Integrated Metabolome and Transcriptome Analysis Provide Insights into the Effects of Grafting on Fruit Flavor of Cucumber with Different Rootstocks
Abstract
:1. Introduction
2. Results
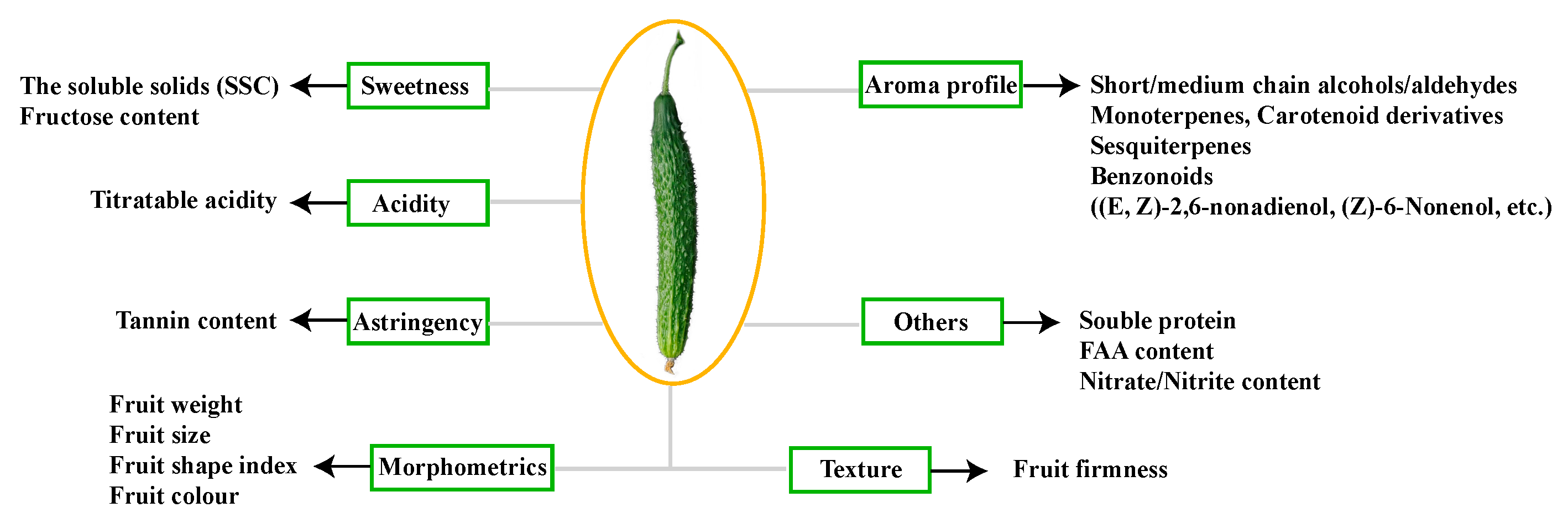
2.1. Phenotype and Physiological Traits of Cucumber Fruit from Plants Grafted on Different Rootstocks
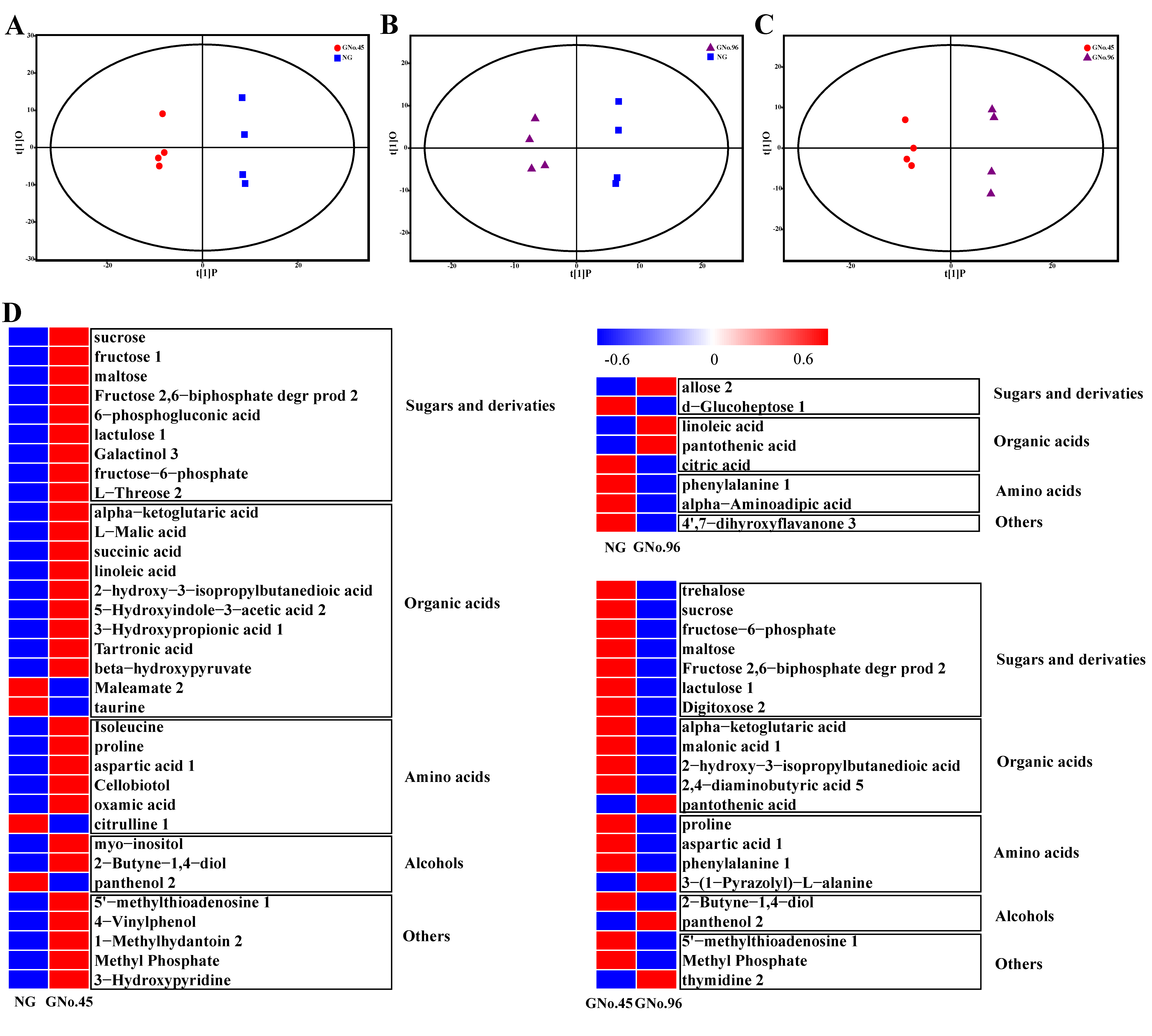
2.2. The Metabolite Profiles Revealed Differences in Metabolic Regulation between Non-Grafted and Grafted Cucumber Plants
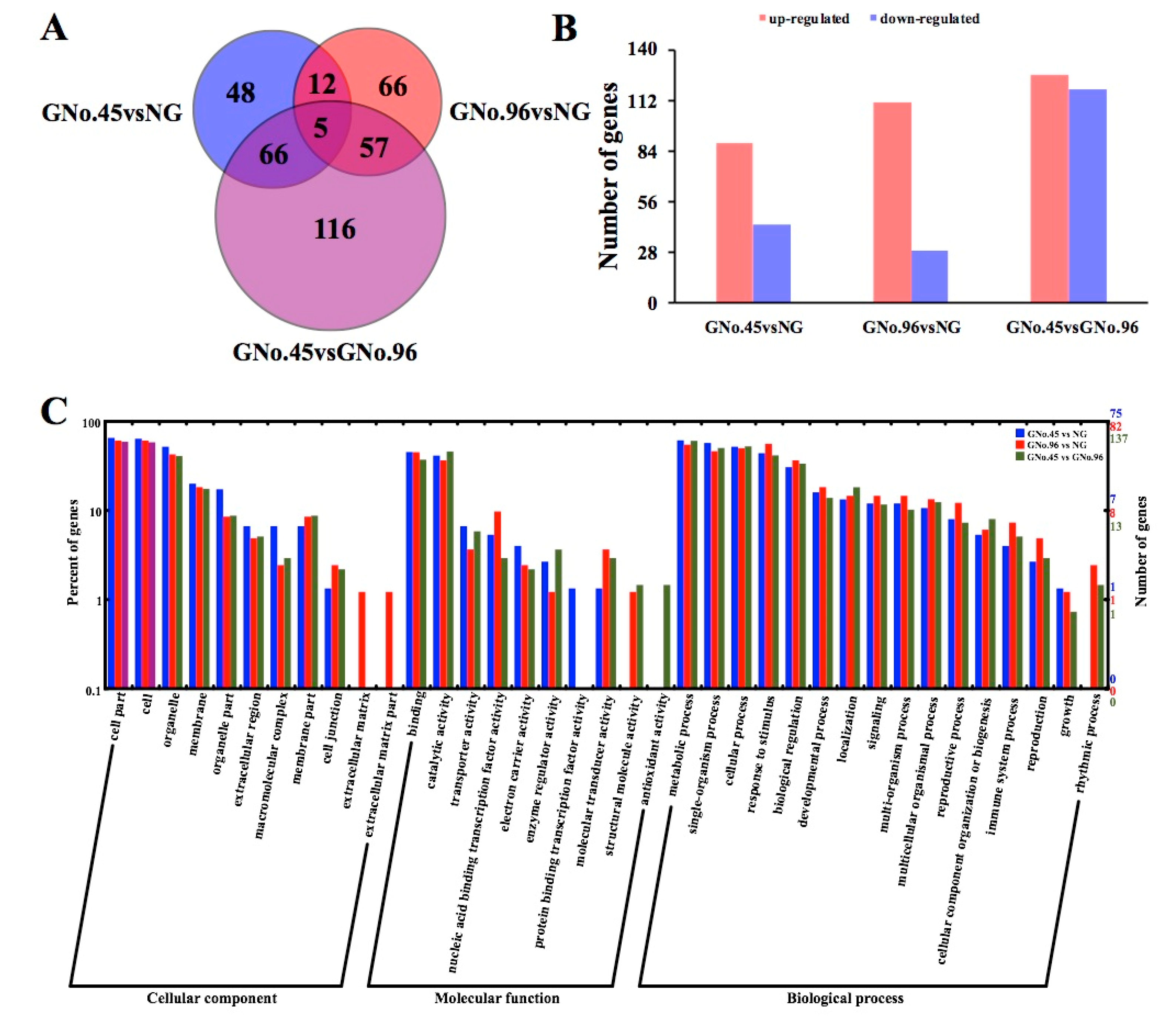
2.3. Identified Differentially Expressed Genes (DEGs) and Their Gene Ontology (GO) Enrichments by Transcriptome Analysis
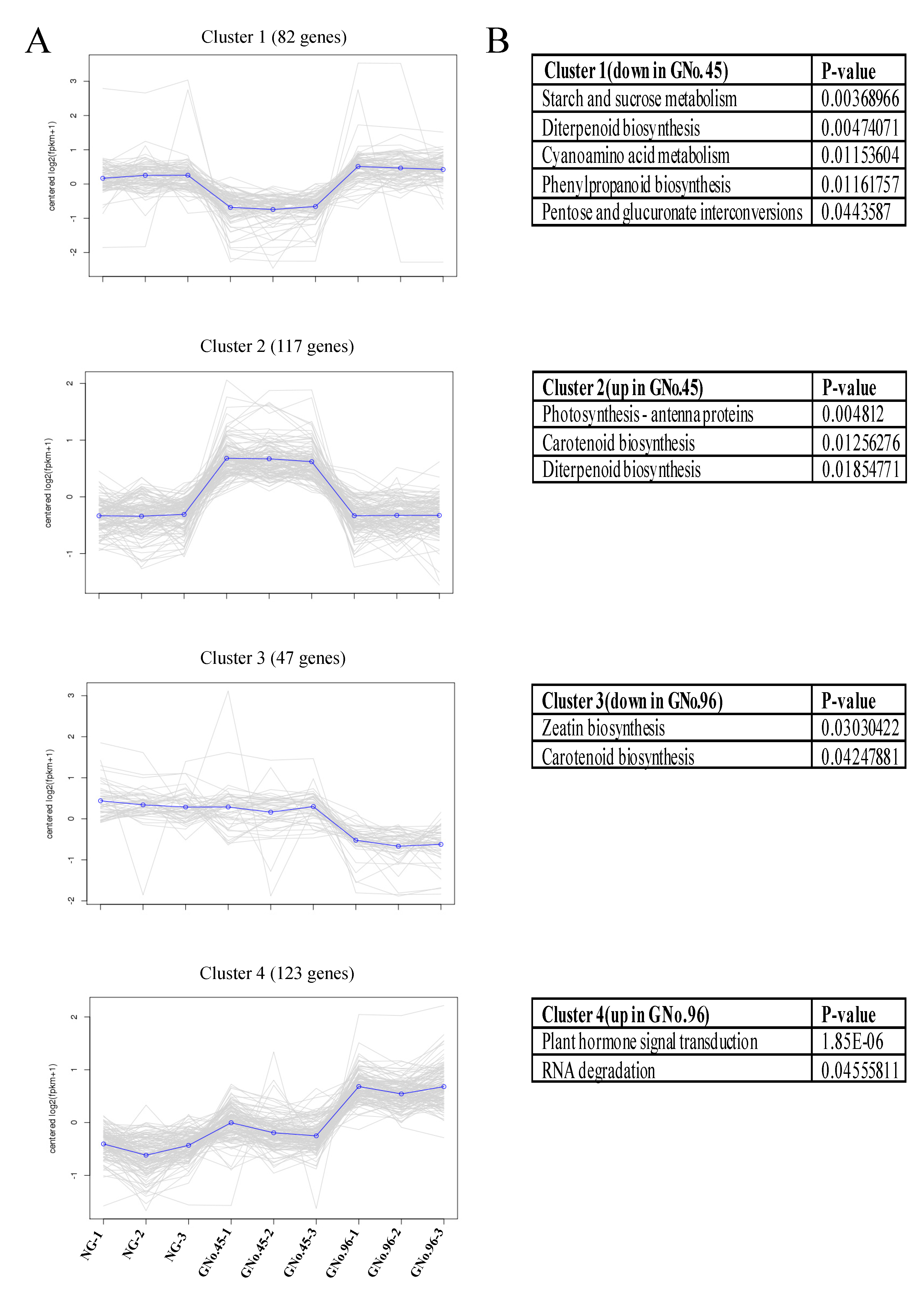
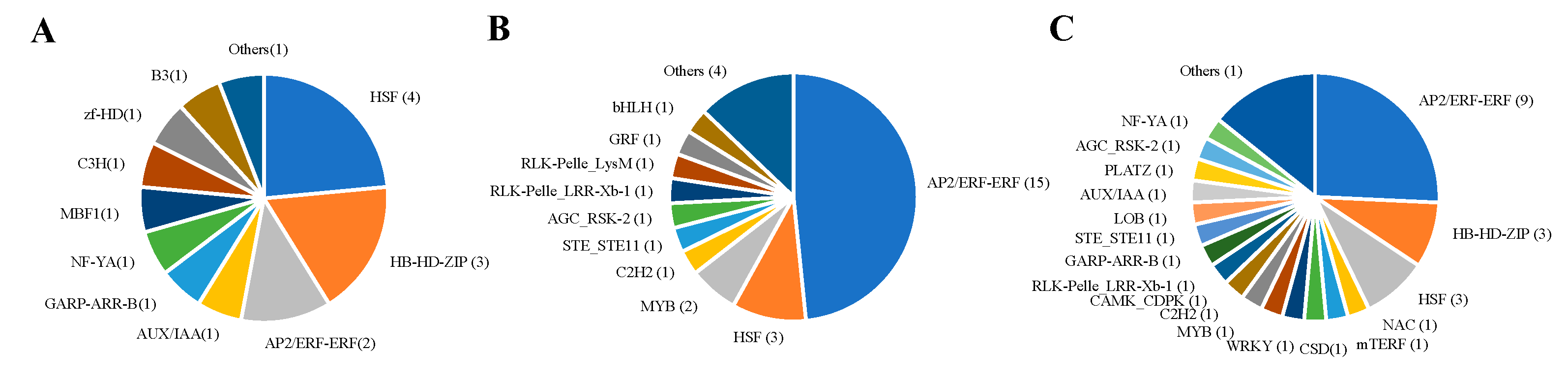
2.4. Clustering of Expression Patterns and Transcription Factors in Analysis of Transcriptome Data
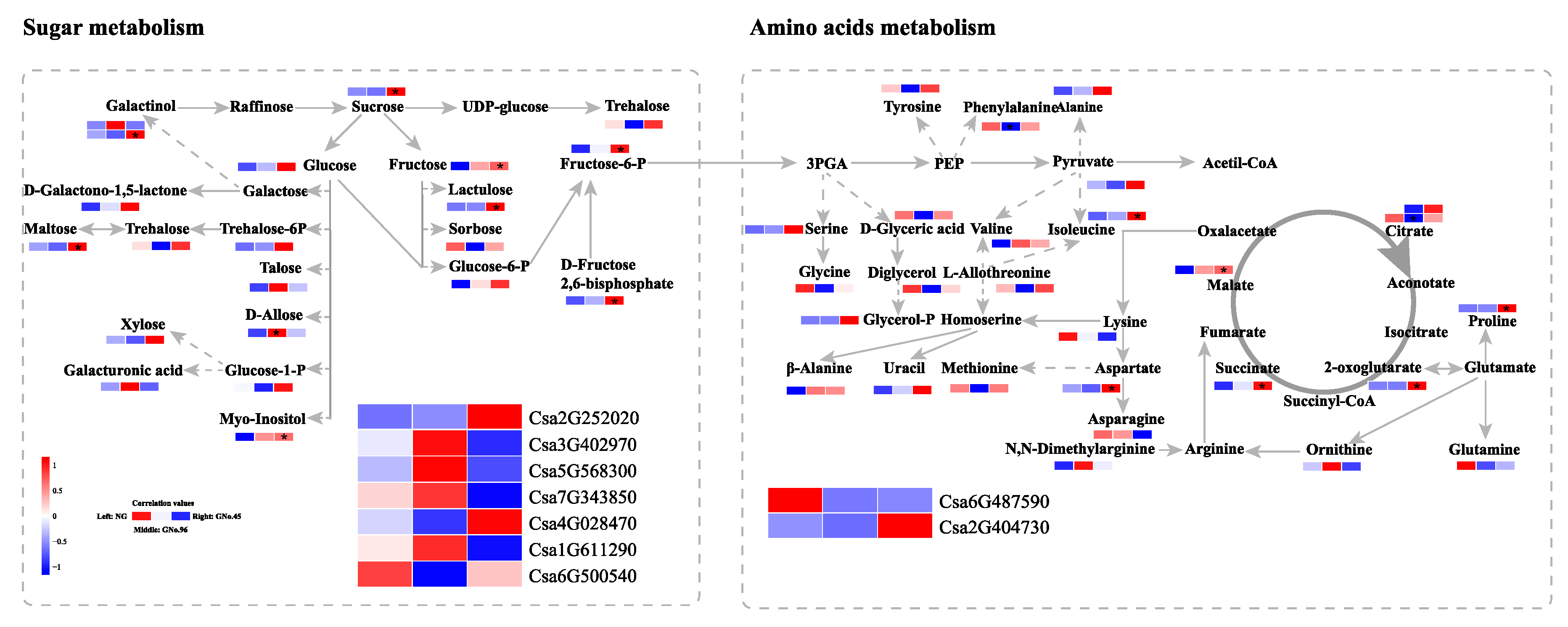
2.5. Candidate Genes for Metabolites of Sugar Metabolism, Linoleic Acid Metabolism, and Amino-Acid Biosynthesis
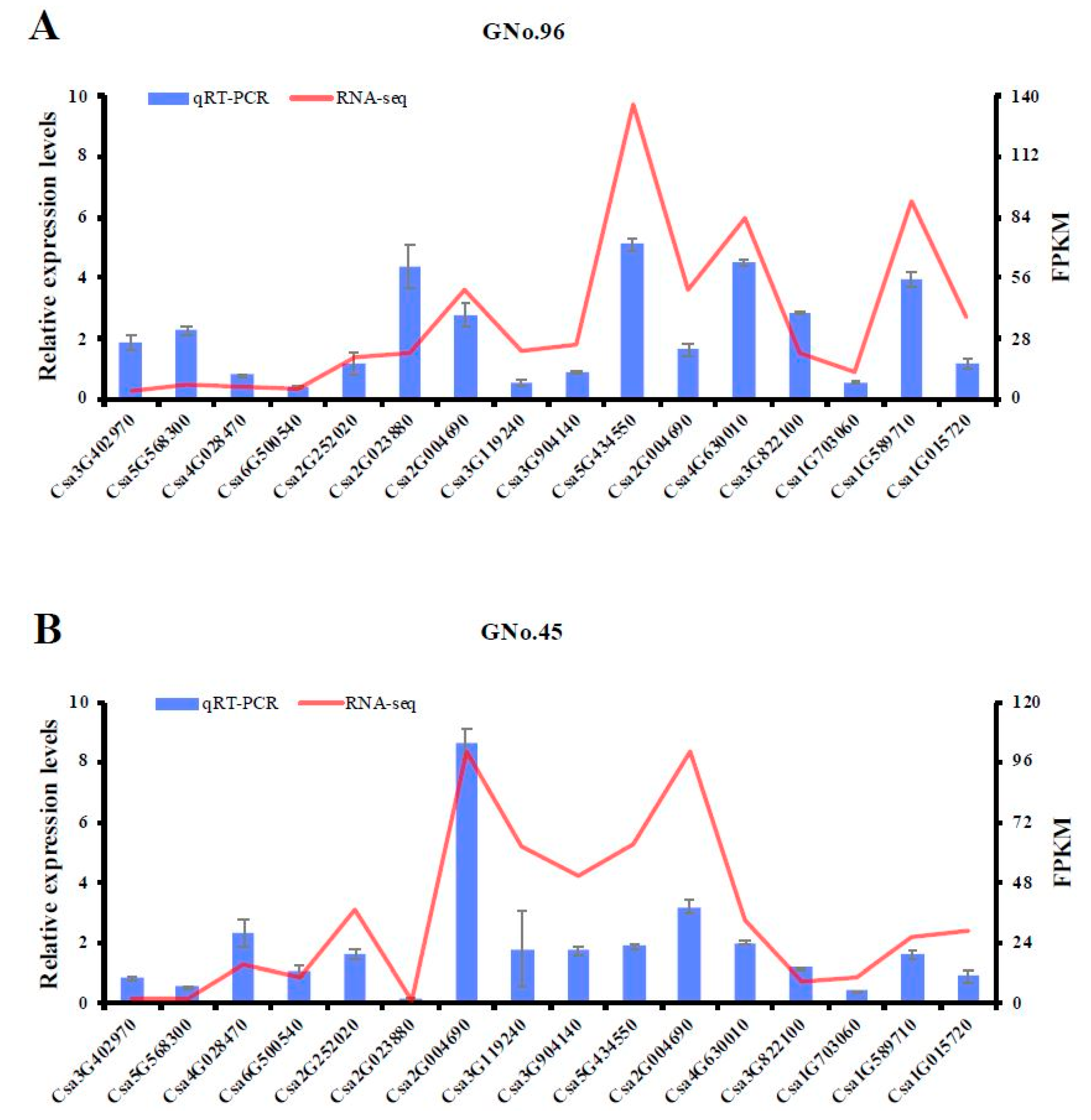
2.6. Validation of Transcriptomic Data Accuracy by qRT-PCR
3. Discussion
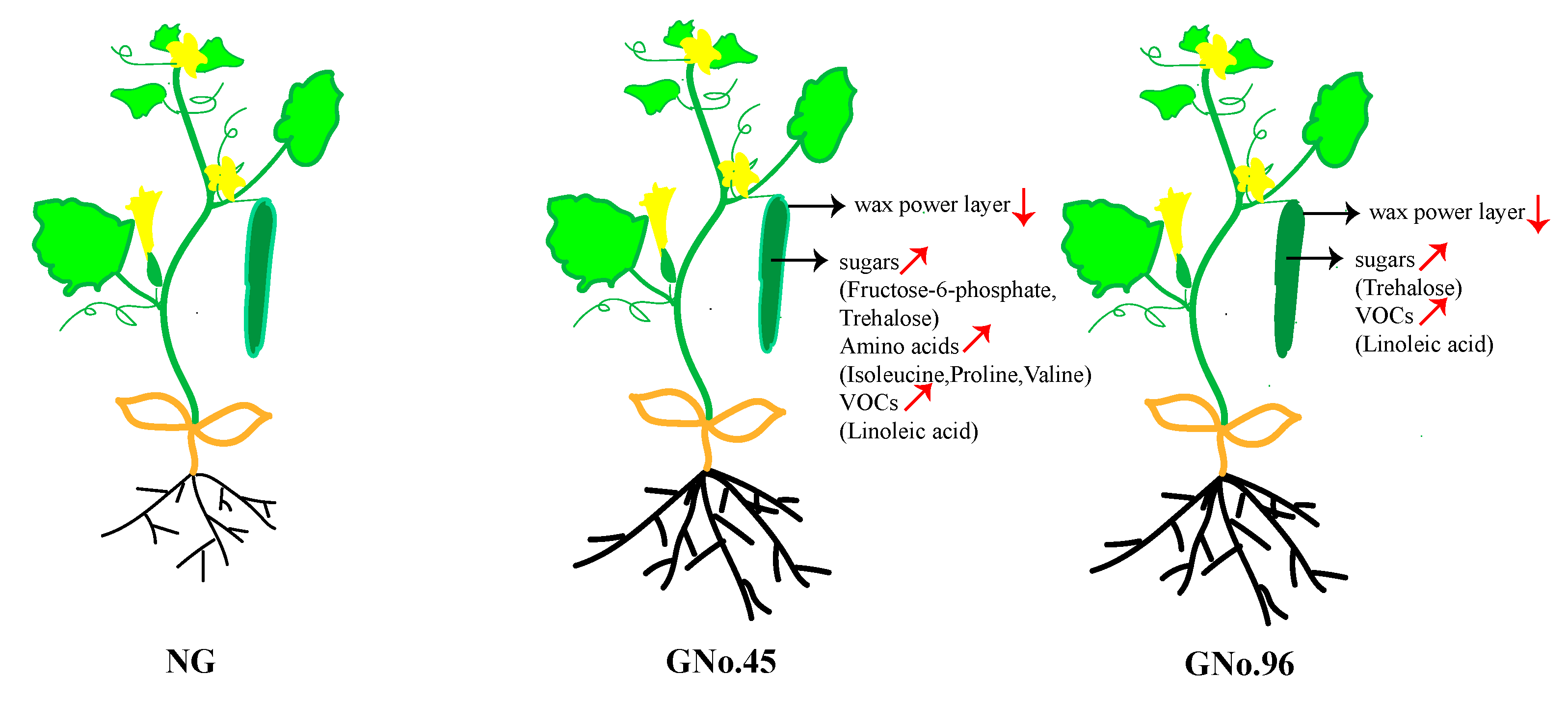
3.1. Fruit Characteristics of Non-Grafted Cucumber and Cucumbers Grafted onto Different Rootstocks
3.2. Different Transcriptomic and Metabolomic Regulation in Grafted Cucumber Fruit for Different Rootstocks
3.3. Volatile Organic Compounds and Transcript Factors Analysis
3.4. Metabolite-Based Determination of Candidate Genes for Fruit in Grafted Cucumber
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Determination of Contents of Soluble Solid, Wax Power, FAA, Soluble Protein, Nitrate, and Acidity
4.3. Sample Extraction and GC-MS Analysis
4.4. RNA Extraction, cDNA Library Preparation, and RNA Sequencing
4.5. Analysis of Differentially Expressed Genes (DEGs) and Functional Annotations
