Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase
Abstract
1. Introduction
2. Results and Discussion
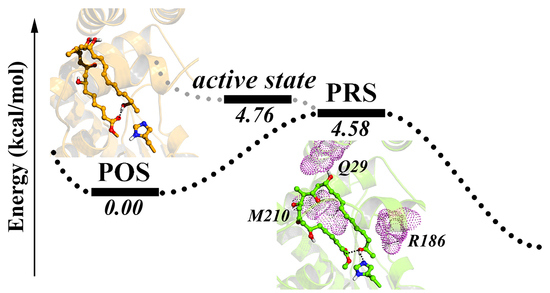
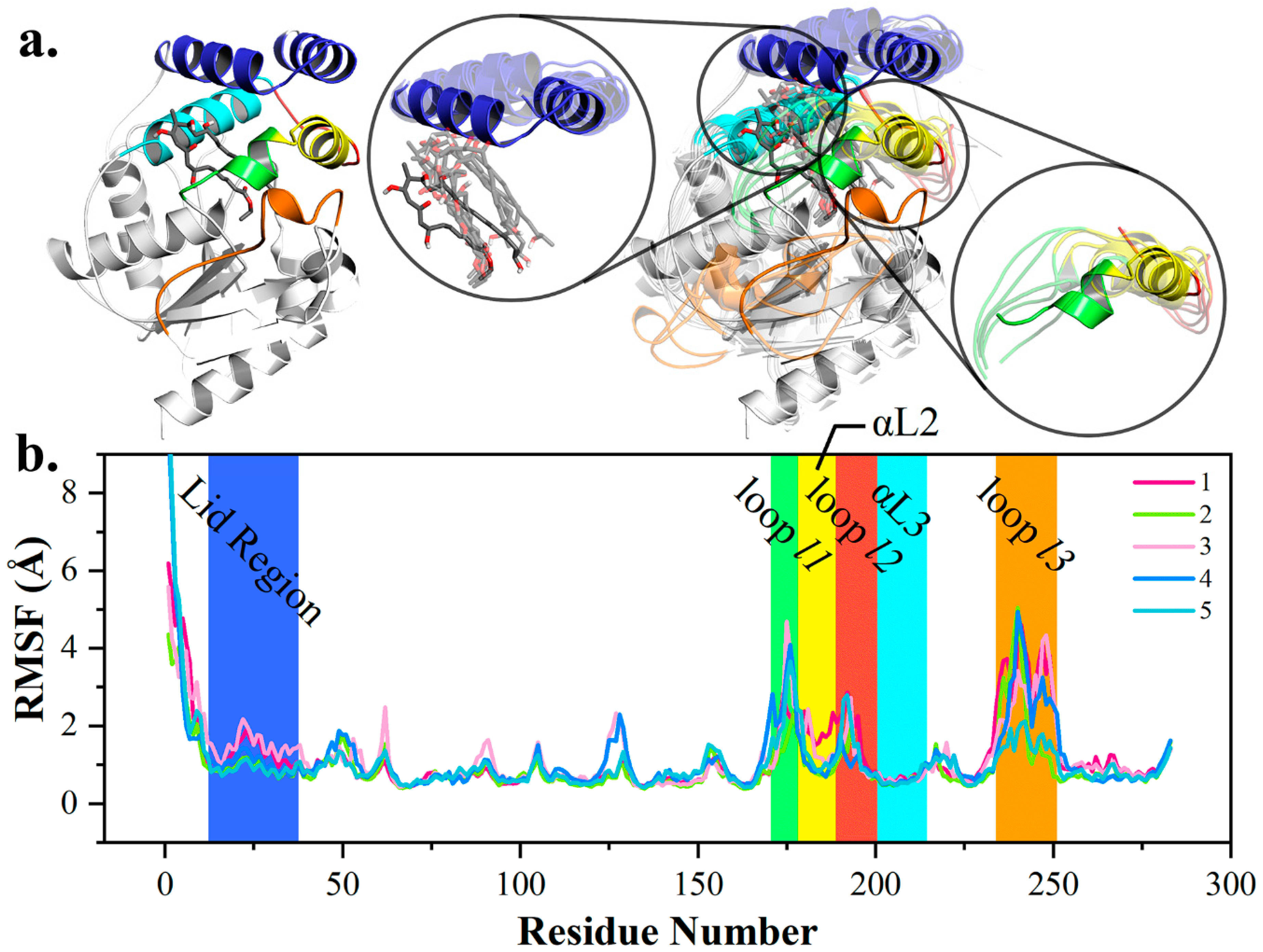
2.1. Key Structural Conformations in MD Simulations
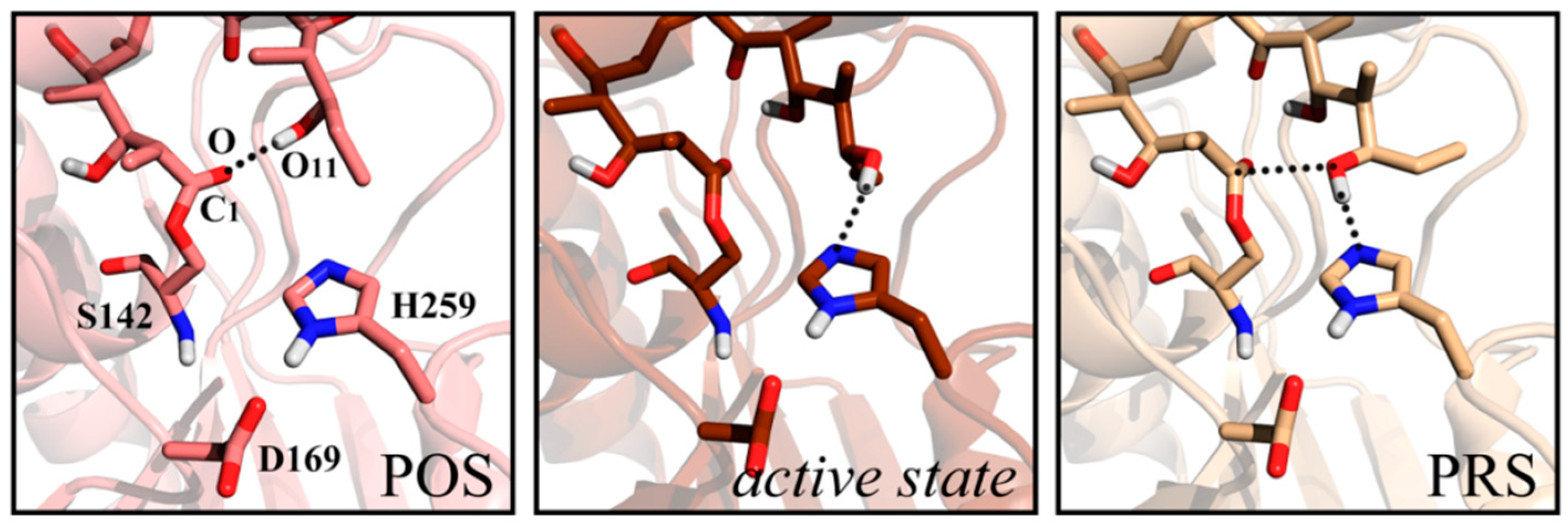
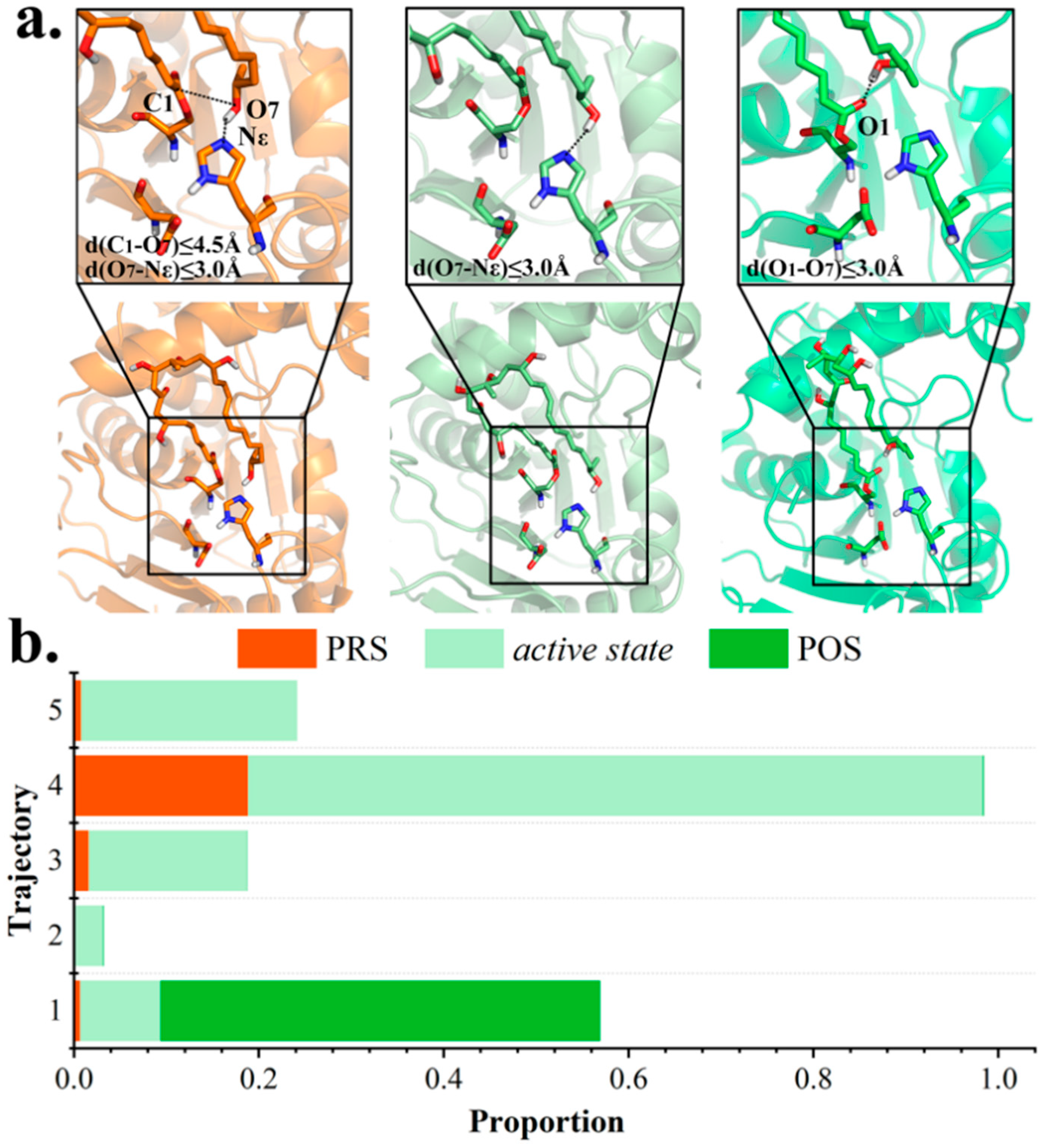
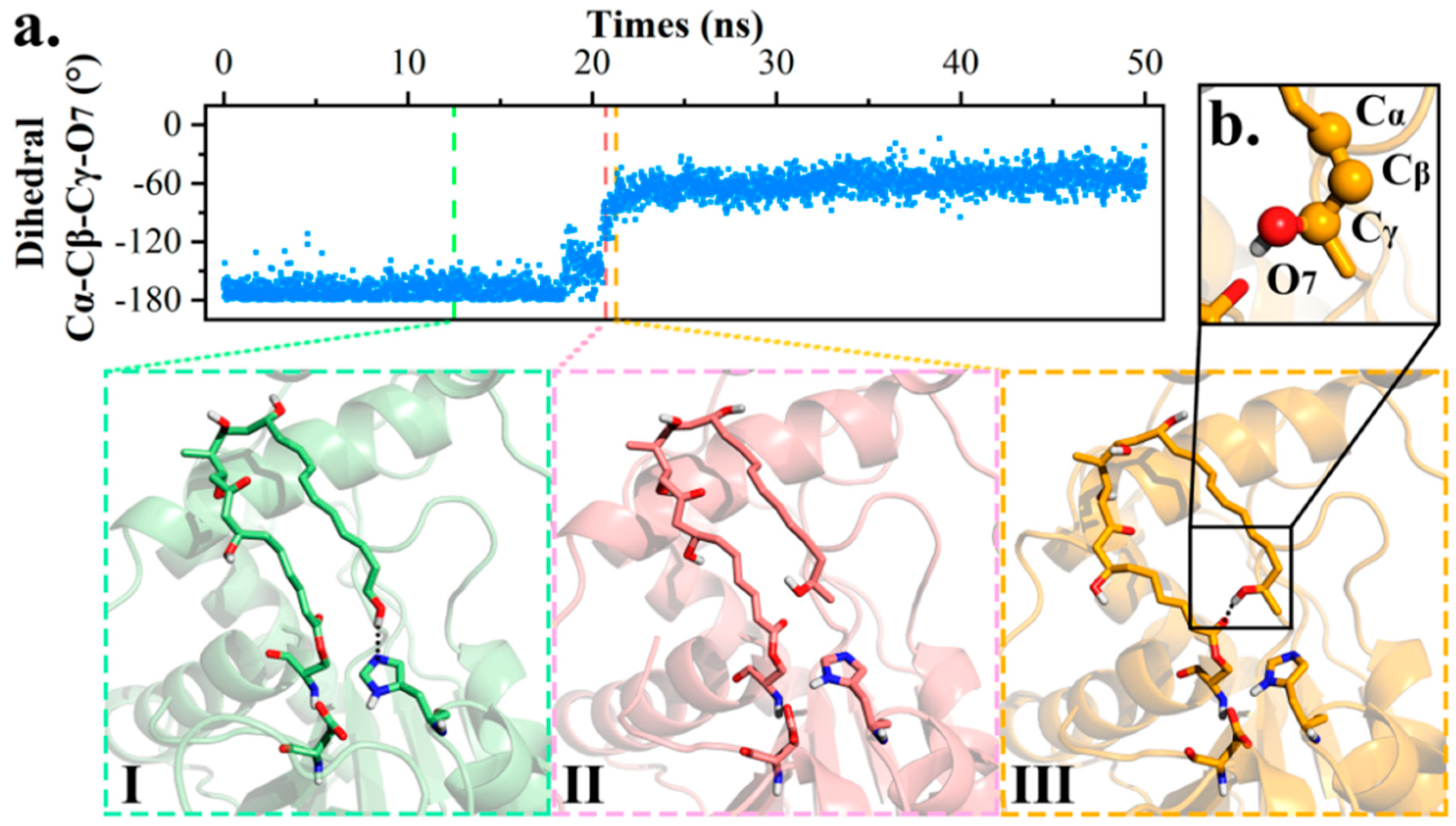
2.2. Conformational Transition between POS and PRS
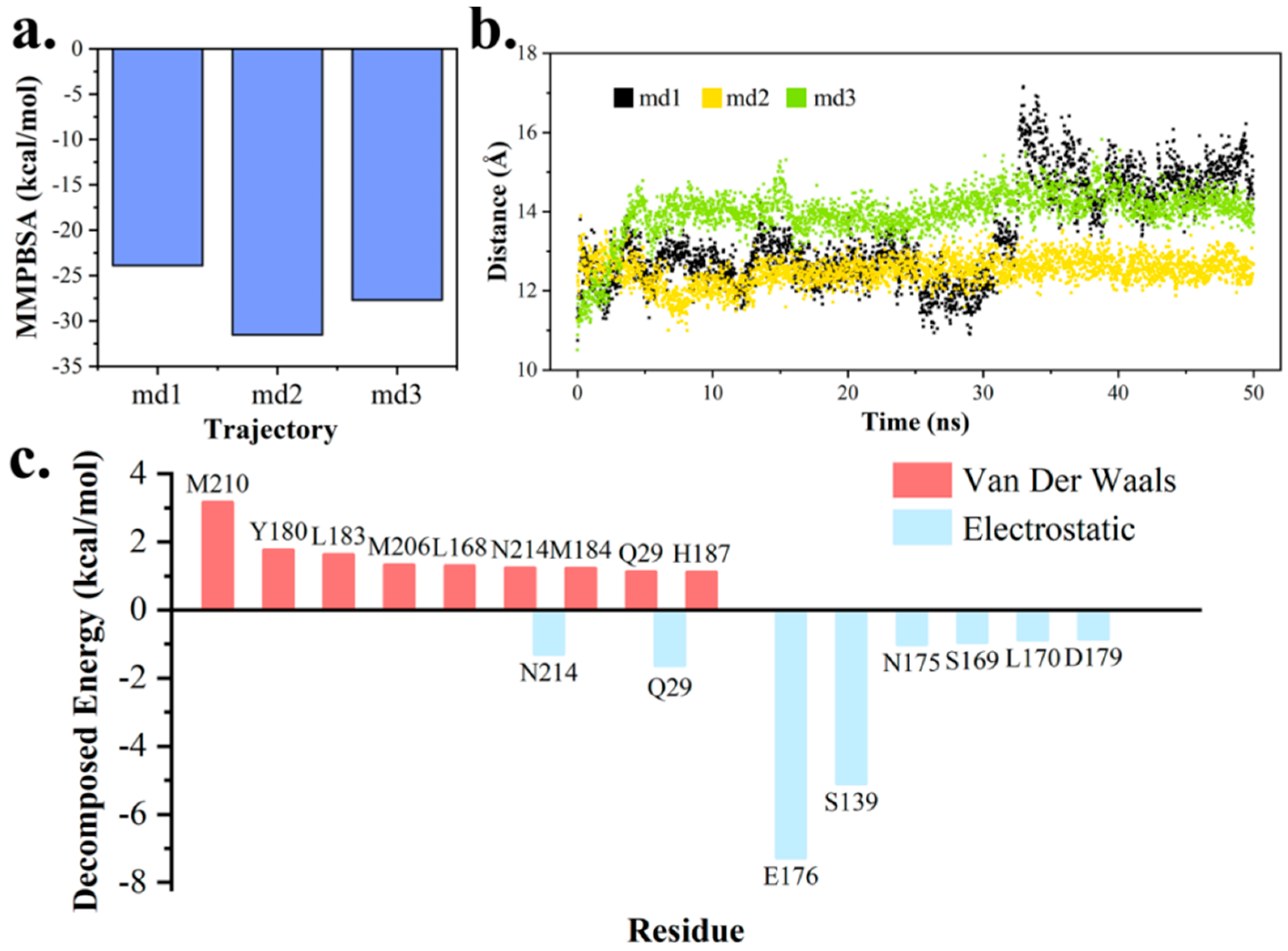
2.3. Hydrophilic and Hydrophobic Interactions in Pima-TE System
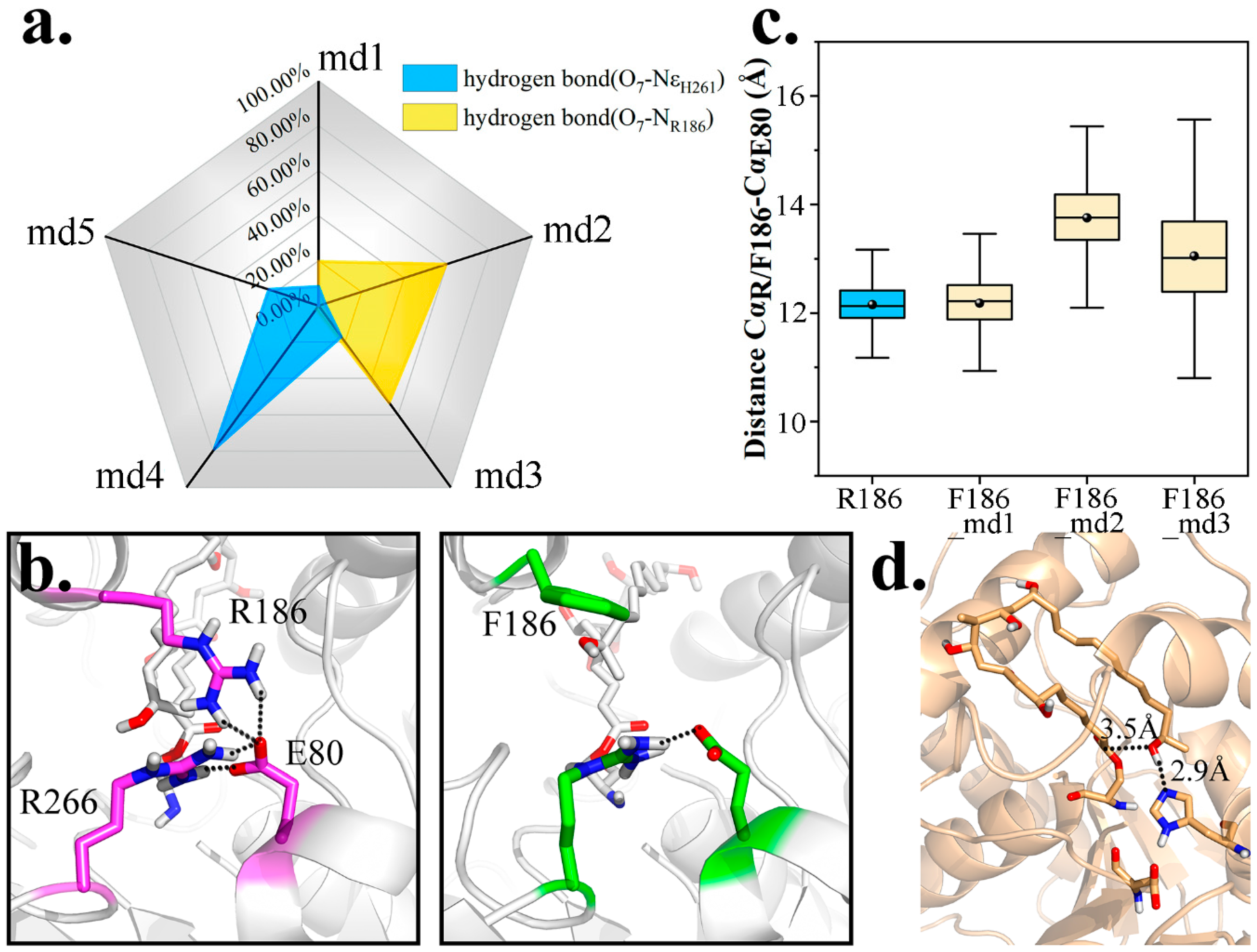
2.4. Key Residues Analyzed Via Mutant Simulations
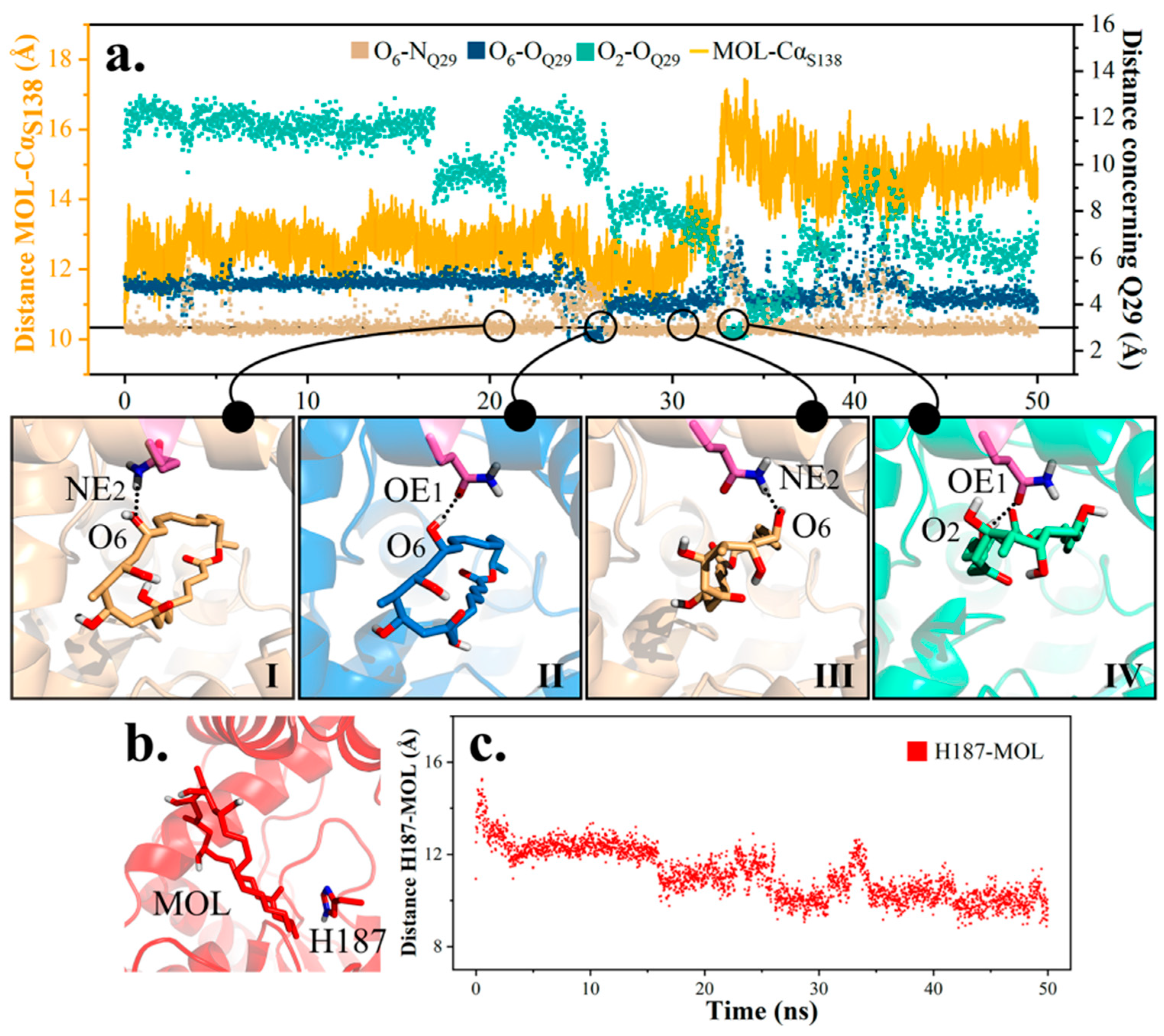
2.4.1. Mutation 1-Q29A
2.4.2. Mutation 2-M210G
2.4.3. Mutation 3-R186F & R186Y
2.4.4. Mutation 4-S138C
2.5. Study on TE’s Effect on the Release of Pimaricin Product
3. Discussion
4. Materials and Methods
4.1. System Preparation
4.2. Molecular Dynamics Simulation
4.3. Quantum Mechanics/Molecular Mechanics) Calculation
4.4. Simulation of Site Mutation Proteins
4.5. Free Energy Calculation and Conformational Stability Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PKS | Polyketide Synthase |
| TE | Thioesterase |
| MD | Molecular Dynamics |
| PRS | Pre-reaction State |
| POS | Pre-organization State |
References
- Gil, J.A.; Martin, J.F. Biotechnology of Antibiotics, 2nd ed.; W. Strohl, M., Ed.; Dekker: New York, NY, USA, 1997. [Google Scholar]
- Aparicio, J.F.; Mendes, M.V.; Anton, N.; Recio, E.; Martin, J.F. Polyene macrolide antibiotic biosynthesis. Curr. Med. Chem. 2004, 11, 1645–1656. [Google Scholar] [CrossRef]
