Survey of SNPs Associated with Total Number Born and Total Number Born Alive in Pig
Abstract
1. Introduction
2. SNPs Associated with Total Number Born Alive
3. SNPs Associated with Total Number Born
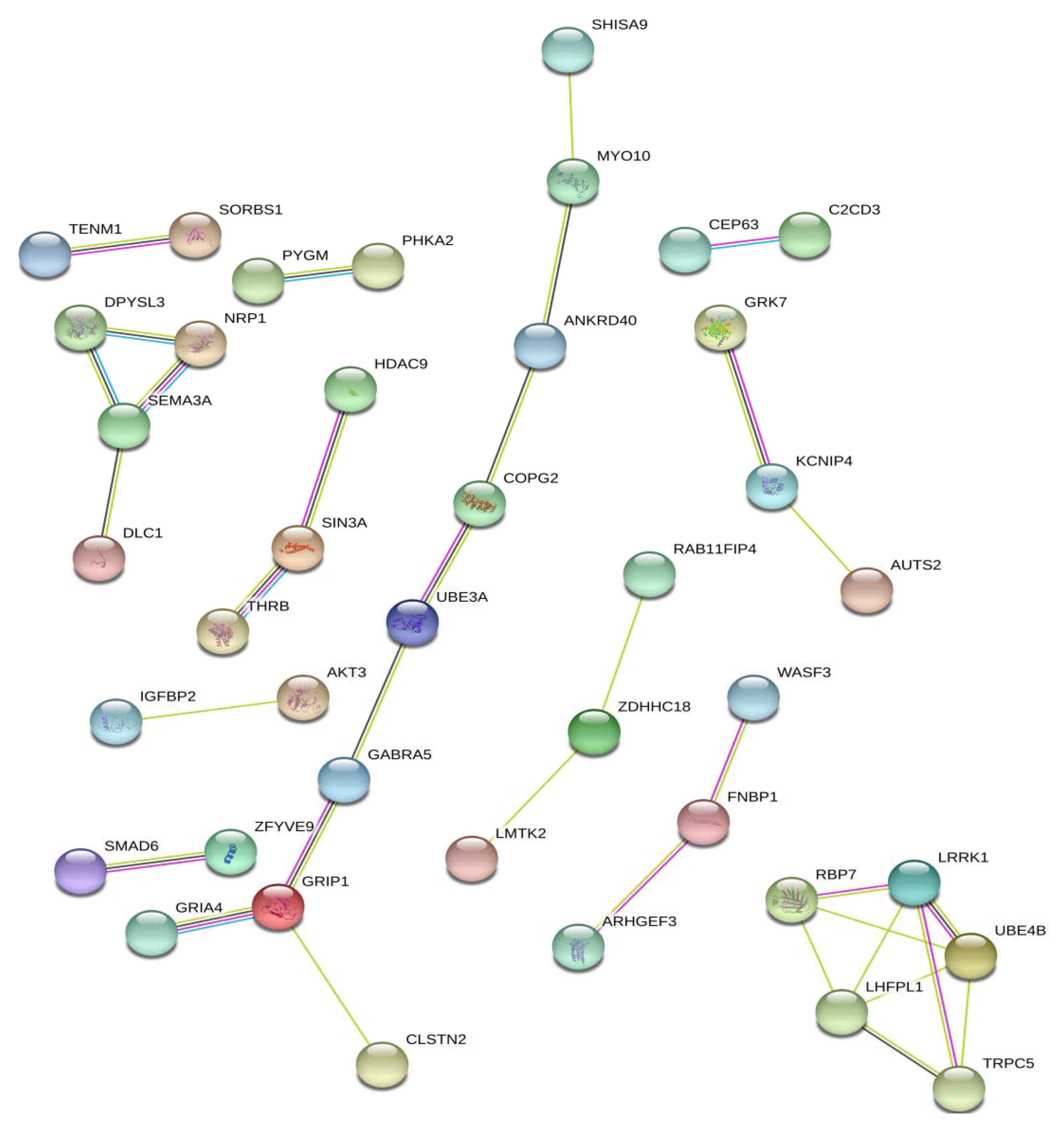
4. Potential Candidate Genes for Litter Traits of Pig
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Ethics
References
- Choi, I.; Steibel, J.P.; Bates, R.O.; Raney, N.E.; Rumph, J.M.; Ernst, C.W. Identification of carcass and meat quality QTL in an F2 Duroc × Pietrain pig resource population using different least-squares analysis models. Front. Genet. 2011, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Dragos-Wendrich, M.; Moser, G.; Bartenschlager, H.; Reiner, G.; Geldermann, H. Linkage and QTL mapping for Sus scrofa chromosome 10. J. Anim. Breed. Genet. 2003, 120, 82–88. [Google Scholar] [CrossRef]
- Tuggle, C.K.; Wang, Y.F.; Couture, O. Advances in Swine Transcriptomics. Int. J. Biol. Sci. 2007, 3, 132–152. [Google Scholar] [CrossRef] [PubMed]
- Pig QTLdb. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/SS/index (accessed on 10 March 2020).
- Rothschild, M.F.; Hu, Z.L.; Jiang, Z. Advances in QTL mapping in pigs. Int J. Biol. Sci. 2007, 3, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Samorè, A.B.; Fontanesi, L. Genomic selection in pigs: State of the art and perspectives. Ital. J. Anim. Sci. 2016, 15, 211–232. [Google Scholar] [CrossRef]
- Bergfelder-Drüing, S.; Grosse-Brinkhaus, C.; Lind, B.; Erbe, M.; Schellander, K.; Simianer, H.; Tholen, E. A Genome-Wide Association Study in Large White and Landrace Pig Populations for Number Piglets Born Alive. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- He, L.C.; Li, P.H.; Ma, X.; Sui, S.P.; Gao, S.; Kim, S.W.; Gu, Y.Q.; Huang, Y.; Ding, N.S.; Huang, R.H. Identification of new single nucleotide polymorphisms affecting total number born and candidate genes related to ovulation rate in Chinese Erhualian pigs. Anim. Genet. 2016, 48, 48–54. [Google Scholar] [CrossRef]
- Knol, E.F.; Nielsen, B.; Knap, P.W. Genomic selection in commercial pig breeding. Anim. Front. 2016, 6, 15–22. [Google Scholar] [CrossRef]
- Lopes, M.S.; Bastiaansen, J.W.M.; Harlizius, B.; Knol, E.F.; Bovenhuis, H.A. Genome-Wide Association Study Reveals Dominance Effects on Number of Teats in Pigs. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Stranger, B.; Stahl, E.; Raj, T. Progress and Promise of Genome-Wide Association Studies for Human Complex Trait Genetics. Genetics 2011, 187, 367–383. [Google Scholar] [CrossRef]
- McEntyre, J.; Ostell, J. The NCBI Handbook—Bethesda (MD); National Center for Biotechnology Information (US): Bethesda, MD, USA, 2015; Chapter 5.