4.6. Validation of Gene Expression by Quantitative Real-Time RCR (qRT-PCR)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rouphael, Y.; Cardarelli, M.; Rea, E.; Colla, G. Improving melon and cucumber photosynthetic activity, mineral composition, and growth performance under salinity stress by grafting onto Cucurbita hybrid rootstocks. Photosynthetica 2012, 50, 180–188. [Google Scholar] [CrossRef]
- Rouphael, Y.; Schwarz, D.; Krumbein, A.; Colla, G. Impact of grafting on product quality of fruit vegetables. Sci. Hortic. 2010, 127, 172–179. [Google Scholar] [CrossRef]
- Klee, H.J.; Tieman, D.M. Genetic challenges of flavor improvement in tomato. Trends Genet. 2013, 29, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Kyriacou, M.C.; Rouphael, Y.; Colla, G.; Zrenner, R.; Schwarz, D. Vegetable Grafting: The Implications of a Growing Agronomic Imperative for Vegetable Fruit Quality and Nutritive Value. Front. Plant Sci. 2017, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Bang, H.; Ham, H. Quality of cucumber fruit as affected by rootstock. In Proceedings of the International Symposium on Vegetable Quality of Fresh and Fermented Vegetables 483, Seoul, Korea, 1 January 1999; pp. 117–124. [Google Scholar] [CrossRef]
- Colla, G.; Rouphael, Y.; Jawad, R.; Kumar, P.; Rea, E.; Cardarelli, M. The effectiveness of grafting to improve NaCl and CaCl2 tolerance in cucumber. Sci. Hortic. 2013, 164, 380–391. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, A.; Song, T.; Jin, Y.; Xu, X.; Gao, Y.; Ye, X.; Qi, H. Transcriptome analysis reveals the effects of grafting on sugar and α-linolenic acid metabolisms in fruits of cucumber with two different rootstocks. Plant Physiol. Biochem. 2018, 130, 289–302. [Google Scholar] [CrossRef]
- Wei, G.; Tian, P.; Zhang, F.; Qin, H.; Miao, H.; Chen, Q.; Hu, Z.; Cao, L.; Wang, M.; Gu, X.; et al. Integrative analyses of nontargeted volatile profiling and transcriptome data provide molecular insight into VOC diversity in cucumber plants (Cucumis sativus). Plant Physiol. 2016, 172, 603–618. [Google Scholar] [CrossRef]