- Szlinder-Richert, J.; Mazerski, J.; Cybulska, B.; Grzybowska, J.; Borowski, E. MFAME, N-methyl-N-d-fructosyl amphotericin B methyl ester, a new amphotericin B derivative of low toxicity: Relationship between self-association and effects on red blood cells. Biochim. Biophys. Acta Gen. Sub. 2001, 1528, 15–24. [Google Scholar] [CrossRef]
- Ogasawara, Y.; Katayama, K.; Minami, A.; Otsuka, M.; Eguchi, T.; Kakinuma, K. Cloning, Sequencing, and Functional Analysis of the Biosynthetic Gene Cluster of Macrolactam Antibiotic Vicenistatin in Streptomyces halstedii. Chem. Biol. 2006, 11, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Cereghetti, D.M.; Carreira, E.M. Amphotericin B: 50 Years of Chemistry and Biochemistry. Synthesis 2006, 37, 914–942. [Google Scholar] [CrossRef]
- Carmody, M.; Murphy, B.; Byrne, B.; Power, P.; Rai, D.; Rawlings, B.; Caffrey, P. Biosynthesis of Amphotericin Derivatives Lacking Exocyclic Carboxyl Groups. J. Biol. Chem. 2005, 280, 34420–34426. [Google Scholar] [CrossRef] [PubMed]
- Gantt, R.W.; Peltierpain, P.; Thorson, J.S. Enzymatic methods for glyco (diversification/randomization) of drugs and small molecules. Nat. Prod. Rep. 2011, 28, 1811–1853. [Google Scholar] [CrossRef]
- Zotchev, S.B. Polyene Macrolide Antibiotics and their Applications in Human Therapy. Curr. Med. Chem. 2003, 10, 211–223. [Google Scholar] [CrossRef]
- Baginski, M.; Czub, J.; Sternal, K. Interaction of amphotericin B and its selected derivatives with membranes: Molecular modeling studies. Chem. Rec. 2010, 6, 320–332. [Google Scholar] [CrossRef]
