1. Introduction
Gramicidin is an antimicrobial peptide that is a byproduct of
Bacillus brevis during sporulation [
1]. Gramicidin contains alternating L- and D-amino acids in its primary sequence and has three isoforms—gramicidin A (gA), gramicidin B (gB) and gramicidin C (gC)—in which residue 11 is Trp for gA, Phe for gB and Tyr for gC [
2,
3]. Gramicidin adopts a number of different conformations in different environmental conditions [
4,
5,
6,
7]. In organic solvents such as methanol, gramicidin forms a β-sheet-like double-strand helical structure, in which two monomers are interwound similarly as DNA double helices [
4,
6,
8]. This double-strand helical dimer has four distinct conformations that can be either parallel or antiparallel, left- or right-handed double-stranded helices [
6,
7,
8,
9]. These double-helical conformers are interconvertible and have distinct circular dichroism (CD) spectra [
6,
8]. On the other hand, in polar solvents such as trifluoroethanol (TFE), gramicidin forms a single-stranded and right-handed helical monomer [
4,
10,
11,
12]. Two of the helical monomers can form a head-to-head channel dimer in a lipid environment [
4,
11,
12]. This channel can specifically translocate small monovalent cations such as H
+, Na
+ and K
+ across membranes [
4,
12,
13,
14].
Gramicidin’s double-stranded helical dimer can interact with monovalent or divalent cations [
15,
16,
17,
18,
19,
20,
21]. The complex of gramicidin A/monovalent cation exists in a left-handed antiparallel double-helical conformation in methanol [
15,
16] and has a distinct CD spectrum compared to that of the ion-free form [
17]. The binding mechanism is a cooperative mode, and the binding affinities are roughly related to the size of the monovalent cations. The order of the binding affinities is Cs > Rb >> K > Li [
17]. Previous studies demonstrated that divalent cations could cause the gramicidin A mixture in methanol to form one specific conformation [
18,
19]. The CD spectrum of the gramicidin A/divalent cation complex is very different from that of the gramicidin A/monovalent cation complex in methanol [
19]. The structure of the gramicidin A/Ca complex determined by the solution NMR technique appears to be a left-handed parallel double helical conformation with Ca bound at the N-terminal mouth of the double helical dimer [
20].
Conversion between conformers has been studied in lipid environments [
21,
22,
23,
24,
25]. In membrane lipid environments, the rate of conversion from the double-stranded helical dimer into the channel dimer is dependent on environmental factors, such as temperature and ionic strength [
21,
22,
23]. Both CD and fluorescence spectroscopies have been used to examine the rates of conversion of the double-stranded helical dimer to the single-stranded helical dimer in synthetic lipid membranes [
21,
23]. However, the conversion between the double-strand helical conformers in organic solvents and different ions has not been studied.
Gramicidin A is a short peptide antibiotic effective against bacteria and fungi [
26]. The antimicrobial activity of gramicidin A has been associated with the disruption of membrane lipids [
27]. We have previously demonstrated that the antimicrobial activity of gramicidin A may also be associated with the formation of free radicals via the disruption of NADH/NAD
+ synthesis [
28]. Recently, the antimicrobial function of gramicidin A was also linked with the formation of ion channels [
29]. However, the relationship between antimicrobial activity and molecular state is not entirely clear.
In the present study, we revisited and characterized the conformational and molecular states of gA in the presence of different calcium and halide ions using circular dichroism (CD), nuclear magnetic resonance (NMR) and state-of-art mass spectroscopy, and found that gramicidin A exists in two molecular states in the presence of calcium halides. In State 1, the main effect of calcium halides on gramicidin A is to convert the conformers into one single conformation. In State 2, calcium halides can induce a dissociation of the gramicidin A dimer into monomers and the denaturation of the gramicidin A monomer. A new finding revealed that the different halide anions may play a role in the molecular state and conformation of gramicidin A. The molecular state and conformation induced by calcium halide can further influence the antimicrobial activity of gramicidin A.
3. Discussion
The effect of calcium cations on gramicidin A’s conformation was previously studied using infrared (IR) and CD spectroscopic techniques [
18,
19,
20,
33]. However, the detailed molecular and conformational states of gramicidin A under the effects of calcium cations and halide anions required further verification. In the present study, we applied state-of-the-art mass, NMR and fluorescence spectroscopy to clarify the detailed molecular and structural states of gramicidin A in the presence of calcium cations and halide anions. We used the obtained structural and molecular information to examine the effect of calcium halides on the antimicrobial activity of gramicidin A.