- Jiang, Z.; Wang, H.; Michal, J.J.; Zhou, X.; Liu, B.; Woods, L.C.S.; Fuchs, R.A. Genome Wide Sampling Sequencing for SNP Genotyping: Methods, Challenges and Future Development. Int. J. Biol. Sci. 2016, 12, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Yang, Q.; Wang, K.; Zhou, J.; Ma, J.; Tang, Q.; Jin, L.; Xiao, W.; Jiang, A.; Jiang, Y.; et al. Single step genome-wide association studies based on genotyping by sequence data reveals novel loci for the litter traits of domestic pigs. Genomics 2018, 110, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Aliloo, H.; Pryce, J.E.; González-Recio, O.; Cocks, B.G.; Hayes, B.J. Accounting for dominance to improve genomic evaluations of dairy cows for fertility and milk production traits. Genet. Sel. Evol. 2016, 48, 186. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, Y.; Hou, L.; Ma, J.; Chen, C.; Ai, H.; Huang, L.; Ren, J. Genome-wide detection of genetic markers associated with growth and fatness in four pig populations using four approaches. Genet. Sel. Evol. GSE 2017, 49, 21. [Google Scholar] [CrossRef] [PubMed]
- Häggman, J.; Uimari, P. Novel harmful recessive haplotypes for reproductive traits in pigs. J. Anim. Breed. Genet. 2017, 134, 129–135. [Google Scholar] [CrossRef]
- Uimari, P.; Sironen, A.; Sevón-Aimonen, M.-L. Whole-genome SNP association analysis of reproduction traits in the Finnish Landrace pig breed. Genet. Sel. Evol. 2011, 43, 42. [Google Scholar] [CrossRef]
- Wen, Y.J.; Zhang, H.; Ni, Y.L.; Huang, B.; Zhang, J.; Feng, J.Y.; Wang, S.B.; Dunwell, J.M.; Zhang, Y.M.; Wu, R. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 2017, 19, 7007–7012. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, L.-G.; Shi, H.-B.; Yan, H.; Zhang, L.-C.; Liu, X.; Lei, P.U.; Liang, J.; Zhang, Y.; Zhao, K.; et al. Hritabilities and genetic and phenotypic correlations of litter uniformity and litter size in Large White sows. J. Integr. Agric. 2016, 15, 848–854. [Google Scholar] [CrossRef]
- Mucha, A.; Piórkowska, K.; Ropka-Molik, K.; Szyndler-Nędza, M. New polymorphic changes in the WNT7A gene and their effect on reproductive traits in pigs. Ann. Anim. Sci. 2018, 18, 375–385. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, X.; Tan, Z.; Xing, K.; Yang, T.; Wang, Y.; Sun, D.; Wang, C. Genome-wide association study for reproductive traits in a Large White pig population. Anim. Genet. 2018, 49, 127–131. [Google Scholar] [CrossRef]
- Coster, A.; Madsen, O.; Heuven, H.C.M.; Dibbits, B.; Groenen, M.A.M.; van Arendonk, J.A.M.; Bovenhuis, H. The Imprinted Gene DIO3 Is a Candidate Gene for Litter Size in Pigs. PLoS ONE 2012, 7, e31825. [Google Scholar] [CrossRef][Green Version]
- An, S.M.; Hwang, J.H.; Kwon, S.; Yu, G.E.; Park, D.H.; Kang, D.G.; Kim, T.W.; Park, H.C.; Ha, J.; Kim, C.W. Effect of Single Nucleotide Polymorphisms in IGFBP2 and IGFBP3 Genes on Litter Size Traits in Berkshire Pigs. Anim. Biotechnol. 2017, 29, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, J.; Han, X.; Qiao, R.; Li, X.; Lv, G.; Wang, K. Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics 2020, 112, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Suwannasing, R.; Duangjinda, M.; Boonkum, W.; Taharnklaew, R.; Tuangsithtanon, K. The identification of novel regions for reproduction trait in Landrace and Large White pigs using a single step genome-wide association study. Asian-Australas. J. Anim. Sci. 2018, 31, 1852–1862. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ye, S.; Teng, J.; Diao, S.; Yuan, X.; Chen, Z.; Zhang, H.; Li, J.; Zhang, Z. Genome-wide association studies for the number of animals born alive and dead in duroc pigs. Theriogenology 2019, 139, 36–42. [Google Scholar] [CrossRef]
- Ma, X.; Li, P.H.; Zhu, M.X.; He, L.C.; Sui, S.P.; Gao, S.; Su, G.S.; Ding, N.S.; Huang, Y.; Lu, Z.Q.; et al. Genome-wide association analysis reveals genomic regions on Chromosome 13 affecting litter size and candidate genes for uterine horn length in Erhualian pigs. Animal 2018, 14, 1–9. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Z.; Ye, S.; He, Y.; Huang, S.; Yuan, X.; Chen, Z.; Zhang, H.; Li, J. Genome-Wide Association Study for Reproductive Traits in a Duroc Pig Population. Animals 2019, 26, 732. [Google Scholar] [CrossRef]
- Sell-Kubiak, E.; Duijvesteijn, N.; Lopes, M.S.; Janss, L.L.G.; Knol, E.F.; Bijmaand, P.; Mulder, H.A. Genome-wide association study reveals novel loci for litter size and its variability in a Large White pig population. BMC Genomics 2015, 16, 1049. [Google Scholar] [CrossRef]
- Onteru, S.K.; Fan, B.; Du, Z.Q.; Garrick, D.J.; Stalder, K.J.; Rothschild, M.F. A whole-genome association study for pig reproductive traits. Anim. Genet. 2012, 43, 18–26. [Google Scholar] [CrossRef]
- Wang, H.; Wu, S.; Wu, J.; Sun, S.; Wu, S.; Bao, W. Association analysis of the SNP (rs345476947) in the FUT2 gene with the production and reproductive traits in pigs. Genes Genom. 2018, 40, 199–206. [Google Scholar] [CrossRef]
- Liu, R.; Deng, D.; Liu, X.; Xiao, Y.; Huang, J.; Wang, F.; Li, X.; Yu, M. A miR-18a binding-site polymorphism in CDC42 3’UTR affects CDC42 mRNA expression in placentas and is associated with litter size in pigs. Mamm. Genom. 2019, 30, 34–41. [Google Scholar] [CrossRef] [PubMed]
- LaSalle, J.M.; Reiter, L.T.; Chamberlain, S.J. Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics 2015, 7, 1213–1228. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.; Kang, L.; Jiang, C.; Jiang, Y. Molecular characterization of porcine NECD, SNRPN and UBE3A genes and imprinting status in the skeletal muscle of neonate pigs. Mol. Biol. Rep. 2012, 39, 9415–9422. [Google Scholar] [CrossRef] [PubMed]