- Atta, H.M.; Selim, S.M.; Zayed, M.S. Natamycin antibiotic produced by Streptomyces sp.: Fermentation, purification and biological activities. J. Ame. Sci. 2012, 8, 469–475. [Google Scholar]
- Stark, J. Natamycin: An effective fungicide for food and beverages. Nat. Antimicrobials Minim. Process. Foods 2003, 82–97. [Google Scholar] [CrossRef]
- Austin, A.; Lietman, T.; Rose-nussbaumer, J. Update on the management of infectious keratitis. Ophthalmology 2017, 124, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
- Priya, A.B.; Kalyan, M. In vitro leishmanicidal effects of the anti-fungal drug natamycin are mediated through disruption of calcium homeostasis and mitochondrial dysfunction. Apoptosis 2018, 23, 420–435. [Google Scholar] [CrossRef]
- Te Welscher, Y.M.; Jones, L.; Van Leeuwen, M.R.; Dijksterhuis, J.; de Kruijff, B.; Eitzen, G.; Breukink, E. Natamycin Inhibits Vacuole Fusion at the Priming Phase via a Specific Interaction with Ergosterol. Antimicrob. Agents Chemother. 2010, 54, 2618–2625. [Google Scholar] [CrossRef] [PubMed]
- Tanner, W. Membrane transport inhibition as mode of action of polyene antimycotics: Recent data supported by old ones. Food Technol. Biotechnol. 2014, 52, 8–12. [Google Scholar] [CrossRef]
- Van Leeuwen, M.R.; Golovina, E.A.; Dijksterhuis, J. The polyene antimycotics nystatin and filipin disrupt the plasma membrane, whereas natamycin inhibits endocytosis in germinating conidia of Penicillium discolor. J. Appl. Microbiol. 2009, 106, 1908–1918. [Google Scholar] [CrossRef] [PubMed]
