Into the Seed: Auxin Controls Seed Development and Grain Yield
Abstract
:1. Introduction
2. Auxin Levels Influence Seed Development
3. Auxin Transport-Mediated Seed Development
4. The Auxin Signaling Pathway Is Involved in Seed Development
5. Auxin Homeostasis-Mediated Regulation of Seed Development
6. Auxin Is a Main Factor Influencing Seed Size and Seed Weight of Crop Species
7. Concluding Remarks: Auxin as a High Potential Target for the Optimization of Crop Yields
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Herridge, R.; Day, R.; Baldwin, S.; Macknight, R. Rapid analysis of seed size in Arabidopsis for mutant and QTL discovery. Plant Methods 2011, 7, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundaresan, V. Control of seed size in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 17887–17888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuma, S.; Salomon, B.; Komatsuda, T. The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol. 2011, 52, 738–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, R.J.; Spielman, M.; Bailey, J.; Dickinson, H.G. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 1998, 125, 3329–3341. [Google Scholar] [PubMed]
- Garcia, D.; Saingery, V.; Chambrier, P.; Mayer, U.; Jurgens, G.; Berger, F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003, 131, 1661–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Shantharaj, D.; Kang, X.; Ni, M. Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 2010, 13, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Chay, P.; Thurling, N. Identification of genes controlling pod length in spring rapeseed, Brassica napus L., and their utilization for yield improvement. Plant Breed. 1989, 103, 54–62. [Google Scholar] [CrossRef]
- King, S.P.; John, E.L.; Robert, T.F. Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol. 1997, 114, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Bennett, E.J.; Roberts, J.A.; Wagstaff, C. The role of the pod in seed development: Strategies for manipulating yield. New Phytol. 2011, 190, 838–853. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Hua, W.; Hu, Z.; Yang, H.; Zhang, L.; Li, R.; Deng, L.; Sun, X.; Wang, X.; Wang, H. Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc. Natl. Acad. Sci. USA 2015, 112, E5123–E5132. [Google Scholar] [CrossRef] [Green Version]
