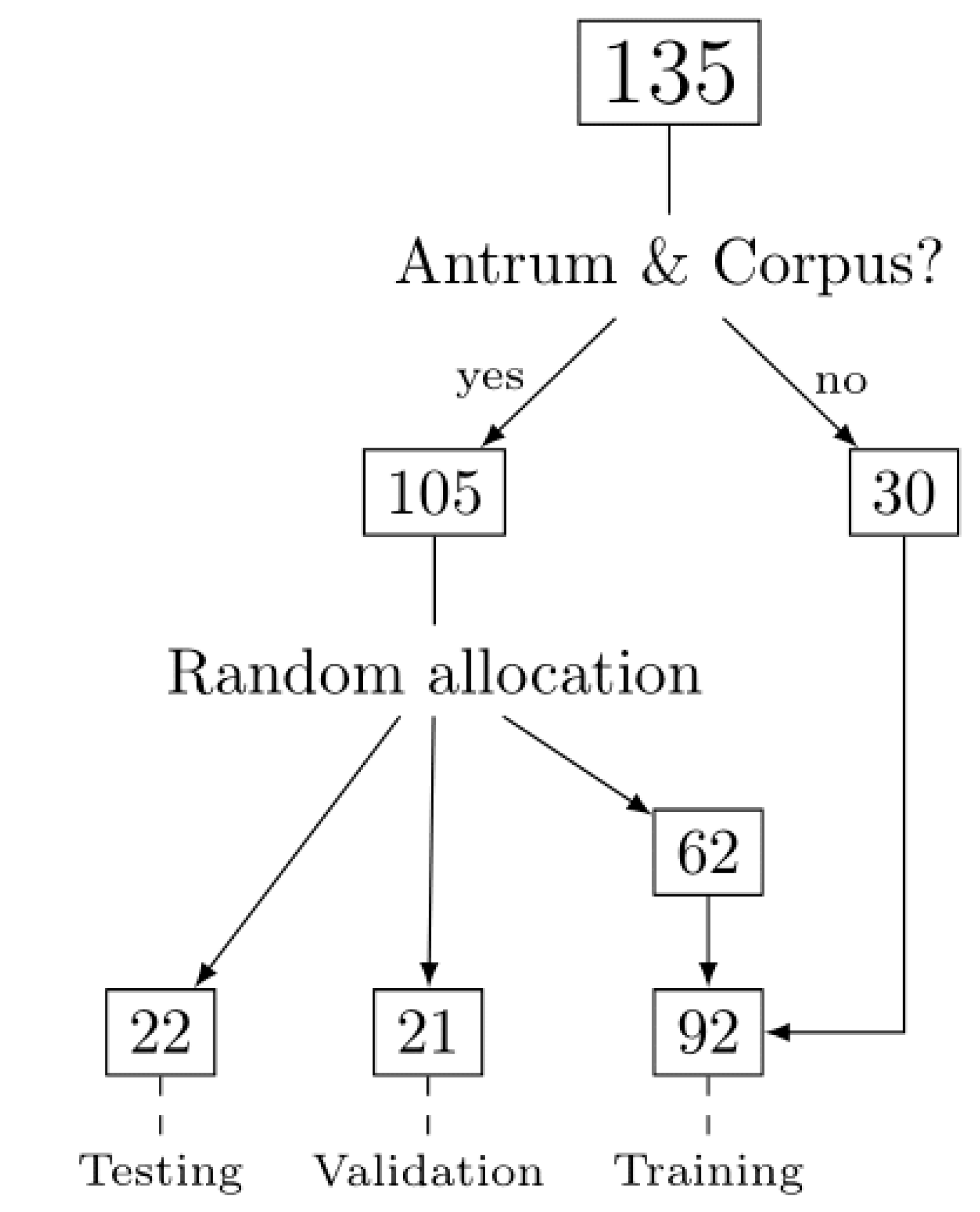
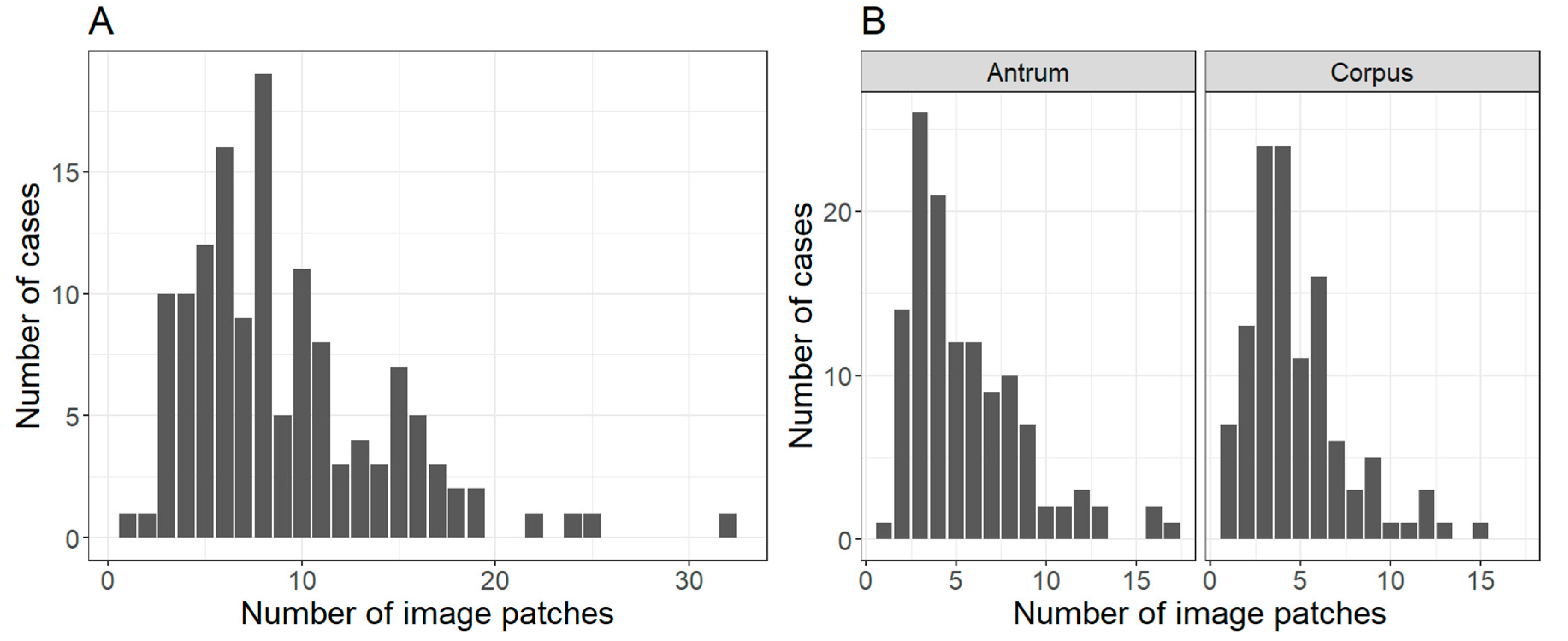
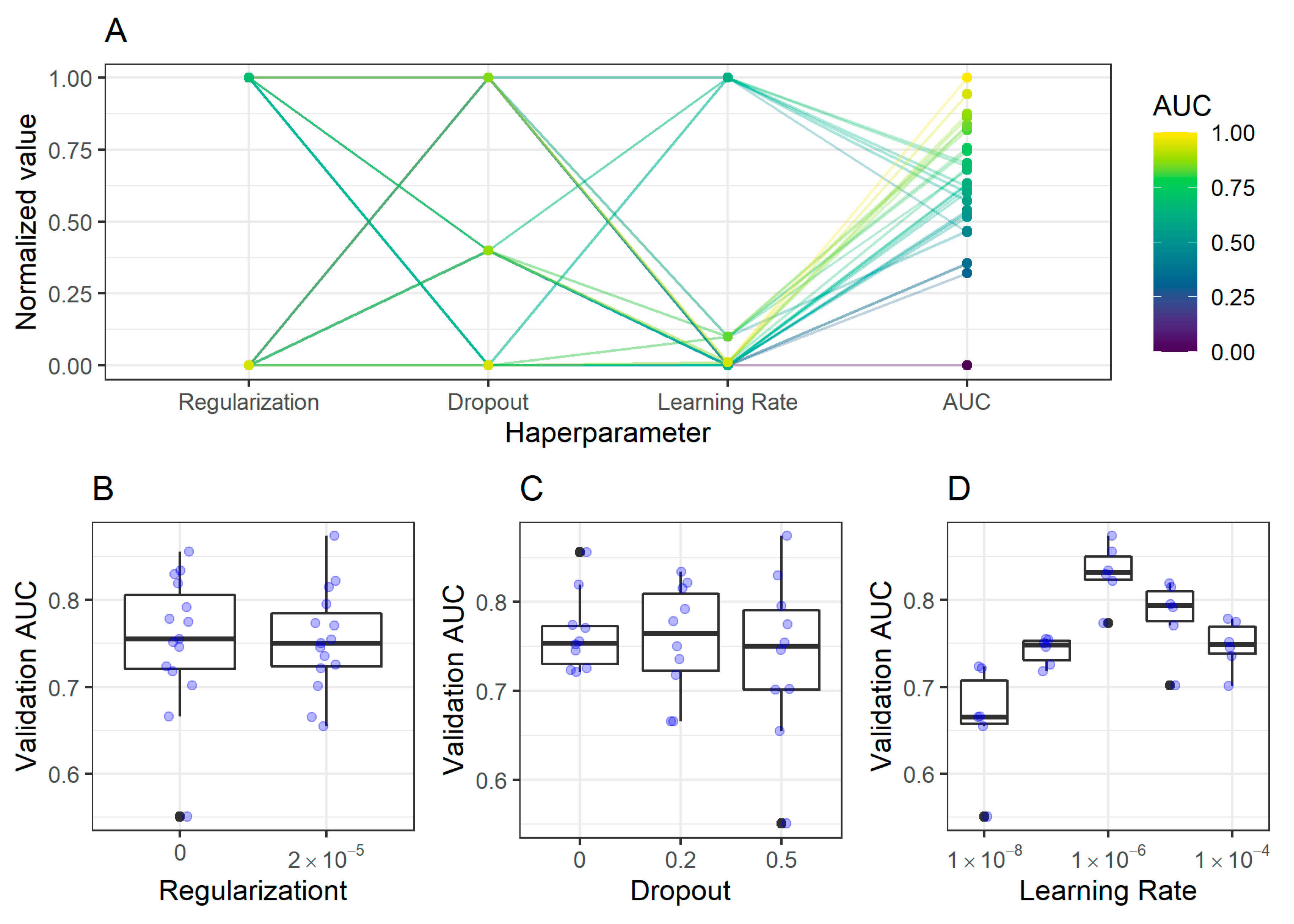
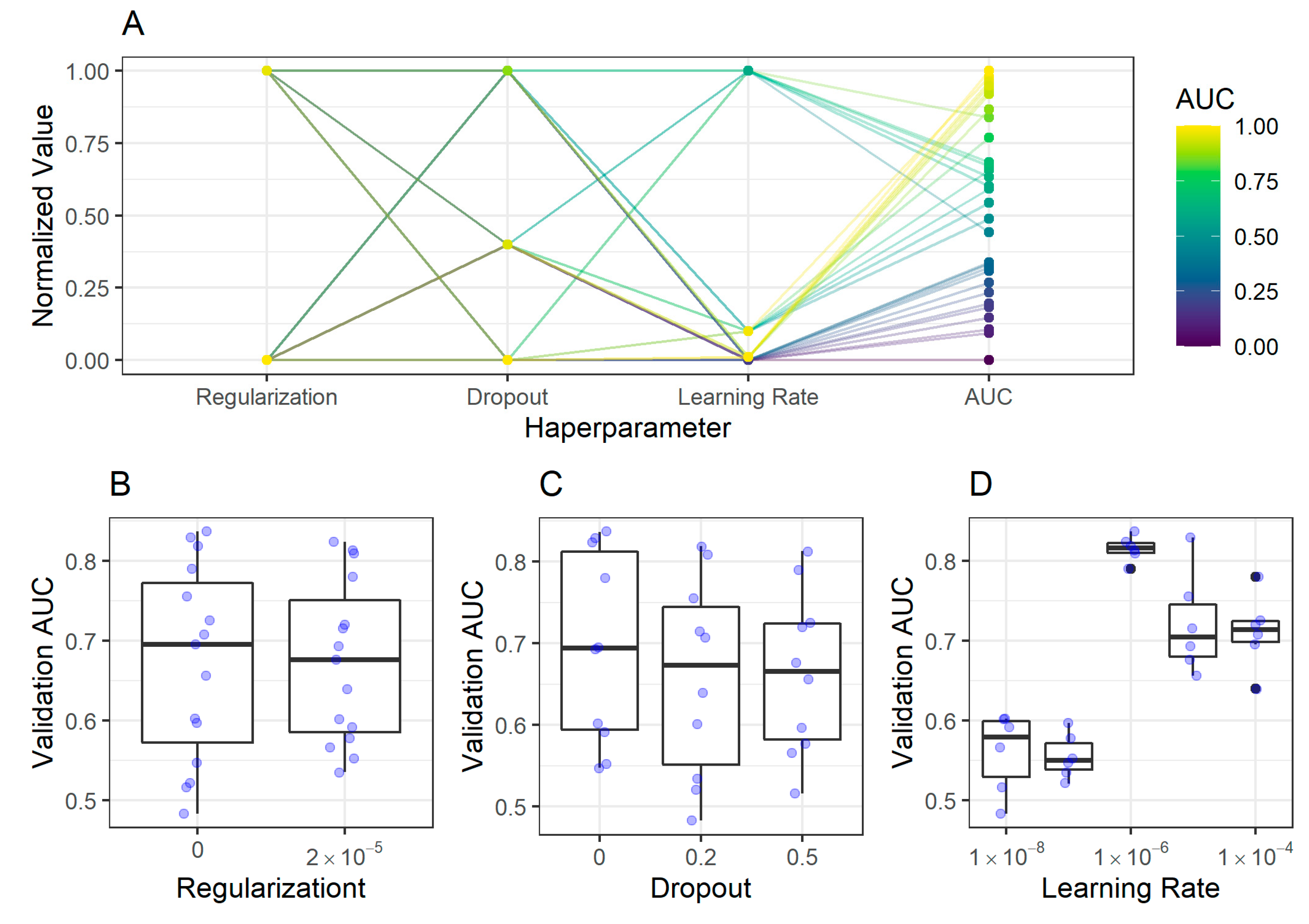
| Set/Region | Antrum Image Patches, n (%) | Corpus Image Patches, n (%) |
|---|---|---|
| Overall (patients n = 135, image patches n = 1230) | 682 | 548 |
| Training (patients n = 62 with both) | (+19 patients with only antrum) | (+11 patients with only corpus) |
| LI | 265 (57) | 133 (37) |
| SI | 197 (43) | 108 (30) |
| A gastritis | / | 122 (34) |
| Validation (patients n = 21) | ||
| LI | 64 (57) | 21 (25) |
| SI | 48 (43) | 29 (35) |
| A gastritis | / | 34 (41) |
| Test (patients n = 22) | ||
| LI | 84 (78) | 38 (38) |
| SI | 24 (22) | 14 (14) |
| A gastritis | / | 49 (49) |