- Hao, L.N.; Chen, S.X.; Wang, C.Y.; Chen, Q.; Wan, X.H.; Shen, X.Q.; Cheng, Z.H.; Meng, H.W. Aroma components and their contents in cucumbers from different genotypes. J. Northwest A F Univ. 2013, 41, 139–146. [Google Scholar] [CrossRef]
- Kemp, T.R.; Knavel, D.E.; Stoltz, L.P. Identification of some volatile compounds from cucumber. J. Agric. Food Chem. 1974, 22, 717–718. [Google Scholar] [CrossRef]
- Güler, Z.; Karaca, F.; Yetişir, H. Volatile compounds in the peel and flesh of cucumber (Cucumis sativus L.) grafted onto bottle gourd (Lagenaria siceraria) rootstocks. J. Hortic. Sci. Biotechnol. 2013, 88, 123–128. [Google Scholar] [CrossRef]
- Spyropoulou, E.A.; Dekker, H.L.; Steemers, L.; Van Maarseveen, J.H.; De Koster, C.G.; Haring, M.A.; Schuurink, R.C.; Allmann, S. Identification and Characterization of (3Z):(2E)-Hexenal Isomerases from Cucumber. Front. Plant Sci. 2017, 8, 1342. [Google Scholar] [CrossRef] [PubMed]
- Tieman, D.; Zhu, G.; Bies, D.; Beltran, K.S.O.; Fisher, J.; Zemach, I.; Monforte, A.J.; Zamir, D.; Rambla, J.L.; Huang, S.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.-W.; Han, J.; Fan, Y.; Tai, Y.; Zhu, B.; Lu, M.; Wang, R.; Wan, X.; Zhang, Z.-Z. Uncovering tea-specific secondary metabolism using transcriptomic and metabolomic analyses in grafts of Camellia sinensis and C. oleifera. Tree Genet. Genomes 2018, 14, 23. [Google Scholar] [CrossRef]
- Negri, A.S.; Prinsi, B.; Failla, O.; Scienza, A.; Espen, L. Proteomic and metabolic traits of grape exocarp to explain different anthocyanin concentrations of the cultivars. Front. Plant Sci. 2015, 6, 603. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-M.; Kubota, C.; Tsao, S.; Bie, Z.; Echevarria, P.H.; Morra, L.; Oda, M. Current status of vegetable grafting: Diffusion, grafting techniques, automation. Sci. Hortic. 2010, 127, 93–105. [Google Scholar] [CrossRef]
