Negative Regulation of ULK1 by microRNA-106a in Autophagy Induced by a Triple Drug Combination in Colorectal Cancer Cells In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Drugs, and Transfection
2.2. Protein Expression Analysis
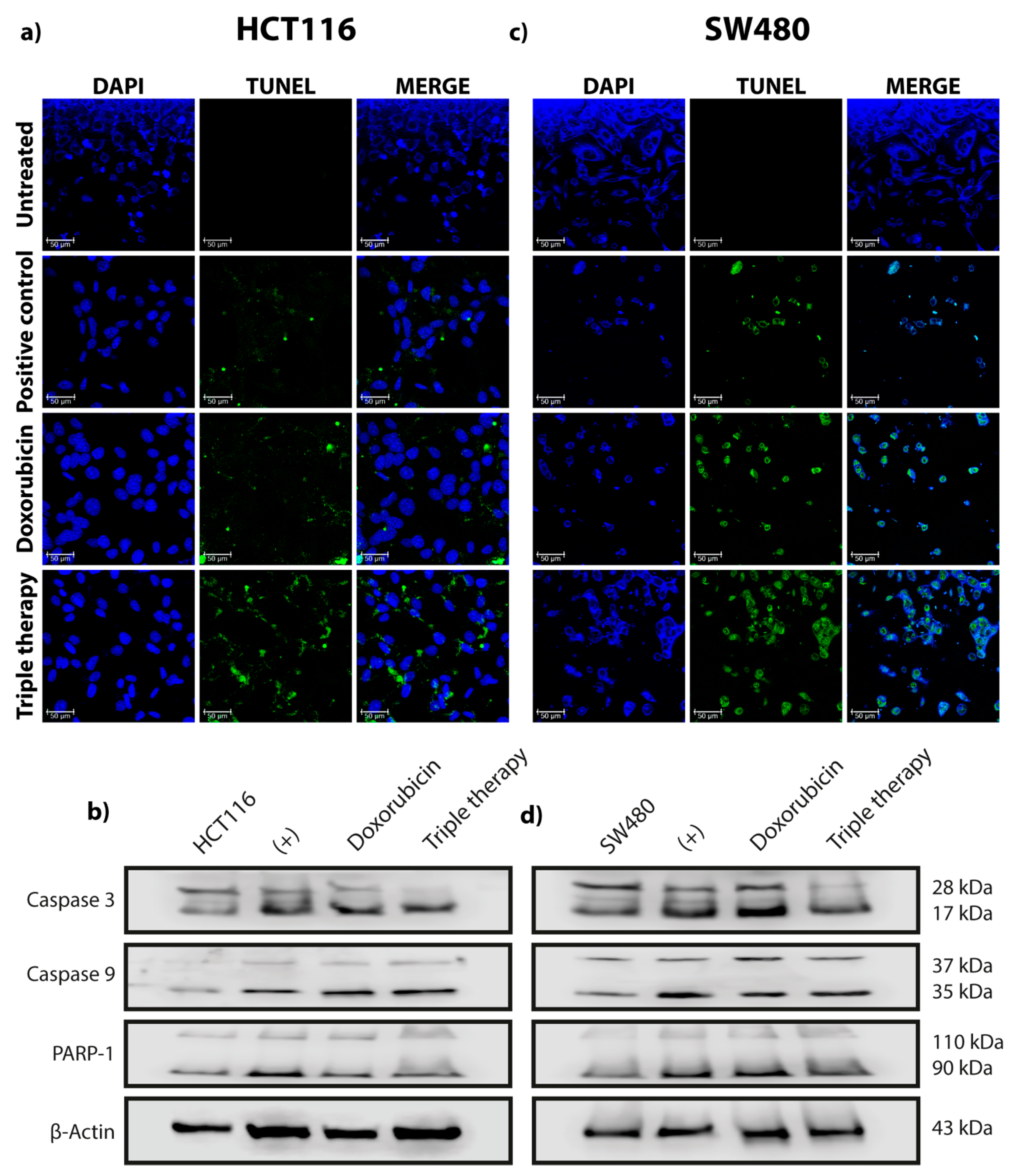
2.3. TUNEL Assay
2.4. qRT-PCR
2.5. Luciferase Reporter Assays
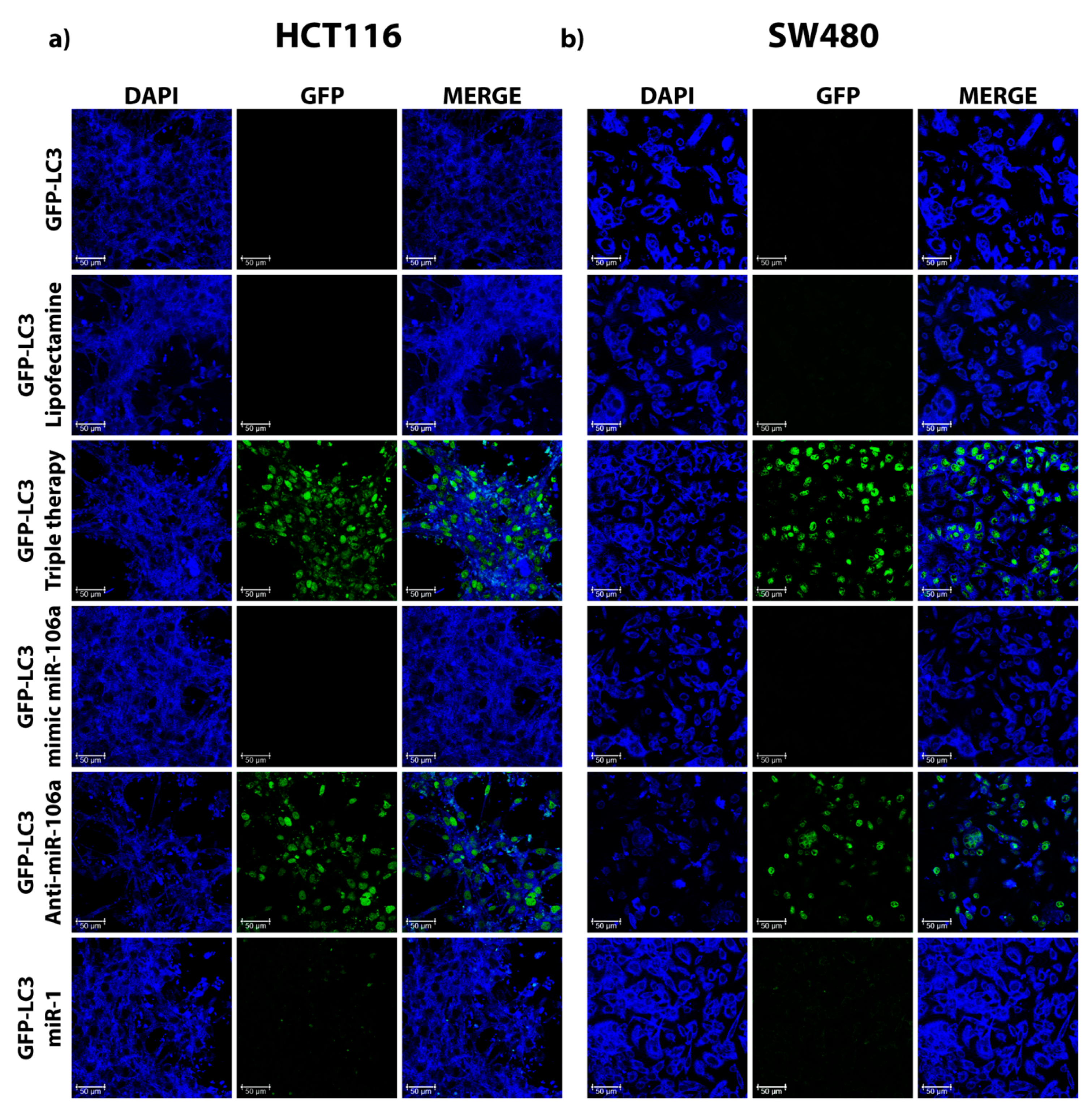
2.6. Monitoring Autophagy by the Formation of GFP-LC3 Puncta
2.7. Statistical Analysis
3. Results
3.1. Triple Therapy Promotes Autophagy in CRC Cells
3.2. ULK1 Is Negatively Regulated by miR-106a in HCT116 CRC Cells
3.3. miR-106a Inhibition Induces the Formation of GFP-LC3 Puncta in HCT116 and SW480 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, C.M.; Yu, L.Y.; Lv, J.F.; Gong, L.; Zhou, H.H.; Chen, X.P.; Fan, L. Effects of immuno-related gene polymorphisms on a bispecific antibody targeting colorectal cancer cell. Per. Med. 2018, 15, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Huo, T.; Canepa, R.; Sura, A.; Modave, F.; Gong, Y. Colorectal cancer stages transcriptome analysis. PLoS ONE 2017, 12, e188697. [Google Scholar] [CrossRef] [PubMed]
- Gelibter, A.J.; Caponnetto, S.; Urbano, F.; Emiliani, A.; Scagnoli, S.; Sirgiovanni, G.; Napoli, V.M.; Cortesi, E. Adjuvant chemotherapy in resected colon cancer: When, how and how long? Surg. Oncol. 2019, 30, 100–107. [Google Scholar] [CrossRef]
- Meyers, B.M.; Cosby, R.; Quereshy, F.; Jonker, D. Adjuvant Chemotherapy for Stage II and III Colon Cancer Following Complete Resection: A Cancer Care Ontario Systematic Review. Clin. Oncol. 2017, 29, 459–465. [Google Scholar] [CrossRef]
- Dehal, A.; Graff-Baker, A.N.; Vuong, B.; Fischer, T.; Klempner, S.J.; Chang, S.C.; Grunkemeier, G.L.; Bilchik, A.J.; Goldfarb, M. Neoadjuvant Chemotherapy Improves Survival in Patients with Clinical T4b Colon Cancer. J. Gastrointest. Surg. 2018, 22, 242–249. [Google Scholar] [CrossRef]
- Clarke, A.J.; Simon, A.K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 2019, 19, 170–183. [Google Scholar] [CrossRef]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Rusmini, P.; Cortese, K.; Crippa, V.; Cristofani, R.; Cicardi, M.E.; Ferrari, V.; Vezzoli, G.; Tedesco, B.; Meroni, M.; Messi, E.; et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy 2019, 15, 631–651. [Google Scholar] [CrossRef] [PubMed]