- Mccall, L.I.; Aroussi, A.E.; Choi, J.Y.; Vieira, D.F.; Muylder, G.D.; Johnston, J.B.; Chen, S.; Kellar, D.; Siqueira-Neto, J.L.; Roush, W.R.; et al. Targeting Ergosterol Biosynthesis in Leishmania donovani: Essentiality of Sterol 14 alpha-demethylase. PLoS Negl. Trop. Dis. 2015, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Whicher, J.R.; Hansen, D.A.; Hansen, W.A.; Chelmer, J.A.; Congdon, G.R.; Alison, R.H.N.; Kristina, H.; Sherman, D.H.; Smith, J.L.; Skiniotis, G. Structure of a modular polyketide synthase. Nature 2014, 510, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Skiba, M.A.; Sikkema, A.P.; Fiers, W.D.; Gerwick, W.H.; Sherman, D.H.; Aldrich, C.C.; Smith, J.L. Domain Organization and Active Site Architecture of a Polyketide Synthase C-methyltransferase. ACS Chem. Biol. 2016, 11, 3319–3327. [Google Scholar] [CrossRef]
- Curran, S.C.; Hagen, A.; Poust, S.; Chan, L.J.G.; Garabedian, B.M.; Rond, T.; Baluyot, M.J.; Vu, J.T.; Lau, A.K.; Yuzawa, S.; et al. Probing the flexibility of an iterative modular polyketide synthase with non-native substrates in vitro. ACS Chem. Biol. 2018, 13, 2261–2268. [Google Scholar] [CrossRef]
- Rittner, A.; Paithankar, K.S.; Vu, K.H.; Grininger, M. Characterization of the polyspecific transferase of murine type I fatty acid synthase (FAS) and implications for polyketide synthase (PKS) engineering. ACS Chem. Biol. 2018, 13, 723–732. [Google Scholar] [CrossRef]