| Antrum Classifier | Corpus Classifier | ||||
|---|---|---|---|---|---|
| Confusion Matrix (by Image Patches) | LI Predicted | SI Predicted | LI Predicted | SI Predicted | A Gastritis Predicted |
| LI true | 54 | 10 | 27 | 3 | 4 |
| SI true | 16 | 32 | 0 | 21 | 0 |
| A gastritis true | / | / | 5 | 16 | 8 |
| Statistics | |||||
| Accuracy (95% CI) | 0.77 (0.68–0.84) | 0.67 (0.55–0.77) | |||
| Classes | LI vs. SI | LI vs. other | SI vs. other | A gastritis vs. other | |
| Sensitivity | 0.77 | 0.53 | 0.67 | 0.84 | |
| Specificity | 0.76 | 1.00 | 0.71 | 0.87 | |
| Positive predictive value | 0.84 | 1.00 | 0.28 | 0.79 | |
| Negative predictive value | 0.67 | 0.70 | 0.93 | 0.90 | |
| Antrum Classifier | Corpus Classifier | ||||
|---|---|---|---|---|---|
| Confusion Matrix (by Image Patches) | LI Predicted | SI Predicted | LI Predicted | SI Predicted | A Gastritis Predicted |
| LI true | 76 | 8 | 27 | 20 | 2 |
| SI true | 8 | 16 | 6 | 26 | 6 |
| A gastritis true | / | / | 7 | 3 | 4 |
| Statistics | |||||
| Accuracy (95% CI) | 0.85 (0.77–0.91) | 0.56 (0.46–0.66) | |||
| Classes | LI vs. SI | LI vs. other | SI vs. other | A gastritis vs. other | |
| Sensitivity | 0.90 | 0.53 | 0.33 | 0.68 | |
| Specificity | 0.67 | 0.77 | 0.89 | 0.64 | |
| Positive predictive value | 0.90 | 0.68 | 0.29 | 0.55 | |
| Negative predictive value | 0.67 | 0.63 | 0.91 | 0.75 | |
| Gastritis Classifier | |||
|---|---|---|---|
| Confusion Matrix (by Patient, n = 17) | A Gastritis Predicted | B Gastritis Predicted | C Gastritis Predicted |