- Cristofani, R.; Montagnani Marelli, M.; Cicardi, M.E.; Fontana, F.; Marzagalli, M.; Limonta, P.; Poletti, A.; Moretti, R.M. Dual role of autophagy on docetaxel-sensitivity in prostate cancer cells. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef]
- Yazdani, H.; Huang, H.; Tsung, A. Autophagy: Dual Response in the Development of Hepatocellular Carcinoma. Cells 2019, 8, 91. [Google Scholar] [CrossRef]
- García-Castillo, V.; López-Urrutia, E.; Villanueva-Sánchez, O.; Ávila-Rodríguez, M.A.; Zentella-Dehesa, A.; Cortés-González, C.; López-Camarillo, C.; Jacobo-Herrera, N.J.; Pérez-Plasencia, C. Targeting metabolic remodeling in triple negative breast cancer in a murine model. J. Cancer 2017, 8, 178–189. [Google Scholar] [CrossRef]
- Sesen, J.; Dahan, P.; Scotland, S.J.; Saland, E.; Dang, V.T.; Lemarié, A.; Tyler, B.M.; Brem, H.; Toulas, C.; Moyal, E.C.J.; et al. Metformin inhibits growth of human glioblastoma cells and enhances therapeutic response. PLoS ONE 2015, 10, e123721. [Google Scholar] [CrossRef]
- Zhao, Z.; Han, F.; Yang, S.; Wu, J.; Zhan, W. Oxamate-mediated inhibition of lactate dehydrogenase induces protective autophagy in gastric cancer cells: Involvement of the Akt-mTOR signaling pathway. Cancer Lett. 2015, 358, 17–26. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z.; Zhang, W.; Zhang, L. MicroRNAs play an essential role in autophagy regulation in various disease phenotypes. BioFactors 2019, 45, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ventura, A.; Jacks, T. MicroRNAs and Cancer: Short RNAs Go a Long Way. Cell 2009, 136, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Han, W.; Sui, X.; Xie, J.; Pan, H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015, 356, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, B.; Wang, H.Y.; Chang, A.; Zheng, X.F.S. Emerging Role of MicroRNAs in mTOR Signaling. Cell. Mol. Life Sci. 2017, 74, 2613–2625. [Google Scholar] [CrossRef]
- Hao, H.; Xia, G.; Wang, C.; Zhong, F.; Liu, L.; Zhang, D. miR-106a suppresses tumor cells death in colorectal cancer through targeting ATG7. Med. Mol. Morphol. 2017, 50, 76–85. [Google Scholar] [CrossRef]