- Ferscht, A. Enzyme Structure and Mechanism, 2nd ed.; W. H. Freeman and Company: New York, NY, USA, 1985. [Google Scholar] [CrossRef]
- Kormana, T.P.; Crawfordb, J.M.; Labonteb, J.W.; Newmanb, A.G.; Wongc, J.; Townsendb, C.A.; Tsaia, S.C. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 6246–6251. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.P.; Shi, T.; Wang, X.L.; Wang, J.T.; Chen, Q.H.; Bai, L.Q.; Zhao, Y.L. Theoretical studies on the Mechanism of Thioesterase-catalyzed Macrocyclization in Erythromycin Biosynthesis. ACS Catal. 2016, 6, 4369–4378. [Google Scholar] [CrossRef]
- Trauger, J.W.; Kohli, R.M.; Walsh, C.T. Cyclization of Backbone-Substituted Peptides Catalyzed by the Thioesterase Domain from the Tyrocidine Nonribosomal Peptide Synthetase. Biochemistry 2001, 40, 7092–7098. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, S.; Murthy, M.R.N. Analysis of temperature factor distribution in high-resolution protein structures. Protein Sci. 1997, 6, 2561–2567. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.Y.; Shen, H.B. Robust Prediction of B-Factor Profile from Sequence Using Two-Stage SVR Based on Random Forest Feature Selection. Protein Peptide Lett. 2009, 16, 1447–1454. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Koch, A.A.; Hansen, D.A.; Shende, V.V.; Furan, L.R.; Houk, K.N.; Gonzalo Jiménez-Osés, G.; Sherman., D.H. A Single Active Site Mutation in the Pikromycin Thioesterase Generates a More Effective Macrocyclization Catalyst. J. Am. Chem. Soc. 2017, 139, 13456–13465. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F.W. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Onur, S.; Pemra, O. gRINN: A tool for calculation of residue interaction energies and protein energy network analysis of molecular dynamics simulations. Nucleic Acids Res. 2018, 46, W554–W562. [Google Scholar] [CrossRef]
- Li, J.; Sun, R.; Wu, Y.H.; Song, M.Z.; Li, J.; Yang, Q.Y.; Chen, X.Y.; Bao, J.K.; Zhao, Q. L1198F Mutation Resensitizes Crizotinib to ALK by Altering the Conformation of Inhibitor and ATP Binding Sites. Int. J. Mol. Sci. 2017, 18, 482. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Gokey, T.; Ting, D.; He, Z.H.; Guliaev, A.B. Dimerization misalignment in human glutamate-oxaloacetate transaminase variants is the primary factor for PLP release. PLOS ONE 2018, 13, e0203889. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.P.; Liu, G.J.; Zhou, H.Y.; Fang, X.; Fang, Y.; Wu, J.H. Computer prediction of paratope on antithrombotic antibody 10B12 and epitope on platelet glycoprotein VI via molecular dynamics simulation. BioMed. Eng. OnLine 2016, 15, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Liu, L.X.; Tao, W.T.; Luo, S.G.; Fan, S.B.; Wang, X.L.; Bai, L.Q.; Zhao, Y.L. Theoretical studies on the Catalytic Mechanism and Substrate Diversity for Macrocyclization of Pikromycin Thioesterase. ACS Catal. 2018, 8, 4323–4332. [Google Scholar] [CrossRef]