- Fang, W.; Wang, Z.; Cui, R.; Li, J.; Li, Y. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 2012, 70, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Fitz Gerald, J.N.; Berger, F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 2005, 17, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohto, M.-A.; Fischer, R.L.; Goldberg, R.B.; Nakamura, K.; Harada, J.J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 2005, 10, 3123–3128. [Google Scholar] [CrossRef] [Green Version]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The Auxin Response Factor 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Adamski, N.M.; Anastasiou, E.; Eriksson, S.; O’Neill, C.M.; Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20115–20120. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, X.; Kang, X.; Zhao, X.; Zhang, X.; Ni, M. Short Hypocotyl Under Blue1 associates with Miniseed3 and Haiku2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 2009, 21, 106–117. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Garcia, D.; Zhang, H.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef]
- Kang, X.; Li, W.; Zhou, Y.; Ni, M. A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet. 2013, 9, e1003347. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Ni, M. Arabidopsis Short Hypocotyl under Blue1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 2006, 18, 921–934. [Google Scholar] [CrossRef] [Green Version]
- Morinaka, Y.; Sakamoto, T.; Inukai, Y.; Agetsuma, M.; Kitano, H.; Ashikari, M.; Matsuoka, M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006, 141, 924–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riefler, M.; Novak, O.; Strnad, M.; Schmulling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.; Opsahl-Sorteberg, H.G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.B.; Huang, H.Y.; Hu, Y.W.; Zhu, S.W.; Wang, Z.Y.; Lin, W.H. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Li, N.; Chen, L.; Xu, Y.; Li, Y.; Zhang, Y.; Li, C.; Li, Y. The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 2014, 26, 665–677. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Du, L.; Xu, R.; Cui, R.; Hao, J.; Sun, C.; Li, Y. Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell 2015, 27, 620–632. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Bai, W.; Zeng, Q.; Song, S.; Zhang, M.; Li, X.; Hou, L.; Xiao, Y.; Luo, M.; Li, D.; et al. Moderately enhancing cytokinin level by down-regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol. Breed. 2015, 35, 60. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Liang, W.; Cui, X.; Chen, M.; Yin, C.; Luo, Z.; Zhu, J.; Lucas, W.J.; Wang, Z.; Zhang, D. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of carbon starved anther, a MYB domain protein. Plant J. 2015, 82, 570–581. [Google Scholar] [CrossRef]