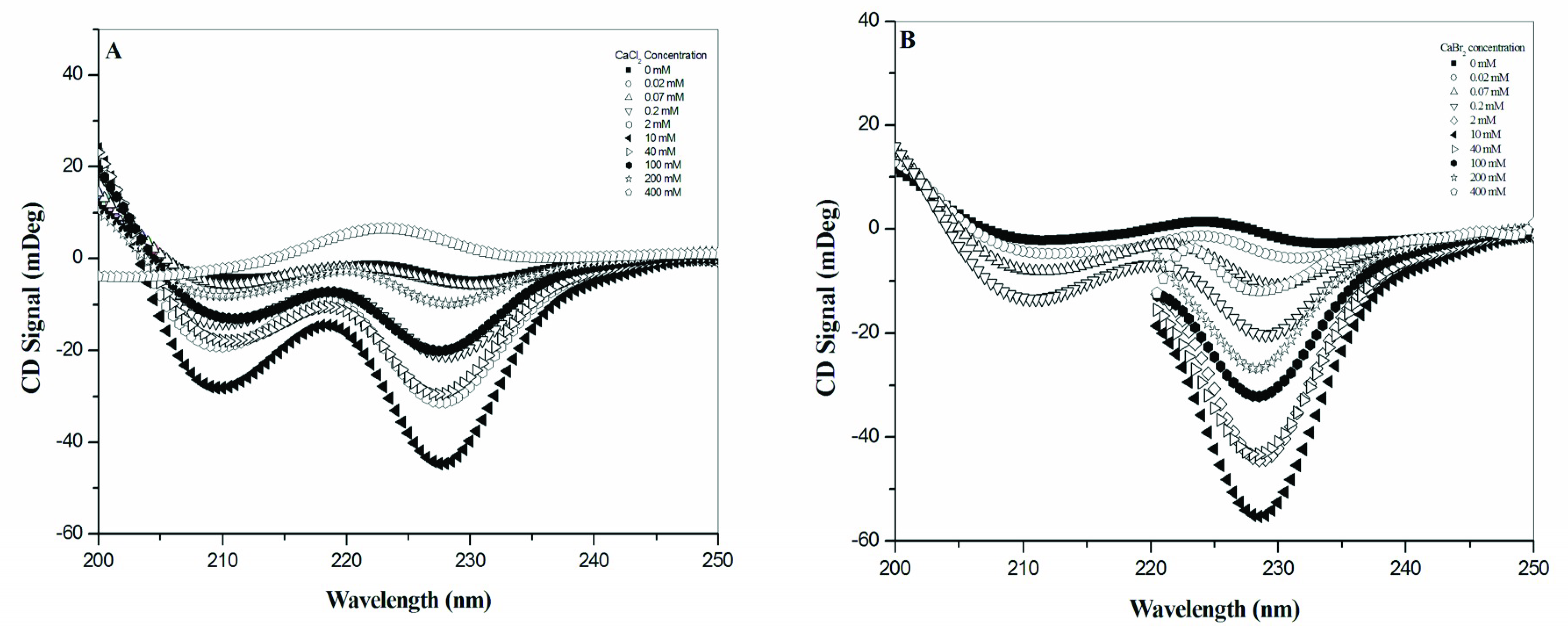
Our results obtained from CD and NMR spectroscopies are similar to those from the studies in CaCl
2 by Wallace and colleagues [
18,
19]. According to the combination of CD and NMR spectra, the effect of calcium halides on the conformation of gramicidin A reached a turning point at the concentration of 10 mM for CaCl
2, CaBr
2 and CaI
2, indicating that gramicidin A may exist in two molecular states, State 1 at salt concentrations ≤ 10 mM and State 2 at salt concentration > 10 mM. The observations obtained with CD and NMR are consistent with the fluorescence spectroscopic studies. In the fluorescence spectra, the intensity increased with an increase in the salt concentration, demonstrated a redshift in wavelength at concentrations ≤ 10 mM, and decreased with an increase in concentration with a blueshift in wavelength at concentrations > 10 mM. These results reinforce that there are two states for gramicidin A upon the titration of calcium halides.