- Cate, J.H. Human eIF3: From ’blobology’ to biological insight. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 19, 1716. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef]
- Zeng, L.; Wan, Y.; Li, D.; Wu, J.; Shao, M.; Chen, J.; Hui, L.; Ji, H.; Zhu, X. The m subunit of murine translation initiation factor EIF3Maintains the integrity of the eIF3 complex and is required for embryonic development, homeostasis, and organ size control. J. Biol. Chem. 2013, 288, 30087–30093. [Google Scholar] [CrossRef]
- Ye, R.-S.; Xi, Q.-Y.; Qi, Q.; Cheng, X.; Chen, T.; Li, H.; Kallon, S.; Shu, G.; Wang, S.; Jiang, Q.; et al. Differentially Expressed miRNAs after GnRH Treatment and Their Potential Roles in FSH Regulation in Porcine Anterior Pituitary Cell. PLoS ONE 2013, 8, e57156. [Google Scholar] [CrossRef]
- Curry, E.; Ellis, S.; Pratt, S. Detection of porcine sperm microRNAs using a heterologous microRNA microarray and reverse transcriptase polymerase chain reaction. Mol. Reprod. Dev. 2009, 76, 218–219. [Google Scholar] [CrossRef]
- Curry, E.; Safranski, T.J.; Pratt, S.L. Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology 2011, 76, 1532–1539. [Google Scholar] [CrossRef]
- Li, M.; Liu, Y.; Wang, T.; Guan, J.; Luo, Z.; Chen, H.; Wang, X.; Chen, L.; Ma, J.; Mu, Z.; et al. Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing. Int. J. Biol. Sci. 2011, 7, 1045. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, J.; Jia, J.; Li, H.; Hu, S.; Lin, Y.; Yan, T. Changes E3 ubiquitin protein ligase 1 gene mRNA expression correlated with IgA1 glycosylation in patients with IgA nephropathy. Renal Fail. 2019, 41, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Haraguchi, S.; Koda, T.; Hashimoto, K.; Nakagawara, A. Muscle atrophy and motor neuron degeneration in human nedl1 transgenic mice. J. Biomed. Biotechnol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.F.; Nonneman, D.J.; Wiedmann, R.T.; Vallet, J.L.; Rohrer, G.A. Genomewide association and identification of candidate genes for ovulation rate in swine. J. Anim. Sci. 2014, 92, 3792–3803. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Suzuki, A.; Hirose, T.; Kitamura, K.; Kutsuzawa, K.; Futaki, M.; Amano, Y.; Ohno, S. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J. Biol. Chem. 2003, 278, 31240–31250. [Google Scholar] [CrossRef] [PubMed]
- Zen, K.; Yasui, K.; Gen, Y.; Dohi, O.; Wakabayashi, N.; Mitsufuji, S.; Itoh, Y.; Zen, Y.; Nakanuma, Y.; Taniwaki, M.; et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene 2009, 28, 2910–2918. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Lee, J.Y.; Park, H.J.; Kim, J.W.; Chung, J.H. Association study between polymorphisms of the PARD3 gene and schizophrenia. Exp. Ther. Med. 2012, 3, 881–885. [Google Scholar] [CrossRef]
- McCole, D.F. IBD candidate genes and intestinal barrier regulation. Inflamm. Bowel Dis. 2014, 20, 1829–1849. [Google Scholar] [CrossRef]
- Spitschak, M.; Hoeflich, A. Potential Functions of IGFBP-2 for Ovarian Folliculogenesis and Steroidogenesis. Front. Endocrinol. 2018, 9, 119. [Google Scholar] [CrossRef]
- Dimmeler, S.; Hermann, C.; Galle, J.; Zeiher, A.M. Upregulation of superoxide dismutase and nitric oxide synthase mediates the apoptosis-suppressive effects of shear stress on endothelial cells. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 656–664. [Google Scholar] [CrossRef]
- Wang, B.Y.; Ho, H.K.; Lin, P.S.; Schwarzacher, S.P.; Pollman, M.J.; Gibbons, G.H.; Tsao, P.S.; Cooke, J.P. Regression of atherosclerosis: Role of nitric oxide and apoptosis. Circulation 1999, 99, 1236–1241. [Google Scholar] [CrossRef]
- Abhary, S.; Burdon, K.P.; Kuot, A.; Javadiyan, S.; Whiting, M.J.; Kasmeridis, N.; Petrovsky, N.; Craig, J.E. Sequence Variation in DDAH1 and DDAH2 Genes Is Strongly and Additively Associated with Serum ADMA Concentrations in Individuals with Type 2 Diabetes. PLoS ONE 2010, 5, e9462. [Google Scholar] [CrossRef] [PubMed]
- Dayoub, H.; Achan, V.; Adimoolam, S.; Jacobi, J.; Stuehlinger, M.C.; Wang, B.; Tsao, P.S.; Kimoto, M.; Vallance, P.; Patterson, A.J.; et al. Dimethylarginine dimethylaminohydrolase regulates nitric oxide synthesis: Genetic and physiological evidence. Circulation 2003, 108, 3042–3047. [Google Scholar] [CrossRef] [PubMed]
- Wojciak-Stothard, B.; Torondel, B.; Zhao, L.; Renne, T.; Leiper, J.M. Modulation of Rac1 activity by ADMA/DDAH regulates pulmonary endothelial barrier function. Mol. Biol. Cell 2009, 20, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Ugi, S.; Maeda, S.; Kawamura, Y.; Kobayashi, M.-A.; Imam-ura, M.; Yoshizaki, T.; Morino, K.; Sekine, O.; Yamamoto, H.; Tani, T.; et al. CCDC3 is specifically upregulated in omental adi-pose tissue in subjects with abdominal obesity. Obesity (Silver Spring) 2014, 22, 1070–1077. [Google Scholar] [CrossRef]
- Santana, M.H.; Ventura, R.V.; Utsunomiya, Y.T.; Neves, H.H.; Alexandre, P.A.; Oliveira Junior, G.A.; Gomes, R.C.; Bonin, M.N.; Coutinho, L.L.; Garcia, J.F.; et al. A genomewide association mapping study using ultrasound-scanned information identifies potential genomic regions and candidate genes affecting carcass traits in Nellore cattle. J. Anim. Breed. Genet. 2015, 132, 420–427. [Google Scholar] [CrossRef]
- Getmantseva, L.; Kolosov, A.; Leonova, M.; Bakoev, S.; Klimenko, A.; Usatov, A. Polymorphism in obesity-related leptin gene and its assotiotion with reproductive traits of sows. Bulg. J. Agric. Sci. 2017, 23, 843–850. [Google Scholar]
- Budak, E.; Fernández Sánchez, M.; Bellver, J.; Cerveró, A.; Simón, C.; Pellicer, A. Interactions of the hormones leptin, ghrelin, adiponectin, resistin, and PYY3–36 with the reproductive system. Fertil. Steril. 2006, 85, 1563–1581. [Google Scholar] [CrossRef]