- Liu, Q.; Wei, M.; Shen, Q.; Wang, X.F.; Yang, F.J.; Shi, Q.H. Effects of Different Rootstocks on Bloom Formation and Absorption and Distribution of Silicon in Grafted Cucumber. Acta Hortic. Sin. 2012, 39, 897–904. [Google Scholar]
- Miao, L.; Duan, Y.; Wang, C.L.; Gao, Y.; Yu, X.C. A Comprehensive Evaluation of Commodity Fruit Production and Tasty in Grafted Cucumber Using Cucurbita Moschata Germplasms as Rootstocks. J. Nucl. Agric. Sci. 2012, 2, 380–390. [Google Scholar]
- Schwechheimer, C.; Bevan, M. The regulation of transcription factor activity in plants. Trends Plant Sci. 1998, 3, 378–383. [Google Scholar] [CrossRef]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Hatanaka, A. The biogeneration of green odour by green leaves. Phytochemistry 1993, 34, 1201–1218. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Cortesi, M.L.; Vollano, L.; Peruzy, M.F.; Marrone, R.; Mercogliano, R. Determination of nitrate and nitrite levels in infant foods marketed in Southern Italy. CyTA J. Food 2015, 13, 629–634. [Google Scholar] [CrossRef]
- Stachniuk, A.; Szmagara, A.; Stefaniak, E.A. Spectrophotometric Assessment of the Differences Between Total Nitrate/Nitrite Contents in Peel and Flesh of Cucumbers. Food Anal. Methods 2018, 11, 2969–2977. [Google Scholar] [CrossRef] [Green Version]
- Elmstrom, G.W.; Davis, P.L. Sugars in developing and mature fruits of several watermelon cultivars. J. Am. Soc. Hortic. Sci. 1981, 106, 330–333. [Google Scholar] [CrossRef]
- Kader, A.A. Flavor quality of fruits and vegetables. J. Sci. Food Agric. 2008, 88, 1863–1868. [Google Scholar] [CrossRef]
- López-Ortega, G.; García-Montiel, F.; Bayo-Canha, A.; Frutos-Ruiz, C.; Frutos-Tomás, D. Rootstock effects on the growth, yield and fruit quality of sweet cherry cv. ‘Newstar’ in the growing conditions of the Region of Murcia. Sci. Hortic. 2016, 198, 326–335. [Google Scholar] [CrossRef]
- Franz, G.; Hassid, W. Biosynthesis of digitoxose and glucose in the purpurea glycosides of Digitalis purpurea. Phytochemistry 1967, 6, 841–844. [Google Scholar] [CrossRef]