- Xiao, Y.G.; Sun, Q.B.; Kang, X.J.; Chen, C.B.; Ni, M. Short Hypocotyl under Blue1 or HAIKU2 mixepression alters canola and Arabidopsis seed development. New Phytol. 2016, 209, 636–649. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Li, Y. Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, F.G. Inducement of fruit development by growth promoting chemicals. Proc. Natl. Acad. Sci. USA 1936, 22, 628–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardi, J.; Lanubile, A.; Li, Q.B.; Kumar, D.; Kladnik, A.; Cook, S.D.; Ross, J.J.; Marocco, A.; Chourey, P.S. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 2012, 160, 1318–1328. [Google Scholar] [CrossRef] [Green Version]
- Lur, H.-S.; Setter, T.L. Role of auxin in maize endosperm development. Plant Physiol. 1993, 103, 273–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front. Plant Sci. 2014, 5, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, D.A.; Wang, L.J.; Ding, C.H.; Xu, Z.H. Auxin distribution and transport during embryogenesis and seed germination of Arabidopsis. Cell Res. 2001, 11, 273–278. [Google Scholar] [CrossRef]
- Uchiumi, T.; Okamoto, T. Rice fruit development is associated with an increased IAA content in pollinated ovaries. Planta 2010, 232, 579–592. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, L.; Shen, Y.; Liu, S.J.; Huang, J.X.; Long, Q.Z.; Wu, W.; Yang, C.Y.; Chen, H.; Guo, X.P.; et al. Loss of function of the cytochrome P450 gene CYP78B5 causes giant embryos in rice. Plant Mol. Biol. Rep. 2015, 33, 69–83. [Google Scholar] [CrossRef]
- Forestan, C.; Meda, S.; Varotto, S. ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 2010, 152, 1373–1390. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.J.; Köhler, C. Auxin production couples endosperm development to fertilization. Nat. Plants 2015, 1, 15184. [Google Scholar] [CrossRef]
- Batista, R.A.; Figueiredo, D.D.; Santos-Gonzalez, J.; Kohler, C. Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev. 2019, 33, 466–476. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.J.; Hennig, L.; Köhler, C. Auxin production in the endosperm drives seed coat development in Arabidopsis. ELife 2016, 5, e20542. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Davidson, S.E.; Davies, N.W.; Smith, J.A.; Dalmais, M.; Bendahmane, A.I.; Quittenden, L.J.; Sutton, L.; Bala, R.K.; Le Signor, C.; et al. Biosynthesis of the halogenated auxin, 4-chloroindole-3-acetic acid. Plant Physiol. 2012, 159, 1055–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef] [Green Version]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Coversion of tryptophan to indole-3-acetic acid by Tryptophan of Arabidopsis and Yuccas in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [Green Version]