| SNP | SSC | Location | Allele | Consequence | SYMBOL | Trait | Breed | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs339929690 | 1 | 59515751 | C | intergenic_variant | - | NBA | Landrace/Yorkshire | [14] |
| 2 | rs329624627 | 1 | 72605122 | C | intron_variant | CRYBG1 | NBA | Landrace/Yorkshire | [14] |
| 3 | rs338462676 | 1 | 74750265 | T | intron_variant | - | NBA | Landrace/Yorkshire | [14] |
| 4 | rs80982567 | 1 | 99668135 | G | intron_variant | - | NBA | Landrace/Yorkshire | [14] |
| 5 | rs329761313 | 1 | 116715803 | C | intron_variant | - | NBA | Landrace/Yorkshire | [14] |
| 6 | rs329145797 | 1 | 116791808 | T | intron_variant | - | NBA | Landrace/Yorkshire | [14] |
| 7 | rs80830052 | 1 | 139481542 | A | intron_variant | ALDH1A3 | NBA | Landace/Large White | [26] |
| 8 | rs80930659 | 1 | 139579572 | G | intron_variant | LRRK1 | NBA | Landace/Large White | [26] |
| 9 | rs80804265 | 1 | 139608452 | A | intron_variant | LRRK1 | NBA | Landace/Large White | [26] |
| 10 | rs80862569 | 1 | 139636710 | C | intergenic_variant | - | NBA | Landace/Large White | [26] |
| 11 | rs80846651 | 1 | 139655026 | A | intergenic_variant | - | NBA | Landace/Large White | [26] |
| 12 | rs81348779 | 1 | 141989297 | G | intron_variant | UBE3A | NBA | Large White | [22] |
| 13 | rs329931325 | 1 | 144835089 | T | intergenic_variant | - | NBA | Landrace/Yorkshire | [14] |
| 14 | rs326961952 | 1 | 145395812 | G | intergenic_variant | - | NBA | Landrace/Yorkshire | [14] |
| 15 | rs336474421 | 1 | 145452726 | T | intergenic_variant | - | NBA | Landrace/Yorkshire | [14] |
| 16 | rs322202112 | 1 | 146334522 | C | intron_variant | - | NBA | Landrace/Yorkshire | [14] |
| 17 | rs332924521 | 1 | 239558816 | C | intron_variant | XPA | NBA | Landrace/Yorkshire | [14] |
| 18 | rs330585697 | 1 | 239775818 | T | intron_variant | TRMO | NBA | Landrace/Yorkshire | [14] |
| 19 | rs334029855 | 1 | NA | NBA | Landrace/Yorkshire | [14] | |||
| 20 | rs81291755 | 2 | 20563683 | G | intergenic_variant | - | NBA | Landrace/Yorkshire | [26] |
| 21 | rs81355894 | 2 | 20637563 | C | intergenic_variant | - | NBA | Landace/Large White | [26] |
| 22 | rs81355903 | 2 | 20665892 | T | intergenic_variant | - | NBA | Landace/Large White | [26] |
| 23 | rs81355915 | 2 | 20717076 | G | intergenic_variant | - | NBA | Landace/Large White | [26] |
| 24 | rs81356698 | 2 | 28380318 | G | intron_variant | EIF3M | NBA | Large White | [22] |
| 25 | rs81265647 | 2 | 125660328 | T | upstream_gene_var | - | NBA | Landace/Large White | [26] |
| 26 | rs328177895 | 2 | NA | NBA | Large White | [23] | |||
| 27 | rs81379421 | 3 | 27307613 | G | intron_variant | XYLT1 | NBA | Landace/Large White | [7] |
| 28 | rs334867206 | 3 | 43312168 | T | intergenic_variant | - | NBA | Large White | [23] |
| 29 | rs319494663 | 3 | 43463318 | C | upstream_gene_var | ssc-let-7a-2 | NBA | Large White | [23] |
| 30 | rs80927364 | 4 | 130597207 | A | intron_variant | DDAH1 | NBA | Erhualian | [28] |
| 31 | rs80890206 | 5 | 31022891 | C | upstream_gene_var | GRIP1 | NBA | Erhualian | [28] |
| 32 | rs80867243 | 5 | 87337099 | C | intron_variant | ELK3 | NBA | Landace/Large White | [7] |
| 33 | rs81279319 | 5 | NA | NBA | Landace/Large White | [7] | |||
| 34 | rs81383147 | 5 | NA | NBA | Landace/Large White | [7] | |||
| 35 | rs81320475 | 6 | 70313133 | A | upstream_gene_var | RBP7 | NBA | Landace/Large White | [26] |
| 36 | rs81285644 | 6 | 70323076 | G | intron_variant | RBP7 | NBA | Landace/Large White | [26] |
| 37 | rs81275494 | 6 | 70408106 | G | intron_variant | UBE4B | NBA | Landace/Large White | [26] |
| 38 | rs81279050 | 6 | 70418172 | A | intron_variant | UBE4B | NBA | Landace/Large White | [26] |
| 39 | rs81270030 | 6 | 70428427 | T | intron_variant | UBE4B | NBA | Landace/Large White | [26] |
| 40 | rs81388947 | 6 | 80229040 | A | intergenic_variant | - | NBA | Duroc | [27] |
| 41 | rs704072370 | 6 | 80388745 | A | intergenic_variant | - | NBA | Duroc | [27] |
| 42 | rs81332505 | 6 | 83142163 | A | intron_variant | MAN1C1 | NBA | Duroc | [27] |
| 43 | rs81476037 | 6 | 83545020 | T | intron_variant | PDIK1L | NBA | Duroc | [27] |
| 44 | rs328276462 | 6 | 83863384 | G | downstream_gene_var | HMGN2 | NBA | Duroc | [27] |
| 45 | rs335265547 | 6 | 83871998 | C | upstream_gene_var | - | NBA | Duroc | [27] |
| 46 | rs81287462 | 6 | 83986459 | A | intergenic_variant | - | NBA | Duroc | [27] |
| 47 | rs81332455 | 6 | 84069079 | G | intron_variant | ARID1A | NBA | Duroc | [27] |
| 48 | rs336670754 | 6 | 84118086 | C | synonymous_variant | ARID1A | NBA | Duroc | [27] |
| 49 | rs342908929 | 6 | 159933806 | T | intron_variant | ZFYVE9 | NBA | Commercia | [25] |
| 50 | rs81345088 | 6 | 168897980 | A | intron_variant | ZMYND12 | NBA | Landace/Large White | [26] |
| 51 | rs81259198 | 6 | 168899114 | A | intron_variant | ZMYND12 | NBA | Landace/Large White | [26] |
| 52 | rs81245903 | 6 | 168916590 | T | intron_variant | RIMKLA | NBA | Landace/Large White | [26] |
| 53 | rs81273774 | 6 | 168931165 | C | intron_variant | RIMKLA | NBA | Landace/Large White | [26] |
| 54 | rs80882306 | 7 | 11003222 | G | upstream_gene_var | - | NBA | Erhualian | [28] |
| 55 | rs325729252 | 7 | 109101108 | C | intergenic_variant | - | NBA | Landace/Large White | [7] |
| 56 | rs81397142 | 7 | NA | NBA | Large White | [22] | |||
| 57 | rs81397215 | 7 | NA | NBA | Large White | [22] | |||
| 58 | rs318557169 | 8 | 95381096 | G | intergenic_variant | - | NBA | Landrace/Yorkshire | [14] |
| 59 | rs327655683 | 8 | 95402330 | A | intergenic_variant | - | NBA | Landrace/Yorkshire | [14] |
| 60 | rs81257618 | 9 | 13492371 | A | intergenic_variant | - | NBA | Landace/Large White | [7] |
| 61 | rs81417393 | 9 | 126541658 | C | intron_variant | - | NBA | Landace/Large White | [7] |