- Tschan, M.P.; Jost, M.; Batliner, J.; Fey, M.F. The Autophagy Gene ULK1 Plays a Role in AML Differentiation and Is Negatively Regulated by the Oncogenic MicroRNA 106a. Blood 2010, 116, 503. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, Q.; Gu, M.; Zhang, K.; Xia, T.; Zhang, S.; Chen, W.; Yin, H.; Yao, H.; Fan, Y.; et al. MIR106A-5p upregulation suppresses autophagy and accelerates malignant phenotype in nasopharyngeal carcinoma. Autophagy 2020, 1–17. [Google Scholar] [CrossRef]
- Peng, Q.; Shen, Y.; Zhao, P.; Cheng, M.; Zhu, Y.; Xu, B. Biomarker roles identification of miR-106 family for predicting the risk and poor survival of colorectal cancer. BMC Cancer 2020, 20. [Google Scholar] [CrossRef]
- Figueroa-González, G.; García-Castillo, V.; Coronel-Hernández, J.; López-Urrutia, E.; León-Cabrera, S.; Arias-Romero, L.E.; Terrazas, L.; Rodríguez-Sosa, M.; Campos-Parra, A.D.; Zúñiga-Calzada, E.; et al. Anti-inflammatory and antitumor activity of a triple therapy for a colitis-related colorectal cancer. J. Cancer 2016, 7, 1632–1644. [Google Scholar] [CrossRef][Green Version]
- ATCC: The Global Bioresource Center. Available online: https://www.atcc.org/ (accessed on 22 October 2020).
- Kleivi, K.; Teixeira, M.R.; Eknaes, M.; Diep, C.B.; Jakobsen, K.S.; Hamelin, R.; Lothe, R.A. Genome signatures of colon carcinoma cell lines. Cancer Genet. Cytogenet. 2004, 155, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; Van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, D92–D97. [Google Scholar] [CrossRef]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknæs, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Skarkova, V.; Kralova, V.; Vitovcova, B.; Rudolf, E. Selected Aspects of Chemoresistance Mechanisms in Colorectal Carcinoma—A Focus on Epithelial-to-Mesenchymal Transition, Autophagy, and Apoptosis. Cells 2019, 8, 234. [Google Scholar] [CrossRef]
- Zhang, G.J.; Li, J.S.; Zhou, H.; Xiao, H.X.; Li, Y.; Zhou, T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J. Exp. Clin. Cancer Res. 2015, 34, 73. [Google Scholar] [CrossRef]