- McAdam, E.L.; Meitzel, T.; Quittenden, L.J.; Davidson, S.E.; Dalmais, M.; Bendahmane, A.I.; Thompson, R.; Smith, J.J.; Nichols, D.S.; Urquhart, S.; et al. Evidence that auxin is required for normal seed size and starch synthesis in Pea. New Phytol. 2017, 216, 193–204. [Google Scholar] [CrossRef] [Green Version]
- Robert, H.S.; Park, C.; Gutièrrez, C.L.; Wójcikowska, B.; Pěnčík, A.; Novák, O.; Chen, J.; Grunewald, W.; Dresselhaus, T.; Friml, J.; et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 2018, 4, 548–553. [Google Scholar] [CrossRef]
- Bernardi, J.; Battaglia, R.; Bagnaresi, P.; Lucini, L.; Marocco, A. Transcriptomic and metabolomic analysis of ZmYUC1 mutant reveals the role of auxin during early endosperm formation in maize. Plant Sci. 2019, 281, 133–145. [Google Scholar] [CrossRef]
- Shi, L.L.; Song, J.R.; Guo, C.C.; Wang, B.; Guan, Z.L.; Yang, P.; Chen, X.; Zhang, Q.H.; King, G.J.; Wang, J.; et al. A CACTA-like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 2019, 98, 524–539. [Google Scholar] [CrossRef]
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156. [Google Scholar] [CrossRef] [Green Version]
- Wójcikowska, B.; Jaskola, K.; Gasiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. Leafy Cotyledon2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.L.; Xue, H.W. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell 2012, 24, 1049–1065. [Google Scholar] [CrossRef] [Green Version]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef] [Green Version]
- Nitsch, J.P. Growth and morphogenesis of the Strawberry as related to auxin. Am. J. Bot. 1950, 37, 211–215. [Google Scholar] [CrossRef]
- Sailer, C.; Schmid, B.; Grossniklaus, U. Apomixis allows the transgenerational fixation of phenotypes in hybrid plants. Curr. Biol. 2016, 26, 331–337. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, D.D.; Köhler, C. Auxin: A molecular trigger of seed development. Genes Dev. 2018, 32, 479–490. [Google Scholar] [CrossRef] [Green Version]
- Carraro, N.; Forestan, C.; Canova, S.; Traas, J.; Varotto, S. ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol. 2006, 142, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Robert, H.S.; Grunewald, W.; Sauer, M.; Cannoot, B.; Soriano, M.; Swarup, R.; Weijers, D.; Bennett, M.; Boutilier, K.; Friml, J. Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 2015, 142, 702–711. [Google Scholar] [CrossRef] [Green Version]
- Petrášek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertová, D.; Wiśniewska, J.; Tadele, Z.; Kubeš, M.; Čovanová, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- De Smet, I.; Jürgens, G. Patterning the axis in plants--auxin in control. Curr. Opin. Genet. Dev. 2007, 17, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Neuhaus, G. Influence of auxin on the establishment of bilateral symmetry in monocots. Plant J. 1996, 9, 659–669. [Google Scholar] [CrossRef]