From the CD and NMR spectra, we demonstrated that the main effect of calcium salt is to drive a mixture of conformers into a single species in State 1. This is consistent with the previous study by Chen and Wallace [
19]. In addition, we further found that this effect on gramicidin A conformation was dependent on the calcium concentration and independent of the types of halide anions. The progress of the NMR spectral change vs. concentration was similar for all different halide anions, indicating that the conformational change of gramicidin A is only induced by Ca
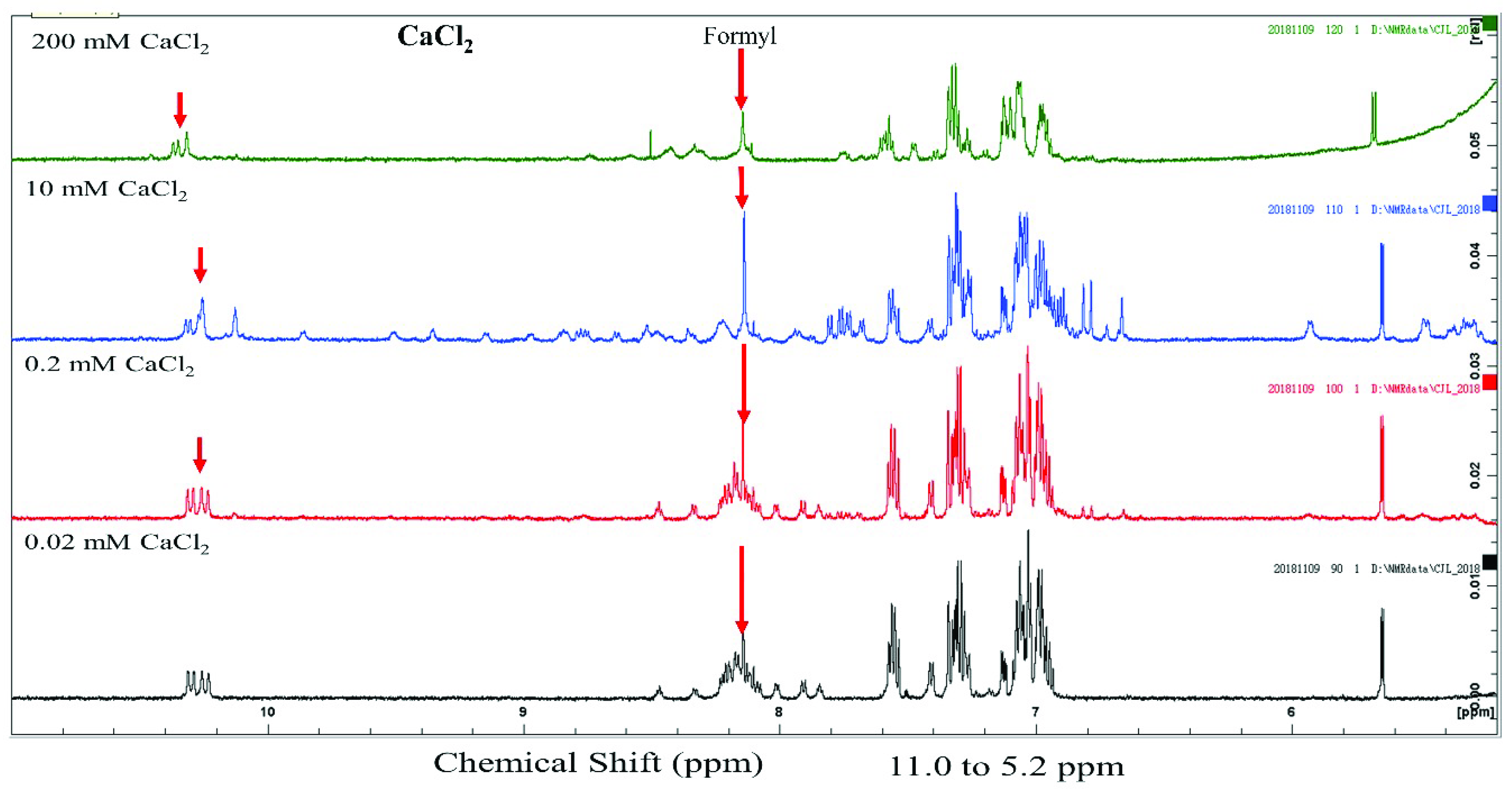
2+ cations. The 1D NMR spectra of gramicidin A at 10 mM showed a similar resonance pattern at the amide proton region for CaCl
2, CaBr
2 and CaI
2. These NMR spectra are similar to those in a previous NMR study [