- Buescher, R.; Buescher, R. Production and Stability of (E, Z)-2, 6-Nonadienal, the Major Flavor Volatile of Cucumbers. J. Food Sci. 2001, 66, 357–361. [Google Scholar] [CrossRef]
- Palma-Harris, C.; McFeeters, R.F.; Fleming, H.P. Solid-Phase Microextraction (SPME) Technique for Measurement of Generation of Fresh Cucumber Flavor Compounds. J. Agric. Food Chem. 2001, 49, 4203–4207. [Google Scholar] [CrossRef]
- Upadhyay, A.; Gaonkar, T.; Upadhyay, A.K.; Jogaiah, S.; Shinde, M.P.; Kadoo, N.Y.; Gupta, V.S. Global transcriptome analysis of grapevine (Vitis vinifera L.) leaves under salt stress reveals differential response at early and late stages of stress in table grape cv. Thompson Seedless. Plant Physiol. Biochem. 2018, 129, 168–179. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Boil. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- Allan, A.C. Domestication: Colour and Flavour Joined by a Shared Transcription Factor. Curr. Boil. 2019, 29, R57–R59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Et Biophys. Acta BBA Bioenergy 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Severo, J.; Tiecher, A.; Pirrello, J.; Regad, F.; Latché, A.; Pech, J.-C.; Bouzayen, M.; Rombaldi, C.V. UV-C radiation modifies the ripening and accumulation of ethylene response factor (ERF) transcripts in tomato fruit. Postharvest Boil. Technol. 2015, 102, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L.; Guotian, L.; et al. Genome-wide Identification and Classification of HSF Family in Grape, and Their Transcriptional Analysis under Heat Acclimation and Heat Stress. Hortic. Plant J. 2018, 4, 133–143. [Google Scholar] [CrossRef]
- Nambara, E. Metabolic Balance and its Outcome: Deficiency of Vitamin B9 and Sucrose Supply Ectopically Induces Starch Synthesis in Etioplasts. Plant Cell Physiol. 2017, 58, 1284–1285. [Google Scholar] [CrossRef]