| 62 | rs81421148 | 10 | 16506621 | C | intron_variant | AKT3 | NBA | Landace/Large White | [7] |
| 63 | rs80978601 | 10 | 25256004 | G | intergenic_variant | - | NBA | Landace/Large White | [7] |
| 64 | rs81255997 | 10 | 28330355 | C | intergenic_variant | - | NBA | Landace/Large White | [7] |
| 65 | rs81274366 | 10 | 57593745 | G | intron_variant | PARD3 | NBA | Erhualian | [28] |
| 66 | rs81236069 | 10 | 58288430 | A | intergenic_variant | - | NBA | Landace/Large White | [7] |
| 67 | rs81302230 | 10 | 58290108 | G | intergenic_variant | - | NBA | Landace/Large White | [7] |
| 68 | rs81430147 | 11 | 836702 | C | downstream_gene_var | - | NBA | Landace/Large White | [7] |
| 69 | rs81430859 | 11 | 4296303 | C | intron_variant | WASF3 | NBA | Landace/Large White | [7] |
| 70 | rs81242222 | 11 | 67129570 | A | intergenic_variant | - | NBA | Landace/Large White | [7] |
| 71 | rs81434499 | 12 | 36207006 | G | intergenic_variant | - | NBA | Erhualian | [28] |
| 72 | rs80962240 | 13 | 52784022 | C | intron_variant | FOXP1 | NBA | Large White | [23] |
| 73 | rs81215583 | 13 | 71905692 | C | synonymous_variant | RPN1 | NBA | Erhualian | [28] |
| 74 | rs81447100 | 13 | 80866929 | A | intron_variant | CLSTN2 | NBA | Erhualian | [28] |
| 75 | rs81447231 | 13 | 82603578 | A | intron_variant | GRK7 | NBA | Erhualian | [28] |
| 76 | rs319258722 | 14 | 123766842 | T | intergenic_variant | - | NBA | Large White | [23] |
| 77 | rs45435330 | 15 | 118847390 | A | intron_variant | IGFBP2 | NBA | Berkshire | [24] |
| 78 | rs324003968 | 15 | NA | NBA | Commercia | [25] | |||
| 79 | rs81459590 | 16 | 6025546 | A | intron_variant | MYO10 | NBA | Landace/Large White | [7] |
| 80 | rs80952566 | 17 | 27123793 | T | intron_variant | - | NBA | Erhualian | [28] |
| 81 | rs81469701 | 18 | 42920811 | T | upstream_gene_var | SCRN1 | NBA | Landace/Large White | [7] |
| 82 | rs81471172 | 18 | 51640938 | T | intron_variant | HECW1 | NBA | Large White | [23] |
| 83 | rs81473442 | X | 91880535 | C | intron_variant | TRPC5 | NBA | Landace/Large White | [26] |
| 84 | rs81323503 | X | 92070342 | A | intergenic_variant | - | NBA | Landace/Large White | [26] |
| 85 | rs81283192 | X | 92244402 | A | intron_variant | RTL4 | NBA | Landace/Large White | [26] |
| 86 | rs337547716 | X | 92330719 | A | intron_variant | RTL4 | NBA | Landace/Large White | [26] |
| 87 | rs80834138 | X | 92447181 | A | intron_variant | LHFPL1 | NBA | Landace/Large White | [26] |
| 88 | rs81339510 | X | 118854990 | T | intergenic_variant | - | NBA | Landace/Large White | [26] |
| NO | SNP | SSC | Location | Allele | Consequence | SYMBOL | Trait | Breed | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs80972494 | 1 | 7262020 | C | intergenic_variant | - | TNB | Large White | [22] |
| 2 | rs80787893 | 1 | 14869534 | C | intron_variant | ZBTB2 | TNB | Large White | [30] |
| 3 | rs80851003 | 1 | 20072414 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 4 | rs80970692 | 1 | 97934013 | A | intron_variant | ZBTB7C | TNB | Large White | [23] |
| 5 | rs80845110 | 1 | 139993309 | A | intron_variant | GABRG3 | TNB | Large White | [22] |
| 6 | rs80933698 | 1 | 140097794 | G | intron_variant | GABRG3 | TNB | Large White | [22] |
| 7 | rs80869858 | 1 | 140178409 | T | intron_variant | GABRG3 | TNB | Large White | [22] |
| 8 | rs80999701 | 1 | 140290677 | A | intron_variant | GABRG3 | TNB | Large White | [22] |
| 9 | rs81348717 | 1 | 140463533 | G | intron_variant | GABRG3 | TNB | Large White | [22] |
| 10 | rs81348724 | 1 | 140550299 | A | intron_variant | GABRA5 | TNB | Large White | [22] |
| 11 | rs81348751 | 1 | 140808020 | A | intron_variant | GABRB3 | TNB | Large White | [22] |
| 12 | rs80989931 | 1 | 140896691 | A | intron_variant | GABRB3 | TNB | Large White | [22] |
| 13 | rs81348779 | 1 | 141989297 | G | downstream_gene_var | UBE3A | TNB | Large White | [22] |
| 14 | rs80912860 | 1 | 164637575 | C | intergenic_variant | - | TNB | Large White | [30] |
| 15 | rs80956812 | 1 | 164674664 | A | intron_variant | SMAD6 | TNB | Large White | [30] |
| 16 | rs81267574 | 1 | 222278217 | G | intron_variant | PIP5K1B | TNB | Erhualian | [8] |
| 17 | rs80938435 | 1 | 245222778 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 18 | rs81296573 | 1 | 263022859 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 19 | rs80913204 | 1 | 270166629 | T | intron_variant | FNBP1 | TNB | Erhualian | [8] |
| 20 | rs81367039 | 1 | NA | TNB | Erhualian | [8] | |||
| 21 | rs81330112 | 2 | 3058633 | A | upstream_gene_var | CTTN | TNB | Erhualian | [8] |
| 22 | rs81214065 | 2 | 7420349 | A | missense_variant | PYGM | TNB | Erhualian | [8] |
| 23 | rs81474834 | 2 | 9252507 | T | intron_variant | - | TNB | Erhualian | [8] |
| 24 | rs81273273 | 2 | 24914867 | G | intron_variant | LDLRAD3 | TNB | Erhualian | [8] |
| 25 | rs81356698 | 2 | 28380318 | G | downstream_gene_var | TNB | Large White | [22] | |
| 26 | rs346316162 | 2 | 84269464 | T | intron_variant | ANKRD31 | TNB | Erhualian | [8] |
| 27 | rs81360234 | 2 | 85003252 | G | intron_variant | SV2C | TNB | Erhualian | [8] |
| 28 | rs81270902 | 2 | 136051929 | G | intron_variant | FSTL4 | TNB | Erhualian | [8] |
| 29 | rs81307772 | 2 | 142675693 | C | intron_variant | PCDHAC2 | TNB | Erhualian | [8] |
| 30 | rs81366836 | 2 | 145019988 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 31 | rs81367208 | 2 | 146880995 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 32 | rs81346993 | 2 | 148668538 | G | intron_variant | DPYSL3 | TNB | Erhualian | [8] |
| 33 | rs328177895 | 2 | NA | TNB | Large White | [23] | |||
| 34 | rs81367039 | 2 | NA | TNB | Erhualian | [8] | |||
| 35 | rs81367039 | 2 | NA | TNB | Erhualian | [8] | |||
| 36 | rs334519198 | 3 | 5376605 | T | intron_variant | LMTK2 | TNB | Large White | [23] |