20], suggesting that the conformation of gramicidin A may form a left-handed parallel double helix under this condition.
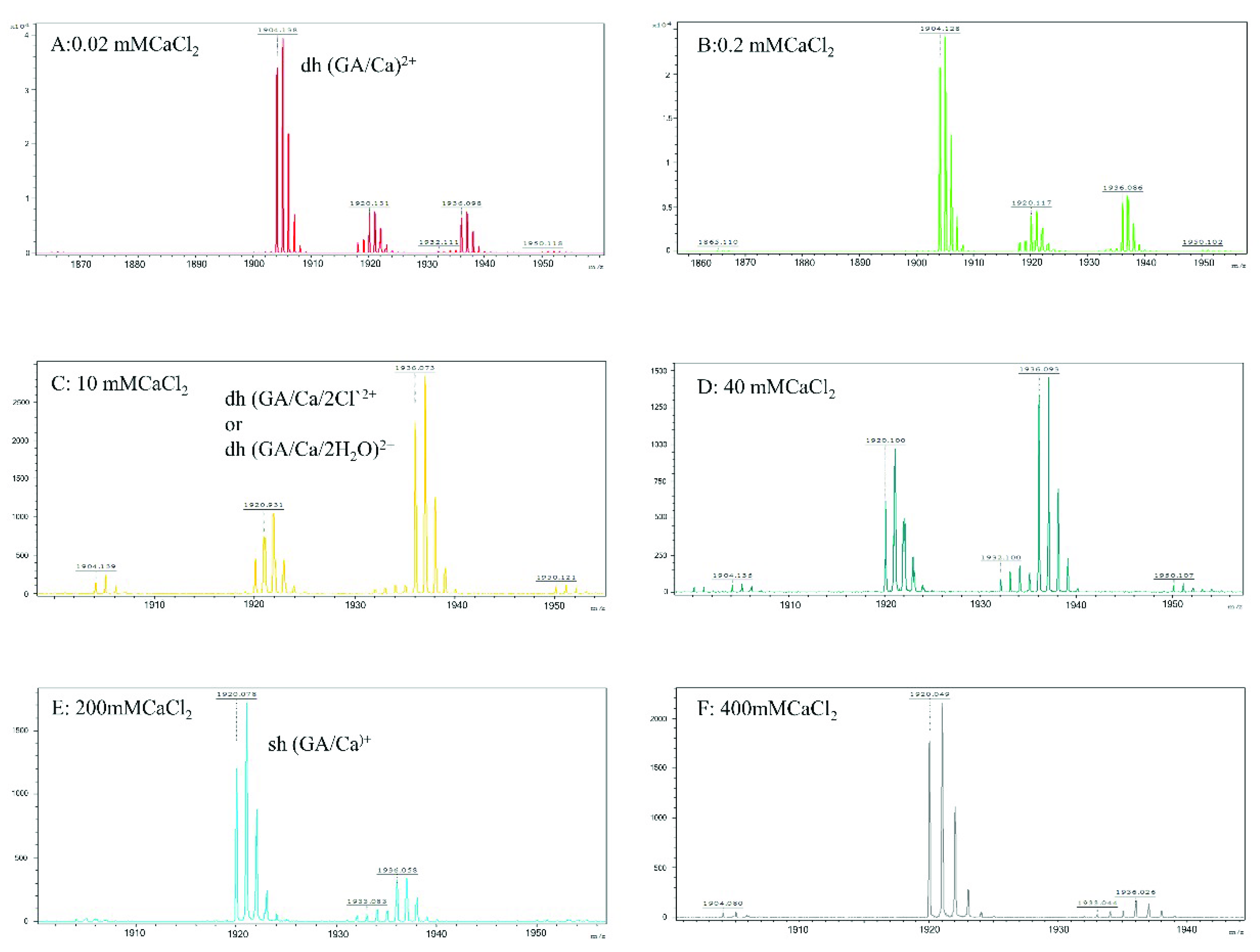
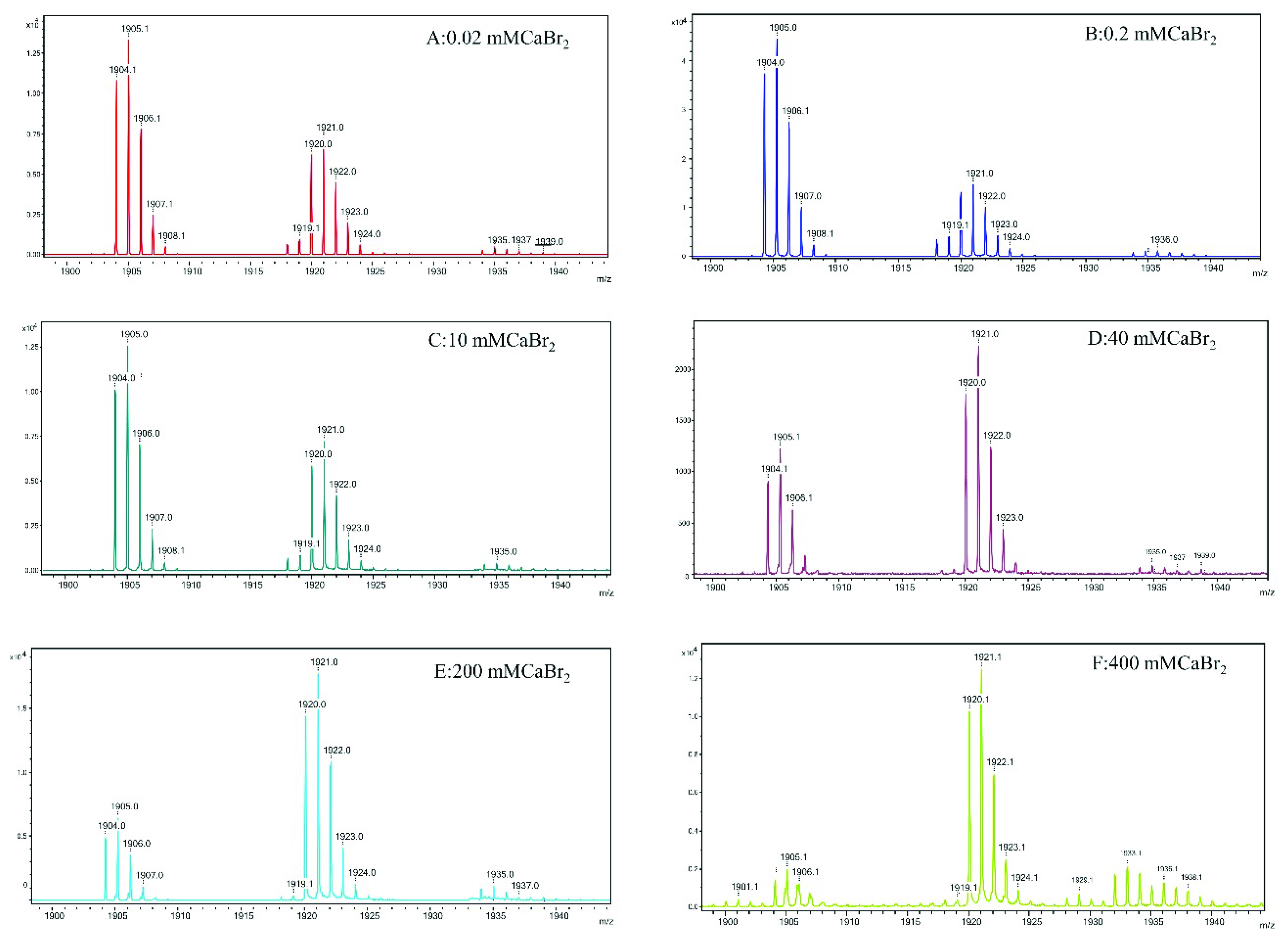
The most interesting findings in the present study are from the mass spectroscopy analyses. The analyses of the mass spectra for all calcium halides suggest that the molecular state of gramicidin A is a mixture of the dimer and monomer in State 1. The major form of gramicidin A exists in a dimeric form for both CaCl2 and CaBr2 but not for CaI2. The dimer content decreased with an increase in the salt concentration, indicating that the addition of the calcium salt drove a dissociation of gramicidin A dimers into monomers. The dissociation of the gramicidin A dimer was more effective with CaI2 or CaBr2 than with CaCl2, suggesting that the dissociation is highly dependent on the type of halide anion. The order by dissociation ability is I− > Br− > Cl−.