| A gastritis true | 7 | 0 | 1 |
| B gastritis true | 1 | 3 | 0 |
| C gastritis true | 0 | 0 | 5 |
| Statistics | |||
| Accuracy (95% CI) | 0.84 (0.64–0.96) | ||
| Classes | A vs. other | B vs. other | C vs. other |
| Sensitivity | 0.88 | 1.00 | 0.83 |
| Specificity | 0.89 | 0.93 | 1.00 |
| Positive predictive value | 0.88 | 0.75 | 1.00 |
| Negative predictive value | 0.89 | 1.00 | 0.92 |
| Overall Gastritis Diagnosis | Antrum Finding/Classifier Result | Corpus Finding/Classifier Result |
|---|---|---|
| A | SI | A gastritis |
| B | SI | SI |
| B | SI | LI |
| A | LI | A gastritis |
| B | LI | SI |
| C | LI | LI |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steinbuss, G.; Kriegsmann, K.; Kriegsmann, M. Identification of Gastritis Subtypes by Convolutional Neuronal Networks on Histological Images of Antrum and Corpus Biopsies. Int. J. Mol. Sci. 2020, 21, 6652. https://doi.org/10.3390/ijms21186652
Steinbuss G, Kriegsmann K, Kriegsmann M. Identification of Gastritis Subtypes by Convolutional Neuronal Networks on Histological Images of Antrum and Corpus Biopsies. International Journal of Molecular Sciences. 2020; 21(18):6652. https://doi.org/10.3390/ijms21186652
Chicago/Turabian StyleSteinbuss, Georg, Katharina Kriegsmann, and Mark Kriegsmann. 2020. "Identification of Gastritis Subtypes by Convolutional Neuronal Networks on Histological Images of Antrum and Corpus Biopsies" International Journal of Molecular Sciences 21, no. 18: 6652. https://doi.org/10.3390/ijms21186652