| 37 | rs81243084 | 3 | 14585248 | T | intron_variant | AUTS2 | TNB | Erhualian | [8] |
| 38 | rs81318451 | 3 | 20859502 | T | intron_variant | HS3ST4 | TNB | Erhualian | [8] |
| 39 | rs81319541 | 3 | 23688143 | G | downstream_gene_var | IGSF6 | TNB | Erhualian | [8] |
| 40 | rs81379942 | 3 | 30034842 | A | intron_variant | SHISA9 | TNB | Erhualian | [8] |
| 41 | rs81369361 | 3 | 43227069 | A | intergenic_variant | - | TNB | Large White | [23] |
| 42 | rs334867206 | 3 | 43312168 | T | intergenic_variant | - | TNB | Large White | [23] |
| 43 | rs319494663 | 3 | 43463318 | C | upstream_gene_variant | ssc-let-7a-2 | TNB | Large White | [23] |
| 44 | rs338135576 | 3 | 52266739 | T | intron_variant | IL1R1 | TNB | Erhualian | [8] |
| 45 | rs81370592 | 3 | 53699303 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 46 | rs81377897 | 3 | 124275280 | G | upstream_gene_variant | - | TNB | Erhualian | [8] |
| 47 | rs81272059 | 3 | 125892592 | G | intron_variant | NOL10 | TNB | Erhualian | [8] |
| 48 | rs81319839 | 4 | 18194352 | A | intergenic_variant | - | TNB | Large White | [23] |
| 49 | rs81312912 | 4 | 18196598 | A | intergenic_variant | - | TNB | Large White | [23] |
| 50 | rs80986621 | 4 | 87990037 | C | missense_variant | UAP1 | TNB | Erhualian | [8] |
| 51 | rs80860510 | 4 | 89154886 | A | intron_variant | SDHC | TNB | Erhualian | [8] |
| 52 | rs80987610 | 4 | 89242132 | G | downstream_gene_var | APOA2 | TNB | Erhualian | [8] |
| 53 | rs80910021 | 4 | 96557774 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 54 | rs80999559 | 4 | 98260992 | C | downstream_gene_var | CERS2 | TNB | Erhualian | [8] |
| 55 | rs80927364 | 4 | 130597207 | A | intron_variant | DDAH1 | TNB | Erhualian | [8] |
| 56 | rs81385465 | 5 | 4131296 | G | intergenic_variant | - | TNB | Large White | [30] |
| 57 | rs336638152 | 5 | 11274122 | A | intron_variant | - | TNB | Duroc | [29] |
| 58 | rs327336155 | 5 | 31752997 | G | intergenic_variant | - | TNB | Large White | [23] |
| 59 | rs80890539 | 5 | 49084918 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 60 | rs80999110 | 5 | 65577418 | T | intergenic_variant | - | TNB | Duroc | [29] |
| 61 | rs328217833 | 5 | 78953555 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 62 | rs81303269 | 5 | 79020148 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 63 | rs81236331 | 6 | 4395948 | T | intron_variant | WFDC1 | TNB | Erhualian | [8] |
| 64 | rs81307446 | 6 | 4817058 | T | intron_variant | CDH13 | TNB | Erhualian | [8] |
| 65 | rs81393472 | 6 | 21728055 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 66 | rs345476947 | 6 | 54042595 | T | intron_variant | FUT2 | TNB | Large White | [32] |
| 67 | rs81322640 | 6 | 74250906 | T | intron_variant | KAZN | TNB | Erhualian | [8] |
| 68 | rs81283746 | 6 | 78467732 | C | downstream_gene_var | UBXN10 | TNB | Erhualian | [8] |
| 69 | rs55618224 | 6 | 80088638 | C | 3_prime_UTR_variant | - | TNB | Yorkshire | [33] |
| 70 | rs81318862 | 6 | 82375634 | A | intergenic_variant | - | TNB | Duroc | [29] |
| 71 | rs329711941 | 6 | 84156205 | C | intron_variant | ZDHHC18 | TNB | Duroc | [29] |
| 72 | rs81391439 | 6 | 129931172 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 73 | rs342908929 | 6 | 159933806 | T | intron_variant | ZFYVE9 | TNB | Commercia | [25] |
| 74 | rs81319462 | 6 | 163598597 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 75 | rs81319428 | 6 | 163598619 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 76 | rs80882306 | 7 | 11003222 | G | upstream_gene_variant | - | TNB | Erhualian | [28] |
| 77 | rs80813007 | 7 | 21583296 | G | downstream_gene_var | - | TNB | Erhualian | [8] |
| 78 | rs80938431 | 7 | 22008244 | G | intron_variant | - | TNB | Erhualian | [8] |
| 79 | rs80933422 | 7 | 27905935 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 80 | rs80797074 | 7 | 29268803 | G | synonymous_variant | DST | TNB | Erhualian | [8] |
| 81 | rs340672537 | 7 | 38439290 | T | intron_variant | ABCC10 | TNB | Erhualian | [8] |
| 82 | rs81398070 | 7 | 58232894 | C | intron_variant | SIN3A | TNB | Erhualian | [8] |
| 83 | rs80824208 | 7 | 63161681 | G | intron_variant | SLC25A21 | TNB | Erhualian | [8] |
| 84 | rs326644823 | 7 | 63618208 | A | downstream_gene_var | - | TNB | Erhualian | [8] |
| 85 | rs80867596 | 7 | 70133128 | G | downstream_gene_var | - | TNB | Erhualian | [8] |
| 86 | rs80891106 | 7 | 73467314 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 87 | rs81295302 | 7 | 74333495 | T | intron_variant | STXBP6 | TNB | Erhualian | [8] |
| 88 | rs81398127 | 7 | 75705436 | G | 3_prime_UTR_variant | CMTM5 | TNB | Erhualian | [8] |
| 89 | rs336977324 | 7 | 81077680 | T | intron_variant | RYR3 | TNB | Erhualian | [8] |
| 90 | rs80927564 | 7 | 108030803 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 91 | rs80942529 | 7 | 110057727 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 92 | rs80969683 | 7 | NA | TNB | Large White | [22] | |||
| 93 | rs81397215 | 7 | NA | TNB | Large White | [22] | |||
| 94 | rs81397142 | 7 | NA | TNB | Large White | [22] | |||
| 95 | rs81403620 | 8 | 15474371 | T | intron_variant | KCNIP4 | TNB | Erhualian | [8] |
| 96 | rs81405013 | 8 | 16400498 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 97 | rs81342198 | 8 | 16560870 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 98 | rs81406385 | 8 | 17783561 | C | intron_variant | - | TNB | Erhualian | [8] |
| 99 | rs81343566 | 8 | 18863902 | C | intron_variant | CCDC149 | TNB | Erhualian | [8] |
| 100 | rs81399474 | 8 | 32370687 | C | downstream_gene_var | UCHL1 | TNB | Erhualian | [8] |
| 101 | rs81399527 | 8 | 32800253 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 102 | rs81399633 | 8 | 33508704 | A | splice_region_variant | ATP8A1 | TNB | Erhualian | [8] |