The reason for this phenomenon is not apparent. A possible reason may be the ionic radii of the halide anion. A peak at
m/
z 1936 assigned as (gA dimer/Ca
++2Cl)
2+ only appeared in the mass spectrum of CaCl
2 but not in the cases of CaBr
2 and CaI
2. A similar observation was reported by Zhou and colleagues [
34]. These results suggest that the gramicidin A dimer may interact only with Cl
−, possibly inside the pore, but not with Br
− or I
−. As the ionic radii of Br
− and I
− are larger than that of Cl
−, both the Br
− and I
− anions are too large to accommodate them inside the dimer pore of gramicidin A. The interaction of Cl
− ions with gramicidin A may stabilize the dimer and reduce the dimer-to-monomer conversion rate. Therefore, the molar ratio of dimer/monomer in CaCl
2 is much higher than the molar ratios in CaBr
2 and CaI
2. However, the exact reason needs to be further verified with a detailed structure.
In State 2, our CD spectra are also similar to those in a study by Chen and Wallace [
19]. The two negative CD peaks gradually converted into a single positive peak in CaCl
2, indicating that gramicidin A may adopt a different conformation. Chen and Wallace proposed that gramicidin A may be a mixture of unstructured and structured monomers in this condition. Analyses of the mass spectra indicate that the molar ratios of dimer/monomer were all less than 1 for most calcium halides except for 40 mM CaCl
2. Our results support the augment that the molecular state of gramicidin A should be a monomer in State 2. Similarly, the formation of monomers is also anion-type dependent.
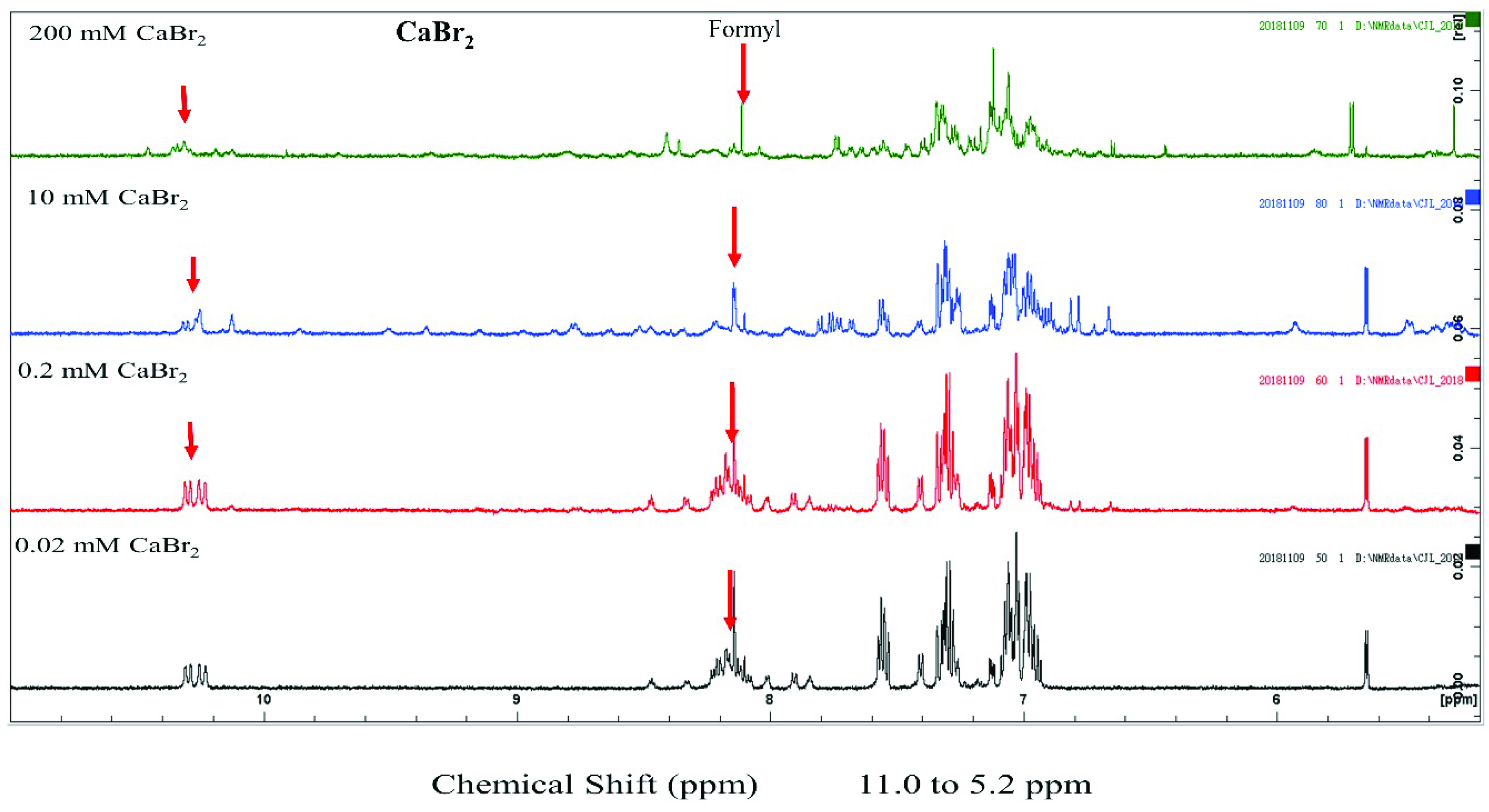