- Fischer-Iglesias, C.; Sundberg, B.; Neuhaus, G.; Jones, A. Auxin distribution and transport during embryonic pattern formation in wheat. Plant J. 2001, 26, 115–129. [Google Scholar] [CrossRef] [Green Version]
- Forestan, C.; Farinati, S.; Varotto, S. The maize PIN gene family of auxin transporters. Front. Plant Sci. 2012, 3, 16. [Google Scholar] [CrossRef] [Green Version]
- Noguero, M.; Signor, C.L.; Vernound, V.; Bandyopadhyay, K.; Sanchez, M.; Fu, C.X.; Torres-Jerez, I.; Wen, J.Q.; Mysore, K.S.; Gallardo, K.; et al. DASH transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J. 2015, 81, 453–466. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A.; Walker, A.R.; Schulz, B.; Feldmann, K.A. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef]
- Yang, Y.; Hammes, U.Z.; Taylor, C.G.; Schachtman, D.P.; Nielsen, E. High-affinity auxin transporter by the aux1 influx carrier protein. Curr. Biol. 2006, 16, 1123–1127. [Google Scholar] [CrossRef] [Green Version]
- Parry, G.; Marchant, A.; May, S. Quick on the uptake: Characterization of a family of plant auxin influx carriers. J. Plant Growth Regul. 2001, 20, 217–225. [Google Scholar] [CrossRef]
- Ugartechea-Chirino, Y.; Swarup, R.; Swarup, K.; Péret, B.; Whitworth, M.; Bennett, M.J.; Bougourd, S. The AUX LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana. Ann. Bot. 2010, 105, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, S.B.; Wang, X.-J.; Hagen, G.; Guifoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef] [Green Version]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jurgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 11, 5844–5849. [Google Scholar] [CrossRef] [Green Version]
- Abel, S.; Theologis, A. Early genes and auxin action. Plant Physiol. 1996, 111, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Hamann, T.; Benkova, E.; Bäurele, I.; Kientz, M.; Jürgens, G. The Arabidopsis Bodenlos gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef] [Green Version]
- Ploense, S.E.; Wu, M.-F.; Nagpal, P.; Reed, J.W. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 2009, 136, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Berleth, T.; Jürgens, G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 1993, 118, 575–587. [Google Scholar]
- Hardtke, C.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [Green Version]