- Pharr, D.M.; Sox, H.N.; Smart, E.L.; Lower, R.L.; Fleming, H.P. Identification and distribution of soluble saccharides in pickling cucumber plants and their fate in fermentation. J. Am. Soc. Hortic. Sci. 1977, 102, 406–409. [Google Scholar]
- Xu, Z.; Zeng, L.; Lalgondar, M.; Bevan, D.; Winkel, B.; Mohamed, A.; Cheng, C.-L.; Shih, M.-C.; Poulton, J.; Esen, A. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Boil. 2004, 55, 343–367. [Google Scholar] [CrossRef]
- Glover, H.; Brady, C. Purification of three pectin esterases from ripe peach fruit. Phytochemistry 1994, 37, 949–955. [Google Scholar] [CrossRef]
- Fils-Lycaon, B.; Buret, M. Changes in glycosidase activities during development and ripening of melon. Postharvest Boil. Technol. 1991, 1, 143–151. [Google Scholar] [CrossRef]
- Iannetta, P.; Valpuesta, V.; Castillejo, C.; De La Fuente, J.I.; Botella, M. Ángel Pectin esterase gene family in strawberry fruit: study of FaPE1, a ripening-specific isoform. J. Exp. Bot. 2004, 55, 909–918. [Google Scholar] [CrossRef]
- Brummell, D.A.; Harpster, M.H. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. In Plant Cell Walls; Springer Science and Business Media: Berlin/Heidelberg, Germany, 2001; pp. 311–340. [Google Scholar]
- Yadav, U.P.; Ivakov, A.; Feil, R.; Duan, G.Y.; Walther, D.; Giavalisco, P.; Piques, M.; Carillo, P.; Hubberten, H.-M.; Stitt, M.; et al. The sucrose–trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 2014, 65, 1051–1068. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Wasternack, C. The Lipoxygenase pathway. Annu. Rev. Plant Boil. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Yemm, E.W.; Cocking, E.C.; Ricketts, R.E. The determination of amino-acids with ninhydrin. Analyst 1955, 80, 209. [Google Scholar] [CrossRef]
- Li, B.X.; Zhang, H.C.; Wang, S.G.; Wang, H.F.; Yang, K.; Liu, W.Q. An improve testing method for determining nitrate contents in fresh vegetable. Plant Physiol. J. 2014, 50, 1749–1752. [Google Scholar] [CrossRef]