We examined the structural state of gramicidin A using NMR spectroscopy. The results showed that the effect of halide anions on the gramicidin A’s conformation was very different. Gramicidin A is a mixture of unstructured and structured monomers in the presence of 200 mM CaCl
2. This is consistent with a previous study by Chen and Wallace [
19]. The conformation of gramicidin A in 200 mM CaBr
2 was more unstructured compared to that in 200 mM CaCl
2. The unstructured effect was even more profound with the addition of 200 mM CaI
2, indicating that Br
− and I
− anions may induce denaturation in gramicidin A. The denaturing effect of halide anions on gramicidin A is well correlated with the trend of the dimer-to-monomer dissociation of gramicidin A: I
− > Br
− > Cl
−.
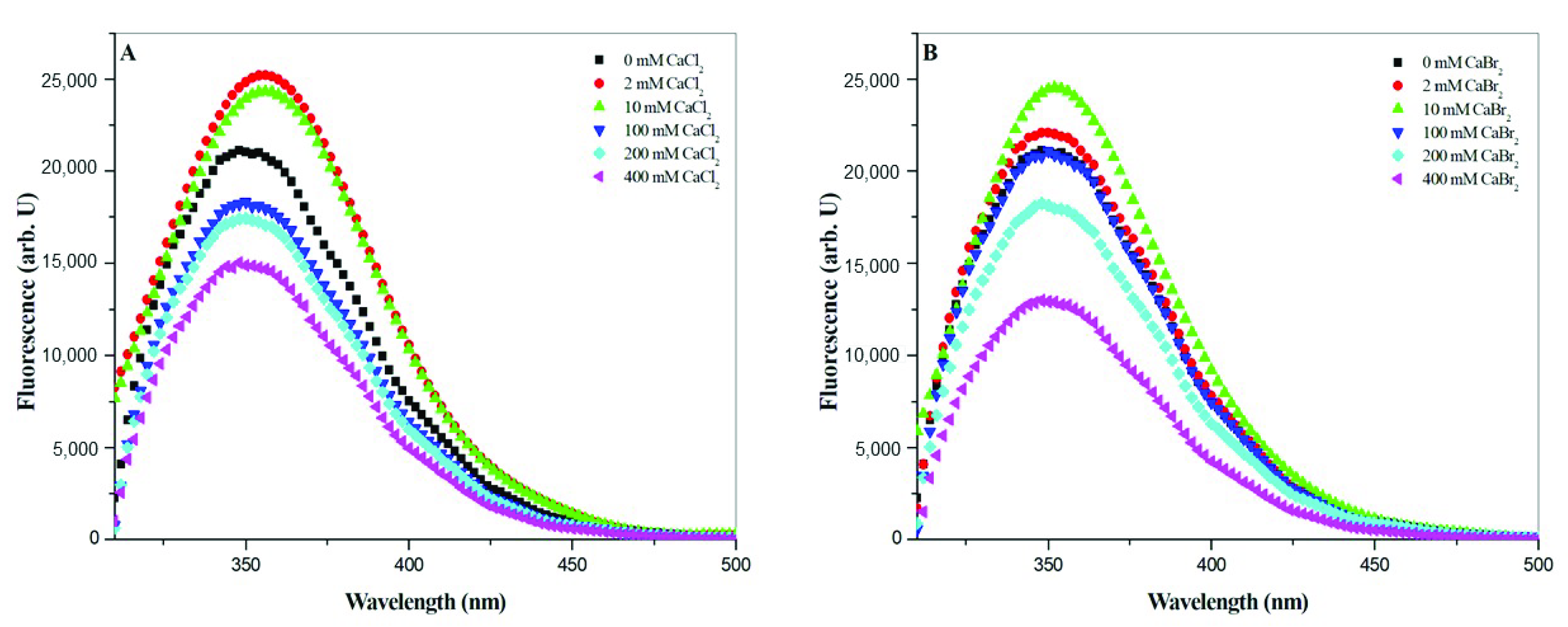
The denaturation of gramicidin A induced by halide was further confirmed by the fluorescence spectroscopy. During protein unfolding, the fluorescence spectrum usually undergoes a blueshift in the maximum wavelength and a decrease in the intensity [
31,
32,
35]. In State 2, the fluorescence intensity decreased with an increase in the salt concentration, and the maximum wavelength underwent a blueshift with an increase in the salt concentration., suggesting that the gramicidin A monomer may be denatured by halide anions. Taken together, these results suggest that the halide anions, particularly Br
− and I
−, may play a role in the denaturation of gramicidin A. The denaturation ability of halide anions, particularly Br
− and I
−, could be taken to account for the nature of the dissociation of gramicidin A.