- Goetz, M.; Vivian-Smith, A.; Johnson, S.D.; Koltunow, A.M. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 2006, 18, 1873–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Gao, X.; Dai, X.; Yang, Z.N.; Zhao, Y. An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 2011, 12, 178. [Google Scholar] [CrossRef] [Green Version]
- Kelley, D.R.; Arreola, A.; Gallagher, T.L.; Gasser, C.S. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 2012, 139, 1105–1109. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Li, B.Y.; Mu, S.Y.; Han, B.; Cui, R.Z.; Xu, M.Y.; You, Z.Z.; Dong, H.S. TTG2-regulated development is related to expression of putative AUXIN RESPONSE FACTOR genes in tobacco. BMC Genomics 2013, 14, 806. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.; Li, B.; Shen, D.; Xie, J.; Long, J.; Dong, H. Tobacco TTG2 regulates vegetative growth and seed production via the predominant role of ARF8 in cooperation with ARF17 and ARF19. BMC Plant Biol. 2016, 16, 126. [Google Scholar] [CrossRef] [Green Version]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Alonso, J.M. Cutting out the middle man in light-hormone interactions. Dev Cell 2016, 39, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2010, 2, a001594. [Google Scholar] [CrossRef]
- Rosquete, M.R.; Barbez, E.; Kleine-Vehn, J. Cellular auxin homeostasis: Gatekeeping is housekeeping. Mol. Plant 2011, 5, 772–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Schölkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; Mu, X.; Grabowski, P.; Schmutz, J.; Lu, C. Enhancing microRNA167A expression in seed decreases the alpha-linolenic acid content and increases seed size in Camelina sativa. Plant J. 2019, 98, 346–358. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.C.; Tong, H.N.; Xiao, Y.H.; Che, R.H.; Xu, F.; Hu, B.; Liang, C.Z.; Chu, J.F.; Li, J.Y.; Chu, C.C. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 11102–11107. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Lu, S.J.; Wang, M.J.; He, H.; Sun, L.; Wang, H.; Liu, X.H.; Jiang, L.; Sun, J.L.; Xin, X.; et al. A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 2018, 11, 736–749. [Google Scholar] [CrossRef] [Green Version]
- Aya, K.; Hobo, T.; Sato-Izawa, K.; Ueguchi-Tanaka, M.; Kitano, H.; Matsuoka, M. A novel AP2-type transcription factor, Small Organ Size1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol. 2014, 55, 897–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, K.; Yoshida, H.; Aya, K.; Kawamura, M.; Hayashi, M.; Hobo, T.; Sato-Izawa, K.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol. Plant 2017, 10, 590–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
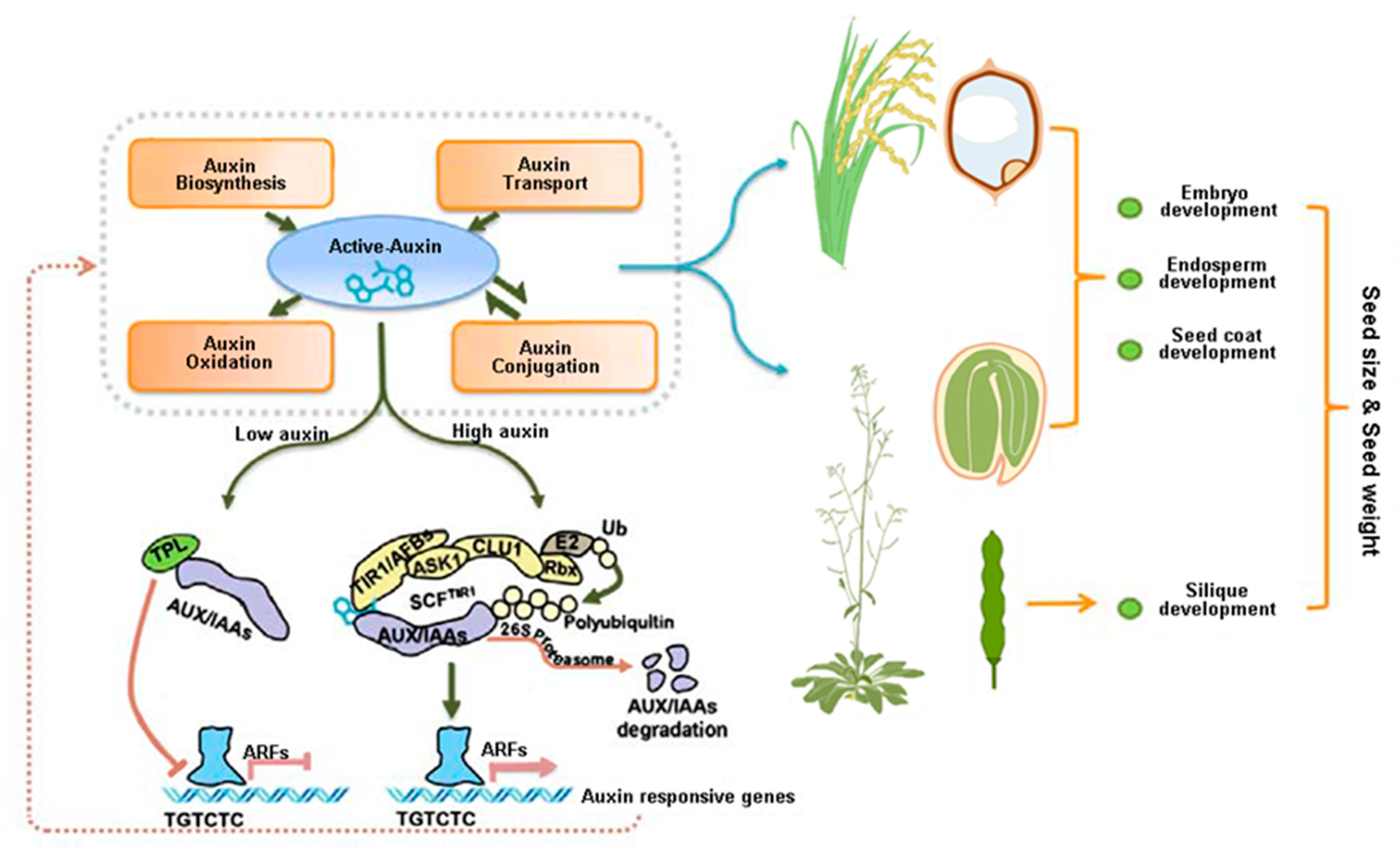
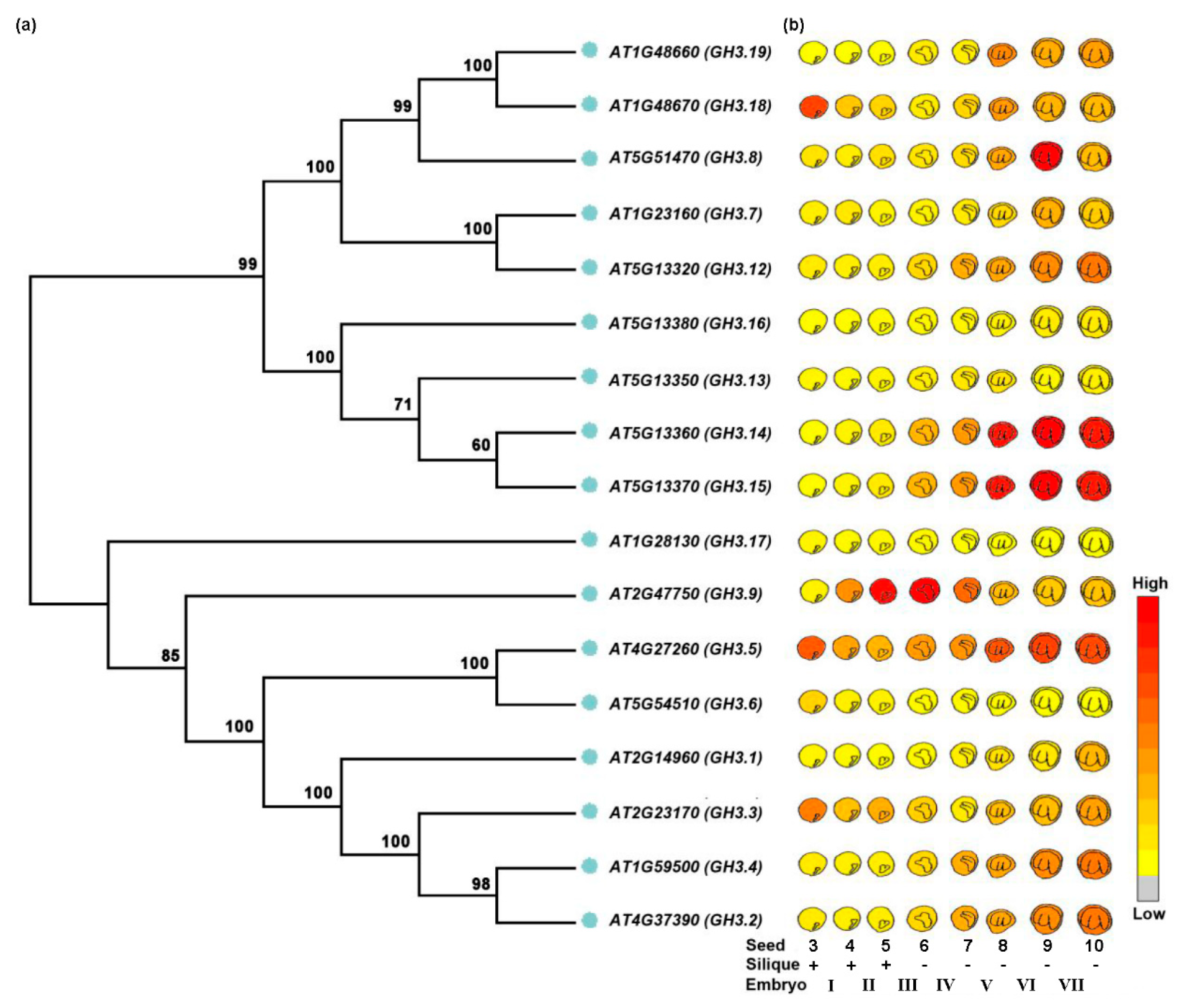