- Giraldes, J.W.; Akey, D.L.; Kittendorf, J.D.; Sherman, D.H.; Smith, J.L.; Fecik, R.A. Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels. Nat. Chem. Biol. 2006, 2, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Accelrys Discovery Studio Visualizer 3.5; Accelerys Software Inc.: San Diego, CA, USA, 2005.
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Amber 2014; Univeristy of California: San Francisco, CA, USA, 2014.
- Jakalian, A.; Bush, B.L.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic Charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000, 21, 132–146. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Gaussian 09; Revision A.02; Gaussian Inc.: Wallingford, CT, USA, 2009.
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.M.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. J. Chem. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Vreven, T.; Byun, K.S.; Komáromi, I.; Dapprich, S.; Montgomery, J.A., Jr.; Morokuma, K.; Frisch, M.J. Combining quantum mechanics methods with molecular mechanics methods in ONIOM. J. Chem. Theory Comput. 2006, 2, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Vreven, T.; Frisch, M.; Kudin, K.; Schlegel, H.; Morokuma, K. Geometry optimization with QM/MM methods II: Explicit quadratic coupling. Mol. Phys. 2006, 104, 701–714. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Rassolov, V.A.; Ratner, M.A.; Pople, J.A.; Redfern, P.C.; Curtiss, L.A. 6-31G* Basis Set for Third-Row Atoms. J. Comput. Chem. 2001, 22, 976–984. [Google Scholar] [CrossRef]