- Moradshahi, A.; Vines, H.M.; Black, C.C. Carbon Dioxide Exchange and Acidity Levels in Detached Pineapple, Ananas comosus (L.), Merr., Leaves during the Day at Various Temperatures, Oxygen and Carbon Dioxide Concentrations. Plant Physiol. 1977, 59, 274–278. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Qin, X.; Gao, L.H.; Li, Q.; Li, S.Z.; He, C.X.; Li, Y.S.; Yu, X.C. Selection of reference genes for quantitative real-time PCR analysis in cucumber (Cucumis sativus L.), pumpkin (Cucurbita moschata Duch.) and cucumber–pumpkin grafted plants. PeerJ 2019, 7, e6536. [Google Scholar] [CrossRef]

| Metabolism Classification | Metabolism Name | Candidate Gene Identifier | Log2FC | Functional Annotation | PCC | Combination |
|---|---|---|---|---|---|---|
| Sugars | Fructose-6-phosphate | Csa2G252020 | 1.04 | Glycolysis/gluconeogenesis | 0.94045252 | GNo.45 vs. NG |
| Csa2G252020 | 1.04 | Biosynthesis of amino acids | 0.94045252 | GNo.45 vs. NG | ||
| Csa2G252020 | 1.04 | Carbon metabolism | 0.94045252 | GNo.45 vs. NG | ||
| Csa2G252020 | 1.04 | Pentose phosphate pathway | 0.94045252 | GNo.45 vs. NG | ||
| Csa2G252020 | 1.04 | Fructose and mannose metabolism | 0.94045252 | GNo.45 vs. NG | ||
| Csa2G252020 | 1.04 | Carbon fixation in photosynthetic organisms | 0.94045252 | GNo.45 vs. NG | ||
| Trehalose | Csa3G402970 | 1.09 | Starch and sucrose metabolism | −0.9259965 | GNo.45 vs. GNo.96 | |
| Csa5G568300 | 2.05 | Starch and sucrose metabolism | −0.9785609 | GNo.45 vs. GNo.96 | ||
| Csa7G343850 | 1.42 | Starch and sucrose metabolism | −0.8501167 | GNo.45 vs. GNo.96 | ||
| Csa4G028470 | −1.54 | Starch and sucrose metabolism | 0.99523926 | GNo.45 vs. GNo.96 | ||
| Fatty acids | Csa1G611290 | 1.12 | Starch and sucrose metabolism | −0.9745222 | GNo.45 vs. GNo.96 | |
| Csa6G500540 | −1.04 | Starch and sucrose metabolism | 0.98015624 | GNo.45 vs. GNo.96 | ||
| Amino acids | Linoleic acid | Csa2G023880 | 1.53 | Linoleic acid metabolism | −0.9748707 | GNo.45 vs. NG |