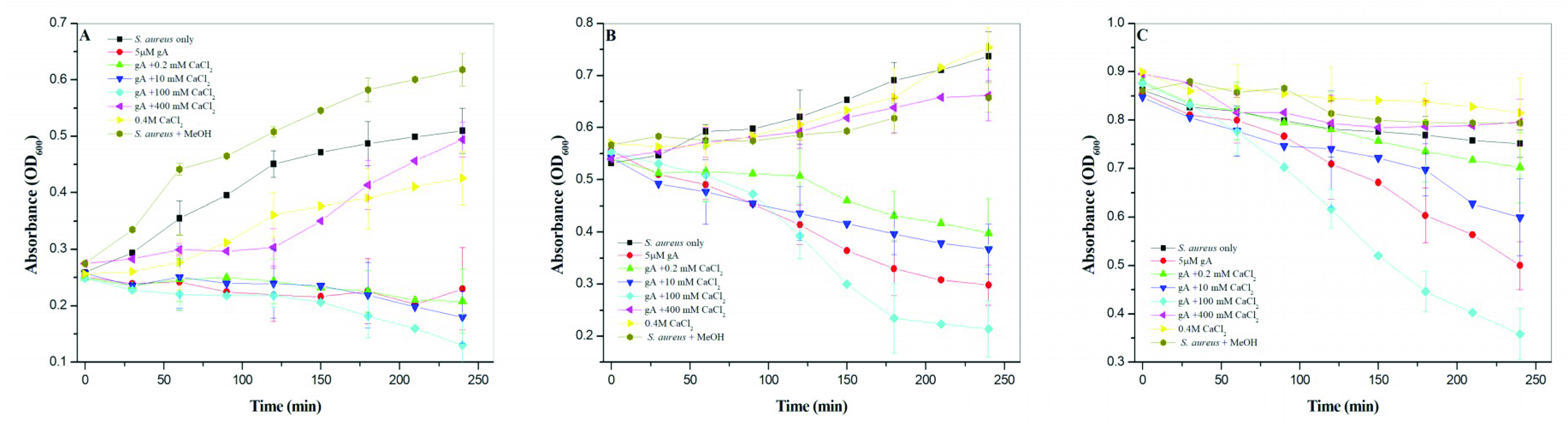
The antimicrobial activity of gramicidin A increased with an increase in the calcium concentration but not for 400 mM CaCl2. At 400 mM CaCl2, the antimicrobial activity of gramicidin A was significantly inhibited. The antimicrobial activity of gramicidin A is in the order 100 mM CaCl2 >> 0 mM CaCl2 > 10 mM CaCl2 ≈ 0.2 mM CaCl2 >> 400 mM CaCl2. The molecular states of gramicidin A/CaCl2 complexes in methanol may be changed when they are added to culture medium, as the addition of the gramicidin A in methanol into culture medium may drive the conversion of dimers into monomers. Therefore, the antimicrobial activity may have been overestimated in the present study. However, this does not affect the present conclusion.
The results obtained for the antimicrobial activity vs. CaCl
2 concentration suggest that the monomeric gramicidin A had the most effective inhibitory ability against bacterial growth. A previous study by Jadhay et al. suggested that gramicidin A in channel form mediated the most effective antimicrobial activity against Gram-positive bacteria [
29]. The formation of gramicidin channels in membranes is highly dependent on the environment [
36,
37]. In 100 mM CaCl
2, the main form of gramicidin A is monomeric. Gramicidin A in the presence of 100 mM CaCl
2 can readily form ion channels in bacterial cell walls and, hence, shows the most effective antimicrobial activity. On the other hand, the major form of gramicidin A is dimeric in 0, 0.2 and 10 mM CaCl
2. When bacteria are treated with gramicidin A in these conditions, it takes time to convert the dimers into monomers. Therefore, gramicidin A in these conditions was less effective in terms of antimicrobial activity when compared to the case at 100 mM CaCl
2.
The case of gramicidin A in 400 mM CaCl
2 was different from that at 100 mM CaCl
2. Gramicidin A may exist largely in the denatured state in 400 mM CaCl
2. This may cause the loss of the antimicrobial function. However, we could not exclude the possibility that the decrease in antimicrobial activity may have been due to the blocking of the gramicidin A channel by the high concentration of calcium cations, as calcium cations have been shown to interact with gramicidin A channels at the mouth sites and block the conduction of monovalent cations across the cell membrane [
22,
33,
38].
In conclusion, we demonstrate that the molecular state of gramicidin A is highly influenced by the concentration of calcium salt and the types of halide anion. Gramicidin A exists in two molecular states. In State 1 (concentrations ≤ 10 mM), the majority of gramicidin A forms a dimer. The addition of calcium salt converts the four conformers into a single species that forms a left-handed parallel double helical structure. In State 2, the addition of calcium salt induces a dissociation of dimers into monomers and a further denaturation of the structured monomers into the unstructured monomers. The abilities of dissociation and denaturation are highly dependent on the type of halide anion. Regarding antimicrobial activity, gramicidin A in the structured monomeric state showed the most effective antimicrobial activity. As the molecular state of gramicidin A is highly dependent on the dissociation ability of the halide anions, the dissociation rate can determine the molecular state and antimicrobial activity. Taken together, our study suggests that the molecular state may play a vital role in the antimicrobial activity.