| 103 | rs81227962 | 8 | 34380373 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 104 | rs81476987 | 8 | 34885027 | C | intron_variant | KCTD8 | TNB | Erhualian | [8] |
| 105 | rs81399897 | 8 | 36628170 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 106 | rs81400131 | 8 | 38946698 | A | downstream_gene_var | CWH43 | TNB | Erhualian | [8] |
| 107 | rs81400868 | 8 | 63262973 | A | downstream_gene_var | U6 | TNB | Erhualian | [8] |
| 108 | rs81401375 | 8 | 73118551 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 109 | rs339466191 | 8 | 80345448 | A | intron_variant | NR3C2 | TNB | Landrace/ Yorkshire | [14] |
| 110 | rs333905163 | 8 | 80589152 | A | intron_variant | NR3C2 | TNB | Landrace/ Yorkshire | [14] |
| 111 | rs81294311 | 8 | 88225561 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 112 | rs319272490 | 8 | 103706053 | C | intergenic_variant | - | TNB | Landrace/ Yorkshire | [14] |
| 113 | rs81403286 | 8 | 107008700 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 114 | rs81403527 | 8 | 112530303 | A | downstream_gene_var | CASP6 | TNB | Erhualian | [8] |
| 115 | rs81403538 | 8 | 112566311 | T | intron_variant | MCUB | TNB | Erhualian | [8] |
| 116 | rs334180816 | 8 | 115856336 | C | intron_variant | NPNT | TNB | Landrace/ Yorkshire | [14] |
| 117 | rs80834695 | 8 | 120291881 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 118 | rs81317149 | 8 | 127090631 | G | intergenic_variant | - | TNB | Large White | [30] |
| 119 | rs81413855 | 9 | 8393770 | G | 3_prime_UTR_variant | C2CD3 | TNB | Erhualian | [8] |
| 120 | rs81416386 | 9 | 11655571 | G | intron_variant | MYO7A | TNB | Erhualian | [8] |
| 121 | rs81407589 | 9 | 21695319 | C | intron_variant | - | TNB | Erhualian | [8] |
| 122 | rs81408950 | 9 | 34248222 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 123 | rs81409102 | 9 | 35438101 | C | upstream_gene_variant | GRIA4 | TNB | Erhualian | [8] |
| 124 | rs81413928 | 9 | 84593678 | A | intron_variant | AGMO | TNB | Landrace | [18] |
| 125 | rs81413949 | 9 | 85057918 | C | intergenic_variant | - | TNB | Landrace | [18] |
| 126 | rs81287478 | 9 | 85128043 | T | intergenic_variant | - | TNB | Landrace | [18] |
| 127 | rs81223525 | 9 | 85362483 | G | intergenic_variant | - | TNB | Landrace | [18] |
| 128 | rs81260290 | 9 | 87932749 | G | intron_variant | HDAC9 | TNB | Erhualian | [28] |
| 129 | rs81414623 | 9 | 96283483 | C | intron_variant | SEMA3A | TNB | Erhualian | [8] |
| 130 | rs81300575 | 9 | 102308556 | T | intron_variant | PHTF2 | TNB | Landrace | [18] |
| 131 | rs81331059 | 9 | 137095797 | G | intron_variant | - | TNB | Erhualian | [8] |
| 132 | rs81419264 | 9 | 137162279 | G | non_coding_transcript | - | TNB | Erhualian | [8] |
| 133 | rs81419315 | 9 | 137216947 | G | upstream_gene_variant | U6 | TNB | Erhualian | [8] |
| 134 | rs81315852 | 9 | 138107624 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 135 | rs81332239 | 9 | 138462123 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 136 | rs81417713 | 9 | NA | TNB | Erhualian | [8] | |||
| 137 | rs81429231 | 10 | 10002753 | G | intron_variant | MARK1 | TNB | Erhualian | [8] |
| 138 | rs80895456 | 10 | 26509297 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 139 | rs341908955 | 10 | 48658434 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 140 | rs81329283 | 10 | 56421545 | A | intron_variant | NRP1 | TNB | Erhualian | [8] |
| 141 | rs81274366 | 10 | 57593745 | G | intron_variant | PARD3 | TNB | Erhualian | [28] |
| 142 | rs81426281 | 10 | 57913635 | T | upstream_gene_variant | - | TNB | Erhualian | [8] |
| 143 | rs81314128 | 10 | 59665035 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 144 | rs80941850 | 11 | 8940153 | T | intron_variant | - | TNB | Erhualian | [8] |
| 145 | rs81477765 | 11 | 19542650 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 146 | rs81332839 | 11 | 19542846 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 147 | rs81285980 | 11 | 19544324 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 148 | rs81237348 | 11 | 22281193 | C | intron_variant | NUFIP1 | TNB | Erhualian | [8] |
| 149 | rs80989787 | 11 | 23362846 | T | intron_variant | ENOX1 | TNB | Large White | [30] |
| 150 | rs81289355 | 11 | 23410214 | C | intron_variant | ENOX1 | TNB | Large White | [30] |
| 151 | rs81293918 | 11 | 27751988 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 152 | rs80813604 | 11 | 48641897 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 153 | rs80950312 | 11 | 48749391 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 154 | rs80869156 | 11 | 64316157 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 155 | rs81329621 | 11 | 66354904 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 156 | rs81343376 | 12 | 6971812 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 157 | rs81332319 | 12 | 7414272 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 158 | rs81439394 | 12 | 9810809 | G | intergenic_variant | - | TNB | Duroc | [29] |
| 159 | rs81433045 | 12 | 26992374 | G | intron_variant | ANKRD40 | TNB | Erhualian | [8] |
| 160 | rs81433877 | 12 | 33577229 | G | intron_variant | MSI2 | TNB | Erhualian | [8] |
| 161 | rs81434044 | 12 | 33799100 | C | intron_variant | MSI2 | TNB | Erhualian | [8] |
| 162 | rs81434064 | 12 | 33903675 | A | intron_variant | MSI2 | TNB | Erhualian | [8] |
| 163 | rs81434489 | 12 | 35993482 | G | intron_variant | VMP1 | TNB | Erhualian | [8] |
| 164 | rs81434499 | 12 | 36207006 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 165 | rs81309004 | 12 | 40519573 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 166 | rs81434931 | 12 | 40974978 | A | intron_variant | ASIC2 | TNB | Erhualian | [8] |