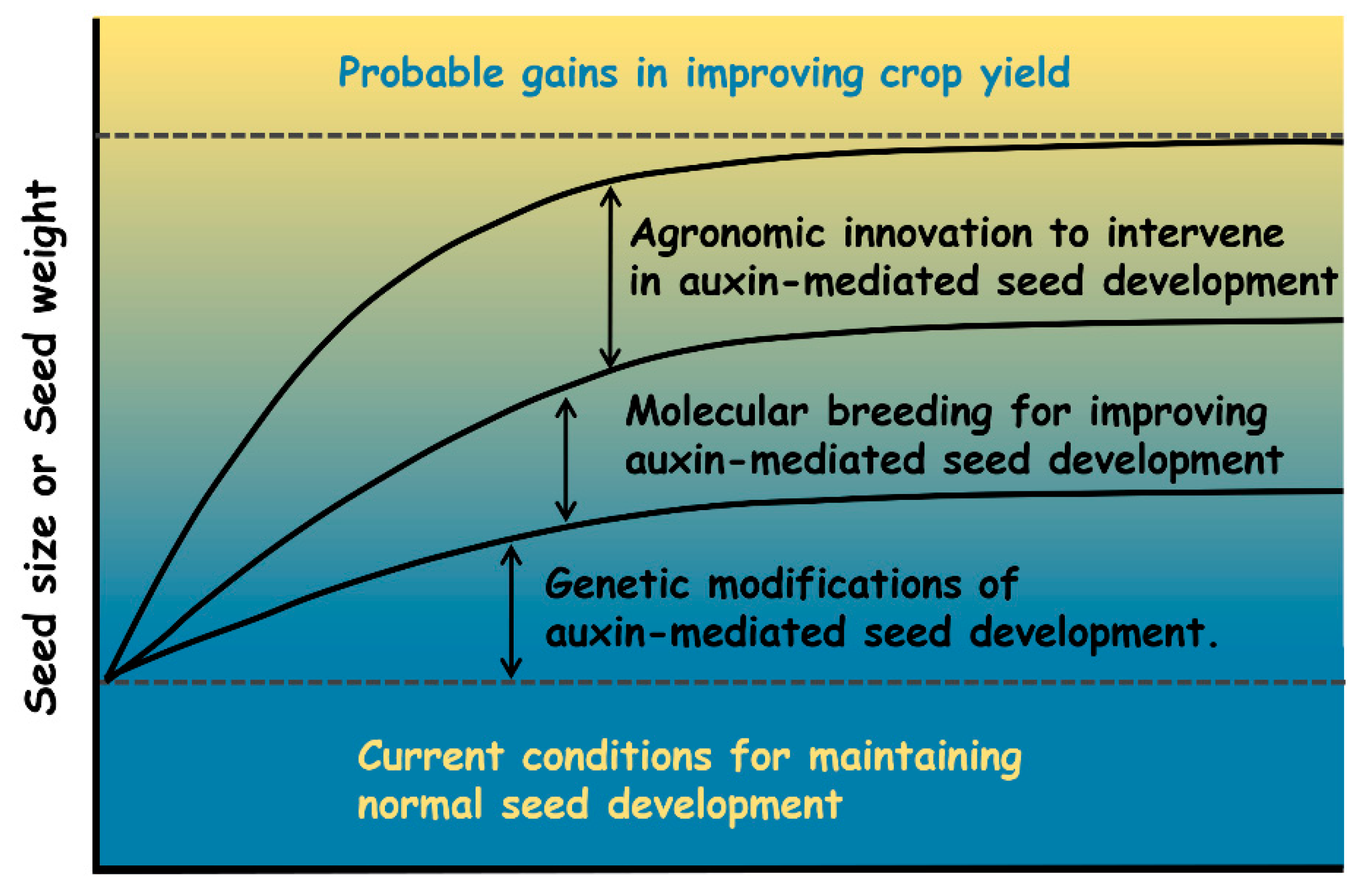
| Botanical Classification | Species | Accession Number | Gene Name | Protein Category | Impact on Seed Development | Possible Role for Auxin | References |
|---|---|---|---|---|---|---|---|
| Monocotyledon | Maize | GRMZM2G091819 | ZmYuc1 | Flavin monooxygenases | Endosperm development | Involved in auxin biosynthesis | Bernardi et al., 2012 |
| Rice | Os06g0623700 | TGW6 | Indole-3-acetic acid IAA-glucose hydrolase | Cell number and grain length, Seed weight | Controls the supply of IAA | Ishimaru et al., 2013 | |
| Rice | Os02g07430 | MADS29 | MADS-box transcription factor | Endosperm development | Induced by auxin | Yin and Xue, 2012 | |
| Rice | Os03g0175800 | BG1 | Novel plasma membrane-associated protein | Grain size | Regulates auxin transport | Liu et al., 2015 | |
| Rice | Os05g32270 | SMOS1 | AP2 transcription factor | Seed size | Induced by exogenous auxin treatment, interacts with ARF | Aya et al., 2014 | |
| Rice | Os07g0603700 | OsGE/CYP78B5 | Cytochrome P450 enzyme | Embryo development | Regulates auxin responsive gene | Chen et al., 2014 | |
| Rice | Os03g62500 | OsSK41 | GLYCOGEN SYNTHASE KINASE 3/SHAGGY-like family | Grain length, Grain weight | Interacts with OsARF4 | Hu et al., 2018 | |
| Rice | Os06g03710 | SMOS2/DLT | GRAS transcription factor | Seed size | Involved in auxin–BR signaling crosstalk | Hirano et al., 2017 | |
| Dicotyledon | Pea | JN990989 | PsTAR2 | Trytophan aminotransferase related protein | Reduced starch content, Seed size | Involved in auxin biosynthesis | Tivendale et al., 2012; McAdam et al., 2017 |
| Rape | BnaA09G55530D | BnaA9.CYP78A9 | Cytochrome P450 enzyme | Silique length, Seed size | Influences auxin metabolism or auxin biosynthesis | Shi et al., 2019 | |
| Arabidopsis | At4G32540, At5G11320, At1G48910, At1G21430 | YUC1, YUC4, YUC10, YUC11 | Flavin monooxygenases | Embryogenesis and post-embryonic organ formation | Involved in auxin biosynthesis | Cheng et al., 2007 | |
| Arabidopsis | At1G28300 | LEC2 | AP2/B3-like transcriptional factor family protein | Embryo development | Regulates the supply of auxin | Stone et al., 2008; Wójcikowska et al., 2013 | |
| Arabidopsis | At1G51950 | IAA18 | Auxin-responsive protein | Cotyledon placement, Embryo growth | Interferes with auxin transport | Ploense et al., 2009 | |
| Arabidopsis | At2G38120, At5G01240, At2G21050 | AUX1, LAX1, LAX2 | Transmembrane amino acid transporter family proteins | Endosperm development, Radicle apex growth | Regulates auxin transport | Robert et al., 2015; Ugartechea-Chirino et al., 2010 | |
| Arabidopsis | At5G60440 | AGL62 | MADS-box transcription factor | Embryo development | Involved in auxin transport | Figueiredo et al., 2015 | |