- Tacar, O.; Dass, C.R. Doxorubicin-induced death in tumour cells and cardiomyocytes: Is autophagy the key to improving future clinical outcomes? J. Pharm. Pharmacol. 2013, 65, 1577–1589. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhou, P.; Xu, K.; Chen, T.; Jiao, J.; Wei, H.; Yang, X.; Xu, W.; Wan, W.; Xiao, J. Metformin induces cell cycle arrest, apoptosis and autophagy through ROS/JNK signaling pathway in human osteosarcoma. Int. J. Biol. Sci. 2020, 16, 74–84. [Google Scholar] [CrossRef]
- Horakova, O.; Kroupova, P.; Bardova, K.; Buresova, J.; Janovska, P.; Kopecky, J.; Rossmeisl, M. Metformin acutely lowers blood glucose levels by inhibition of intestinal glucose transport. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Seliger, C.; Leukel, P.; Moeckel, S.; Jachnik, B.; Lottaz, C.; Kreutz, M.; Brawanski, A.; Proescholdt, M.; Bogdahn, U.; Bosserhoff, A.K.; et al. Lactate-modulated induction of THBS-1 activates Transforming growth factor (TGF)-beta2 and migration of glioma cells in vitro. PLoS ONE 2013, 8, e78935. [Google Scholar] [CrossRef]
- Yang, Y.; Su, D.; Zhao, L.; Zhang, D.; Xu, J.; Wan, J.; Fan, S.; Chen, M. Different effects of LDH-A inhibition by oxamate in non-small cell lung cancer cells. Oncotarget 2014, 5, 11886–11896. [Google Scholar] [CrossRef] [PubMed]
- Valvona, C.J.; Fillmore, H.L. Oxamate, but not selective targeting of LDH-A, inhibits medulloblastoma cell glycolysis, growth and motility. Brain Sci. 2018, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Qi, J.; Sun, X.; Wang, W.; Wei, G.; Wu, Y.; Gao, Q.; Zheng, J. MicroRNA-181a promotes cell proliferation and inhibits apoptosis in gastric cancer by targeting RASSF1A. Oncol. Rep. 2018, 40, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Li, H.; Zhang, T.; Lu, J.; Zhong, L. MicroRNA-4530 suppresses cell proliferation and induces apoptosis by targeting RASA1 in human umbilical vein endothelial cells. Mol. Med. Rep. 2019, 49, 3393–3402. [Google Scholar] [CrossRef]
- Mens, M.M.J.; Ghanbari, M. Cell Cycle Regulation of Stem Cells by MicroRNAs. Stem Cell Rev. Rep. 2018, 14, 309–322. [Google Scholar] [CrossRef]