| 167 | rs81435036 | 12 | 41338150 | C | intron_variant | ASIC2 | TNB | Erhualian | [8] |
| 168 | rs81477883 | 12 | 43427846 | T | intron_variant | RAB11FIP4 | TNB | Erhualian | [8] |
| 169 | rs81447979 | 13 | 11362350 | A | upstream_gene_variant | THRB | TNB | Erhualian | [8] |
| 170 | rs80837222 | 13 | 13802573 | T | intron_variant | NEK10 | TNB | Erhualian | [8] |
| 171 | rs81445072 | 13 | 38880472 | C | intron_variant | ARHGEF3 | TNB | Erhualian | [8] |
| 172 | rs80964445 | 13 | 75637294 | G | intron_variant | CEP63 | TNB | Erhualian | [8] |
| 173 | rs81447100 | 13 | 80866929 | A | intron_variant | CLSTN2 | TNB | Erhualian | [8] |
| 174 | rs81441751 | 13 | 182451198 | C | intron_variant | CHODL | TNB | Large White | [30] |
| 175 | rs81442196 | 13 | 190614395 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 176 | rs80961068 | 13 | 200778735 | T | intron_variant | TTC3 | TNB | Large White | [23] |
| 177 | rs80841768 | 14 | 12118215 | T | intron_variant | PNOC | TNB | Erhualian | [8] |
| 178 | rs330621374 | 14 | 15126027 | A | intergenic_variant | - | TNB | Landrace/ Yorkshire | [14] |
| 179 | rs339777110 | 14 | 25374812 | G | intron_variant | TMEM132D | TNB | Duroc | [29] |
| 180 | rs334650508 | 14 | 26675179 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 181 | rs318344052 | 14 | 26907303 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 182 | rs80979042 | 14 | 62040147 | T | intron_variant | BICC1 | TNB | Duroc | [29] |
| 183 | rs80825112 | 14 | 62071531 | A | intron_variant | BICC1 | TNB | Duroc | [29] |
| 184 | rs80892145 | 14 | 73289391 | T | intron_variant | LRRC20 | TNB | Erhualian | [8] |
| 185 | rs80960182 | 14 | 84555165 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 186 | rs80873788 | 14 | 85481113 | A | upstream_gene_variant | RGR | TNB | Erhualian | [8] |
| 187 | rs80799654 | 14 | 98901034 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 188 | rs80800806 | 14 | 104041994 | T | intron_variant | IDE | TNB | Erhualian | [8] |
| 189 | rs80968475 | 14 | 107031142 | T | intron_variant | SORBS1 | TNB | Erhualian | [8] |
| 190 | rs80959797 | 14 | 107336340 | G | intron_variant | ENTPD1 | TNB | Erhualian | [8] |
| 191 | rs80938898 | 14 | 107377294 | C | intron_variant | ENTPD1 | TNB | Erhualian | [8] |
| 192 | rs80971725 | 14 | 107584405 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 193 | rs80843720 | 14 | 107597908 | T | intron_variant | ZNF518A | TNB | Erhualian | [8] |
| 194 | rs80987149 | 14 | 107725686 | T | intergenic_variant | - | TNB | Erhualian | [8] |
| 195 | rs81449479 | 14 | 107794160 | G | 3_prime_UTR_variant | OPALIN | TNB | Erhualian | [8] |
| 196 | rs80867623 | 14 | 129792523 | G | intron_variant | SEC23IP | TNB | Large White | [22] |
| 197 | rs80983641 | 14 | 137193623 | G | intron_variant | PTPRE | TNB | Erhualian | [8] |
| 198 | rs80892643 | 14 | NA | TNB | Erhualian | [8] | |||
| 199 | rs80782668 | 14 | NA | TNB | Erhualian | [8] | |||
| 200 | rs80947288 | 14 | NA | TNB | Duroc | [29] | |||
| 201 | rs80868389 | 15 | 9150979 | A | intergenic_variant | - | TNB | Erhualian | [8] |
| 202 | rs81452018 | 15 | 26337909 | G | intergenic_variant | - | TNB | Landace/Large White | [31] |
| 203 | rs80893476 | 15 | 82075550 | C | intron_variant | MTX2 | TNB | Erhualian | [8] |
| 204 | rs45435330 | 15 | 118847390 | A | intron_variant | IGFBP2 | TNB | Berkshire | [24] |
| 205 | rs324003968 | 15 | NA | TNB | Commercia | [25] | |||
| 206 | rs81459026 | 16 | 41470716 | G | intron_variant | IPO11 | TNB | Erhualian | [8] |
| 207 | rs81289648 | 16 | 74903886 | A | intergenic_variant | - | TNB | Erhualian | [28] |
| 208 | rs81463092 | 16 | 74971494 | G | intergenic_variant | - | TNB | Erhualian | [8] |
| 209 | rs81463053 | 16 | NA | TNB | Erhualian | [8] | |||
| 210 | rs331119584 | 17 | 1486143 | T | intron_variant | DLC1 | TNB | Erhualian | [8] |
| 211 | rs328230332 | 17 | 34329730 | T | intergenic_variant | - | TNB | Duroc | [29] |
| 212 | rs81472303 | 18 | 18253338 | C | intron_variant | COPG2 | TNB | Large White | [22] |
| 213 | rs81471172 | 18 | 51640938 | T | intron_variant | HECW1 | TNB | Large White | [23] |
| 214 | rs81471381 | 18 | 53672799 | G | intron_variant | SUGCT | TNB | Large White | [30] |
| 215 | rs332595701 | 18 | 53697787 | G | intron_variant | SUGCT | TNB | Large White | [30] |
| 216 | rs81225238 | X | 15335129 | T | intron_variant | PHKA2 | TNB | Erhualian | [8] |
| 217 | rs80891661 | X | 101710962 | C | intergenic_variant | - | TNB | Erhualian | [8] |
| 218 | rs322114058 | X | 101975674 | A | intron_variant | TENM1 | TNB | Erhualian | [8] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakoev, S.; Getmantseva, L.; Bakoev, F.; Kolosova, M.; Gabova, V.; Kolosov, A.; Kostyunina, O. Survey of SNPs Associated with Total Number Born and Total Number Born Alive in Pig. Genes 2020, 11, 491. https://doi.org/10.3390/genes11050491
Bakoev S, Getmantseva L, Bakoev F, Kolosova M, Gabova V, Kolosov A, Kostyunina O. Survey of SNPs Associated with Total Number Born and Total Number Born Alive in Pig. Genes. 2020; 11(5):491. https://doi.org/10.3390/genes11050491
Chicago/Turabian StyleBakoev, Siroj, Lyubov Getmantseva, Faridun Bakoev, Maria Kolosova, Valeria Gabova, Anatoly Kolosov, and Olga Kostyunina. 2020. "Survey of SNPs Associated with Total Number Born and Total Number Born Alive in Pig" Genes 11, no. 5: 491. https://doi.org/10.3390/genes11050491
APA StyleBakoev, S., Getmantseva, L., Bakoev, F., Kolosova, M., Gabova, V., Kolosov, A., & Kostyunina, O. (2020). Survey of SNPs Associated with Total Number Born and Total Number Born Alive in Pig. Genes, 11(5), 491. https://doi.org/10.3390/genes11050491