| Arabidopsis | At1G73590, At1G70940, At2G01420, At1G23080 | PIN1, PIN3, PIN4, PIN7 | PIN-FORMED proteins | Embryo development | Regulates auxin transport | Friml et al., 2003 | |
| Medicago truncatula | Medtr2g014060 | DASH | DOF transcription factor | Endosperm development | Affects auxin export | Noguero et al., 2015 | |
| Arabidopsis | At5G16560 | KANADI | Homeodomain-like superfamily protein | Integument development | Regulated by auxin | Kelley et al., 2012 | |
| Arabidopsis | At3G62980 | TIR1 | F-box protein | Embryo development | Response to auxin | Dharmasiri et al., 2005 | |
| Arabidopsis | At4G03190, At3G26810, At1G12820 | AFB1, AFB2, AFB3 | F-box proteins | Embryo development | Response to auxin | Dharmasiri et al., 2005 | |
| Arabidopsis | At5G62000 | ARF2 | AUXIN RESPONSE FACTOR (ARF) transcription factor | Integument development, Seed size | Response to auxin | Schruff et al., 2006 | |
| Arabidopsis | At2G33860 | ETT/ARF3 | AUXIN RESPONSE FACTOR (ARF) transcription factor | Integument development | Response to auxin | Kelley et al., 2012 | |
| Arabidopsis | At1G04550 | BDL/IAA12 | AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) transcriptional repressors | Embryo development | Response to auxin | Hamann et al., 2002 | |
| Arabidopsis | At1G19850 | MONOPTEROS/ARF5 | AUXIN RESPONSE FACTOR (ARF) transcription factor | Embryo development | Response to auxin | Berleth and Jürgens, 1993 | |
| Arabidopsis | At3G22886 | MIR167A | microRNA | Ovule development, Embryos growth, Endosperms development | Response to auxin | Yao et al., 2019; Na et al., 2019 | |
| Rape | BnaA09G55580D | ARF18 | AUXIN RESPONSE FACTOR (ARF) transcription factor | Silique development | Response to auxin | Liu et al., 2015 | |
| Tobacco | LOC107800718 | NtTTG2 | WRKY transcription factor | Seed production, Seed development | Impacts the nuclear import of NtARF8 | Zhu et al., 2013; Ge et al., 2016 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Li, G.; Qu, D.; Li, X.; Wang, Y. Into the Seed: Auxin Controls Seed Development and Grain Yield. Int. J. Mol. Sci. 2020, 21, 1662. https://doi.org/10.3390/ijms21051662
Cao J, Li G, Qu D, Li X, Wang Y. Into the Seed: Auxin Controls Seed Development and Grain Yield. International Journal of Molecular Sciences. 2020; 21(5):1662. https://doi.org/10.3390/ijms21051662
Chicago/Turabian StyleCao, Jinshan, Guoji Li, Dejie Qu, Xia Li, and Youning Wang. 2020. "Into the Seed: Auxin Controls Seed Development and Grain Yield" International Journal of Molecular Sciences 21, no. 5: 1662. https://doi.org/10.3390/ijms21051662