| 4.32 | Linoleic acid metabolism | 0.99350244 | GNo.96 vs. NG | |||
| Isoleucine | Csa6G487590 | −1.04 | Biosynthesis of amino acids | −0.864884 | GNo.45 vs. NG | |
| Csa2G404730 | 1.11 | Biosynthesis of amino acids | 0.96014285 | GNo.45 vs. NG | ||
| Csa2G252020 | 1.04 | Biosynthesis of amino acids | 0.94045252 | GNo.45 vs. NG | ||
| Proline | Csa6G487590 | −1.04 | Biosynthesis of amino acids | −0.864884 | GNo.45 vs. NG | |
| Csa2G404730 | 1.11 | Biosynthesis of amino acids | 0.96014285 | GNo.45 vs. NG | ||
| Csa2G252020 | 1.04 | Biosynthesis of amino acids | 0.94045252 | GNo.45 vs. NG | ||
| Valine | Csa6G487590 | −1.04 | Biosynthesis of amino acids | −0.864884 | GNo.45 vs. NG | |
| Csa2G404730 | 1.11 | Biosynthesis of amino acids | 0.96014285 | GNo.45 vs. NG | ||
| Csa2G252020 | 1.04 | Biosynthesis of amino acids | 0.94045252 | GNo.45 vs. NG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, L.; Di, Q.; Sun, T.; Li, Y.; Duan, Y.; Wang, J.; Yan, Y.; He, C.; Wang, C.; Yu, X. Integrated Metabolome and Transcriptome Analysis Provide Insights into the Effects of Grafting on Fruit Flavor of Cucumber with Different Rootstocks. Int. J. Mol. Sci. 2019, 20, 3592. https://doi.org/10.3390/ijms20143592
Miao L, Di Q, Sun T, Li Y, Duan Y, Wang J, Yan Y, He C, Wang C, Yu X. Integrated Metabolome and Transcriptome Analysis Provide Insights into the Effects of Grafting on Fruit Flavor of Cucumber with Different Rootstocks. International Journal of Molecular Sciences. 2019; 20(14):3592. https://doi.org/10.3390/ijms20143592
Chicago/Turabian StyleMiao, Li, Qinghua Di, Tianshu Sun, Yansu Li, Ying Duan, Jun Wang, Yan Yan, Chaoxing He, Changlin Wang, and Xianchang Yu. 2019. "Integrated Metabolome and Transcriptome Analysis Provide Insights into the Effects of Grafting on Fruit Flavor of Cucumber with Different Rootstocks" International Journal of Molecular Sciences 20, no. 14: 3592. https://doi.org/10.3390/ijms20143592