- Swanson, J.M.J.; Henchman, R.H.; McCammon, J.A. Revisiting free energy calculations: A theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophys. J. 2004, 86, 67–74. [Google Scholar] [CrossRef]




| Substrate Type | Name | System | No. of Runs Per Complex | Length Per Run (ns) |
|---|---|---|---|---|
| Polyketide Chain | wild type | pima-TEWT + polyketide chain | 5 | 50 |
| M210G | pima-TEM210G + polyketide chain | 3 | 30 | |
| Q29A | pima-TEQ29A + polyketide chain | 3 | 30 | |
| R186F | pima-TER186F + polyketide chain | 3 | 30 | |
| R186Y | pima-TER186Y + polyketide chain | 3 | 30 | |
| S138C | pima-TES138C + polyketide chain | 3 | 50 | |
| Product | ring | pima-TEWT + product | 3 | 50 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Wang, R.; Li, C.; Bai, L.; Zhao, Y.-L.; Shi, T. Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase. Int. J. Mol. Sci. 2019, 20, 877. https://doi.org/10.3390/ijms20040877
Fan S, Wang R, Li C, Bai L, Zhao Y-L, Shi T. Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase. International Journal of Molecular Sciences. 2019; 20(4):877. https://doi.org/10.3390/ijms20040877
Chicago/Turabian StyleFan, Shuobing, Rufan Wang, Chen Li, Linquan Bai, Yi-Lei Zhao, and Ting Shi. 2019. "Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase" International Journal of Molecular Sciences 20, no. 4: 877. https://doi.org/10.3390/ijms20040877
APA StyleFan, S., Wang, R., Li, C., Bai, L., Zhao, Y.-L., & Shi, T. (2019). Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase. International Journal of Molecular Sciences, 20(4), 877. https://doi.org/10.3390/ijms20040877