- Pourhanifeh, M.H.; Mahjoubin-Tehran, M.; Karimzadeh, M.R.; Mirzaei, H.R.; Razavi, Z.S.; Sahebkar, A.; Hosseini, N.; Mirzaei, H.; Hamblin, M.R. Autophagy in cancers including brain tumors: Role of MicroRNAs. Cell Commun. Signal. 2020, 18, 88. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, N.; Liu, P.; Chen, Q.; Situ, H.; Xie, T.; Zhang, J.; Peng, C.; Lin, Y.; Chen, J. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget 2014, 5, 7013–7026. [Google Scholar] [CrossRef]
- Wu, H.; Wang, F.; Hu, S.; Yin, C.; Li, X.; Zhao, S.; Wang, J.; Yan, X. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell. Signal. 2012, 24, 2179–2186. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chuang, A.Y.; Ratovitski, E.A. Phospho-ΔNp63α/miR-885-3p axis in tumor cell life and cell death upon cisplatin exposure. Cell Cycle 2011, 10, 3938–3947. [Google Scholar] [CrossRef] [PubMed]
- Tormo, E.; Pineda, B.; Serna, E.; Guijarro, A.; Ribas, G.; Fores, J.; Chirivella, E.; Climent, J.; Lluch, A.; Eroles, P. MicroRNA Profile in Response to Doxorubicin Treatment in Breast Cancer. J. Cell. Biochem. 2015, 116, 2061–2073. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, S.; Zhang, L.; Xie, T.; Song, S.; Huang, J.; Zhang, Y.; Ouyang, L.; Liu, B. Identification of ULK1 as a novel biomarker involved in MIR-4487 and MIR-595 regulation in neuroblastoma SH-SY5Y cell autophagy. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gozuacik, D.; Akkoc, Y.; Gulfem Ozturk, D.; Kocak, M. Autophagy-Regulating microRNAs and Cancer. Front. Oncol. 2017, 7. [Google Scholar] [CrossRef]
- Rothschild, S.I.; Gautschi, O.; Batliner, J.; Gugger, M.; Fey, M.F.; Tschan, M.P. MicroRNA-106a targets autophagy and enhances sensitivity of lung cancer cells to Src inhibitors. Lung Cancer 2017, 107, 73–83. [Google Scholar] [CrossRef]
- Li, N.; Li, X.; Li, S.; Zhou, S.; Zhou, Q. Cisplatin-induced downregulation of SOX1 increases drug resistance by activating autophagy in non-small cell lung cancer cell. Biochem. Biophys. Res. Commun. 2013, 439, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-C.; Huang, F.-Z.; Xu, H.-B.; Sun, J.-C.; Wang, C.-F. MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic cancer cells by targeting ATG5. Int. J. Biochem. Cell Biol. 2019, 111, 63–71. [Google Scholar] [CrossRef]