4. Materials and Methods
4.1. Materials
Gramicidin A was purchased from Merck (Darmstadt, Germany). Spectrograde methanol and methyl-d3-alcohol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Calcium chloride, calcium bromide and calcium iodide were purchased from Sigma-Aldrich (St. Louis, MO, USA). All chemicals were reagent grade and used without further purification.
4.2. Circular Dichroism (CD) Spectroscopy
Circular dichroism spectra were recorded using a J-815 CD spectrometer (JASCO International Co. Ltd., Tokyo, Japan) or a Chirascan-plus qCD spectrometer (Applied Photophysics, Surrey, UK). All measurements were performed in a quartz cell with a pathlength of 0.1 mm. The CD spectra of 100 μM gramicidin A in the presence of calcium salts with a designated concentration were collected from 200 to 250 nm with a 0.5 nm interval at 25 °C. The reported circular dichroism spectra were corrected with baseline using methanol containing the same concentration of salt and smoothed using a Savitsky–Golay function in Origin 6.0.
4.3. Mass Spectroscopy
Gramicidin A samples (100 µM each) were mixed with CaCl2, CaBr2 or CaI2 at ten different concentrations (0, 0.02, 0.07, 0.2, 2, 10, 40, 100, 200 and 300 mM) dissolved in either D2O or methanol. These samples were analyzed using MALDI-TOF mass spectrometry.
The matrix used for the analysis was α-cyano-4-OH cinnamic acid (CHCA). A 10 mg amount of CHCA was weighed and dissolved in 1.0 mL of 50% acetonitrile, 50% distilled H2O and 0.1% trifluoroacetic acid (TFA). The gramicidin A samples were then mixed with the CHCA matrix by adding 1 µL of the sample with 1 µL of the CHCA matrix. After mixing the sample and matrix well, 1 µL of the resulting mixtures were then spotted on to the MALDI plate for analysis. The MALDI spectra in the reflection mode were recorded with a Bruker New ultrafleXtremeTM MALDI-TOF/TOF mass spectrometer from Bruker Daltonik, Bremen, Germany, at the Academia Sinica Institute of Chemistry Mass Spectrometry Center.
4.4. Nuclear Magnetic Resonance (NMR) Spectroscopy
We analyzed gramicidin A (100 μM) with CaCl2, CaBr2 or CaI2 (0.02, 0.2, 10 and 200 mM) dissolved in 90% d4-methanol/10% methanol using solution NMR. The NMR spectra were obtained using a Bruker DRX-500 NMR spectrometer equipped with a TXI z-gradient (1H, 13C, 15N) probe at 300 K (Bruker Biospin GmbH, Rheinstetten, Germany). In these spectra, the OH signal of methanol was suppressed using the presaturation method. All the NMR spectra were phased and baseline-corrected using the Topspin software (version 3.2.2; Bruker Biospin GmbH, Rheinstetten, Germany) and referenced to the chemical shift of methanol at 3.3 ppm.
4.5. Steady-State Fluorescence Measurements
All steady-state fluorescence measurements were performed with a Jasco FP-6500 fluorescence spectrometer (Tokyo, Japan) equipped with a water circulator and stirring accessory. The emission spectra of 100 μM gramicidin A in the presence of calcium halide salts at designed concentrations were recorded from 310 to 500 nm with an excitation wavelength of 290 nm. The bandwidths for excitation and emission were 3 and 5 nm, respectively. All measurements were performed in a quartz cell with a path length of 1 cm. The background spectra of the salts alone were taken first for later subtraction. The reported spectra were the averages obtained from at least three individual samples and three repeated measurements of each sample. All measurements were carried out at 25.0 ± 0.5 °C.
4.6. Bacterial Growth Assay
Staphylococcus aureus (S. aureus) (ATCC-25923) was kindly provided by Dr. J.W. Liou in Tzu Chi University. The S. aureus was grown in Luria–Bertani (LB) broth medium in a 250 mL flask at 37 °C overnight. This overnight S. aureus culture was then diluted to OD600 = 0.1 with LB medium. This S. aureus was then grown to lag phase (OD600 = 0.2), exponential phase (OD600 = 0.6) and late-exponential-to-stationary phase (OD600 = 1.5). At the designated growth phase, 5 mL of S. aureus culture was treated at 5% (V/V) with a stock solution containing gramicidin A (a final concentration of 5 μM) and CaCl2. The stock solution was prepared in the same way as used for the spectroscopic studies, in which a final concentration of 100 μM gramicidin A was dissolved in methanol containing 0, 0.2, 10, 100 and 400 mM of CaCl2. The S. aureus samples treated with or without gramicidin A/CaCl2 were incubated at 37 °C. The optical density at a wavelength of 600 nm, (OD600), was used to determine the growth curve of S. aureus using a microplate reader (FlexStation 3, MD) every 30 min.