- Mitrakas, A.G.; Kalamida, D.; Giatromanolaki, A.; Pouliliou, S.; Tsolou, A.; Kyranas, R.; Koukourakis, M.I. Autophagic flux response and glioblastoma sensitivity to radiation. Cancer Biol. Med. 2018, 15, 260–274. [Google Scholar] [CrossRef]
- Qadir, M.A.; Kwok, B.; Dragowska, W.H.; To, K.H.; Le, D.; Bally, M.B.; Gorski, S.M. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res. Treat. 2008, 112, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Samaddar, J.S.; Gaddy, V.T.; Duplantier, J.; Thandavan, S.P.; Shah, M.; Smith, M.J.; Browning, D.; Rawson, J.; Smith, S.B.; Barrett, J.T.; et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol. Cancer Ther. 2008, 7, 2977–2987. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salgado-García, R.; Coronel-Hernández, J.; Delgado-Waldo, I.; Cantú de León, D.; García-Castillo, V.; López-Urrutia, E.; Gutiérrez-Ruiz, M.C.; Pérez-Plasencia, C.; Jacobo-Herrera, N. Negative Regulation of ULK1 by microRNA-106a in Autophagy Induced by a Triple Drug Combination in Colorectal Cancer Cells In Vitro. Genes 2021, 12, 245. https://doi.org/10.3390/genes12020245
Salgado-García R, Coronel-Hernández J, Delgado-Waldo I, Cantú de León D, García-Castillo V, López-Urrutia E, Gutiérrez-Ruiz MC, Pérez-Plasencia C, Jacobo-Herrera N. Negative Regulation of ULK1 by microRNA-106a in Autophagy Induced by a Triple Drug Combination in Colorectal Cancer Cells In Vitro. Genes. 2021; 12(2):245. https://doi.org/10.3390/genes12020245
Chicago/Turabian StyleSalgado-García, Rebeca, Jossimar Coronel-Hernández, Izamary Delgado-Waldo, David Cantú de León, Verónica García-Castillo, Eduardo López-Urrutia, Ma. Concepción Gutiérrez-Ruiz, Carlos Pérez-Plasencia, and Nadia Jacobo-Herrera. 2021. "Negative Regulation of ULK1 by microRNA-106a in Autophagy Induced by a Triple Drug Combination in Colorectal Cancer Cells In Vitro" Genes 12, no. 2: 245. https://doi.org/10.3390/genes12020245
APA StyleSalgado-García, R., Coronel-Hernández, J., Delgado-Waldo, I., Cantú de León, D., García-Castillo, V., López-Urrutia, E., Gutiérrez-Ruiz, M. C., Pérez-Plasencia, C., & Jacobo-Herrera, N. (2021). Negative Regulation of ULK1 by microRNA-106a in Autophagy Induced by a Triple Drug Combination in Colorectal Cancer Cells In Vitro. Genes, 12(2), 245. https://doi.org/10.3390/genes12020245








