M2UNet: Multi-Scale Feature Acquisition and Multi-Input Edge Supplement Based on UNet for Efficient Segmentation of Breast Tumor in Ultrasound Images
Abstract
1. Introduction
- This paper introduced a novel multi-scale feature acquisition module named MFA. This module adeptly harnesses semantic information from various receptive fields to facilitate the precise segmentation of tumors;
- To address the challenges of boundary blurring and edge irregularities in breast ultrasound imaging, we have introduced a novel MES module. This module effectively mitigates these issues by complementing ultrasound tumor edges with features extracted from the decoder output;
- We validated the effectiveness of our method by constructing a breast ultrasound dataset using all malignant tumors in collaboration with Fujian Cancer Hospital (FCBU) and performed experimental validation on this dataset and the publicly available dataset (BUSI).
2. Materials and Methods
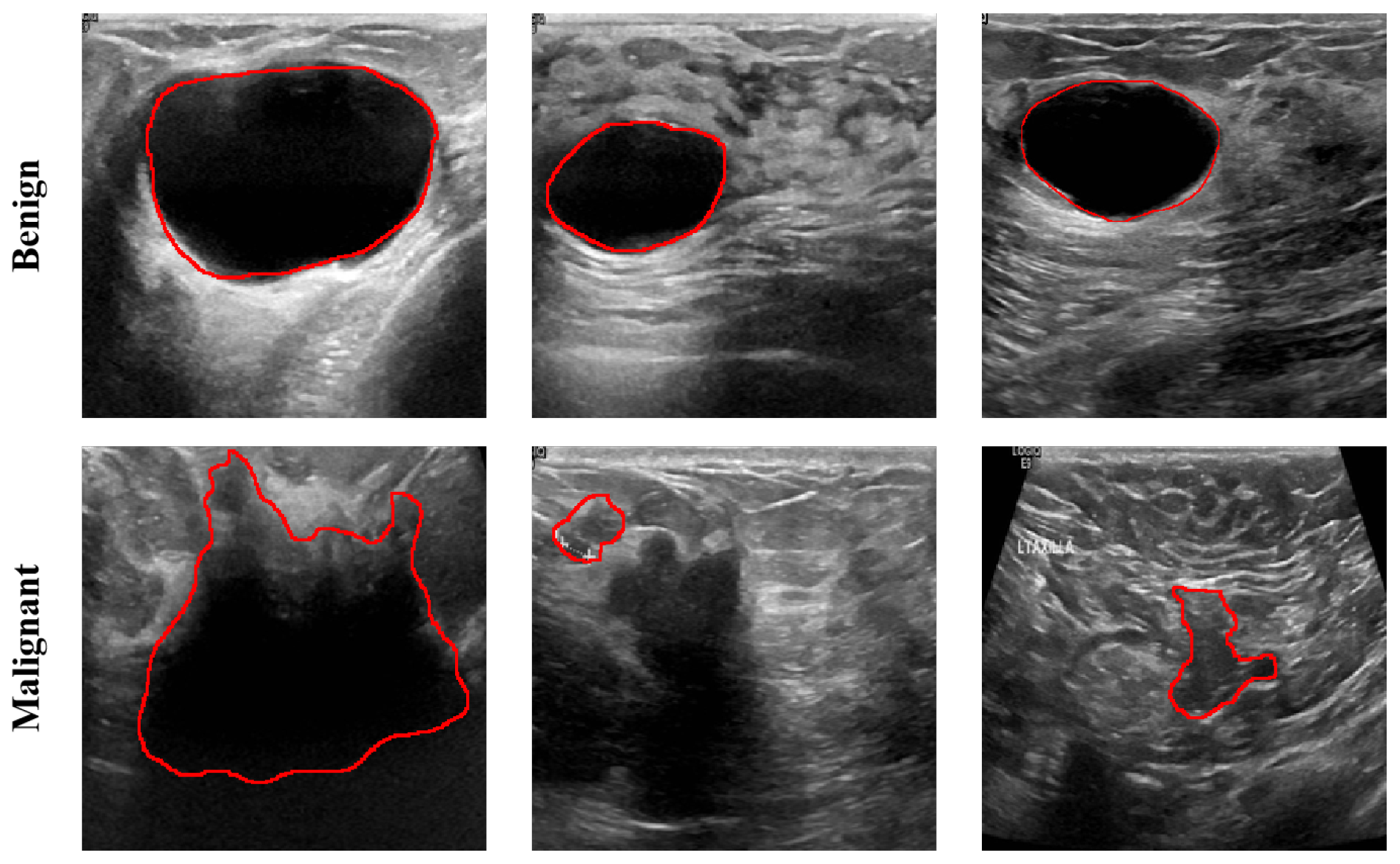
2.1. Dataset Description
2.2. Methods
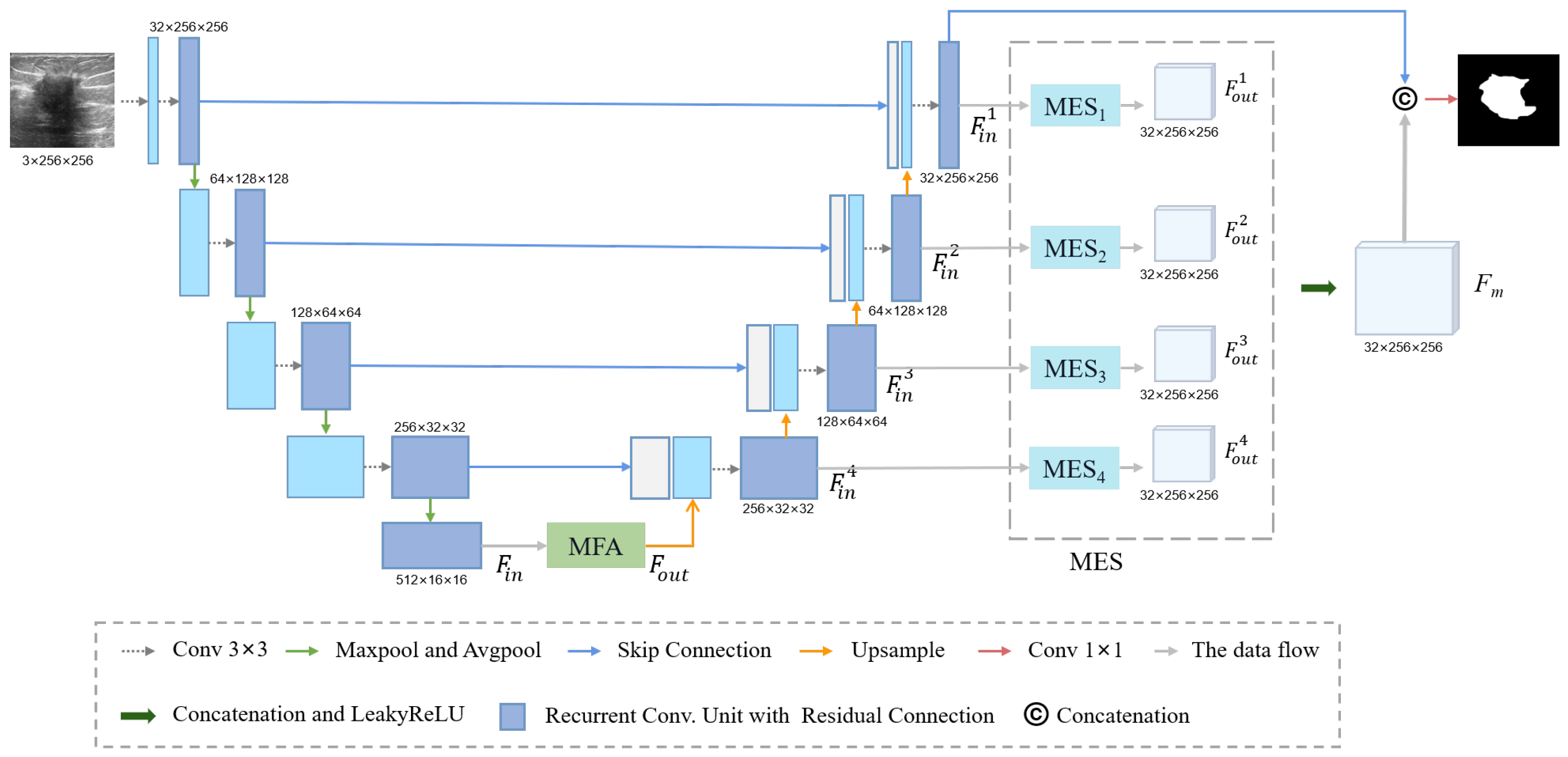
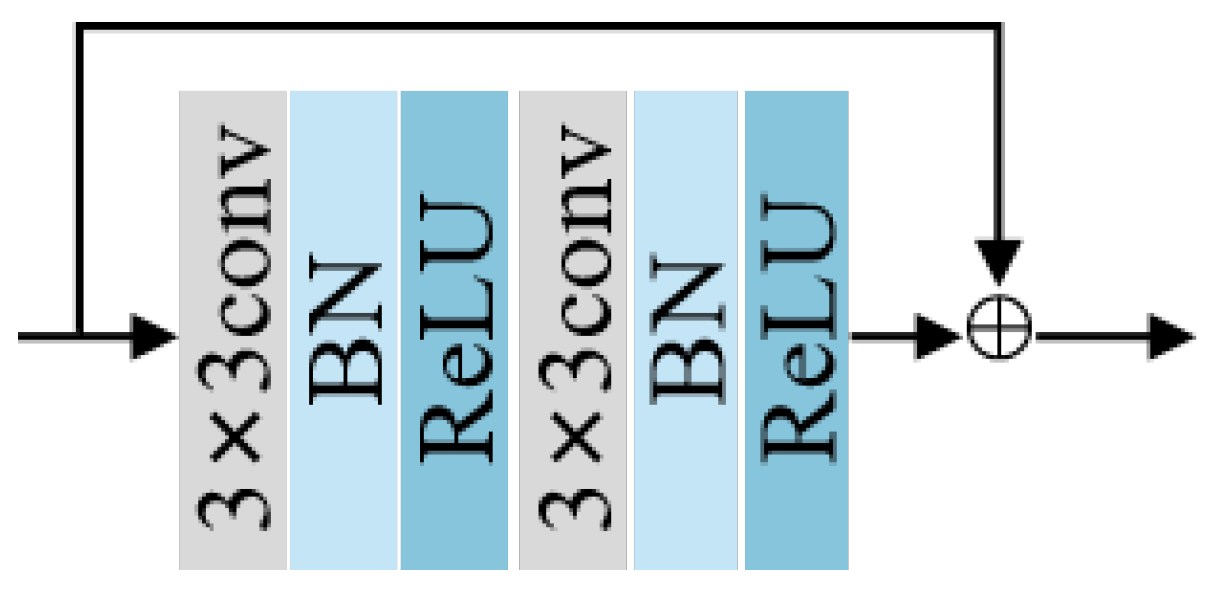
2.2.1. Residual Block and Down-Sampling Blocks
2.2.2. Multi-Scale Feature Acquisition Module
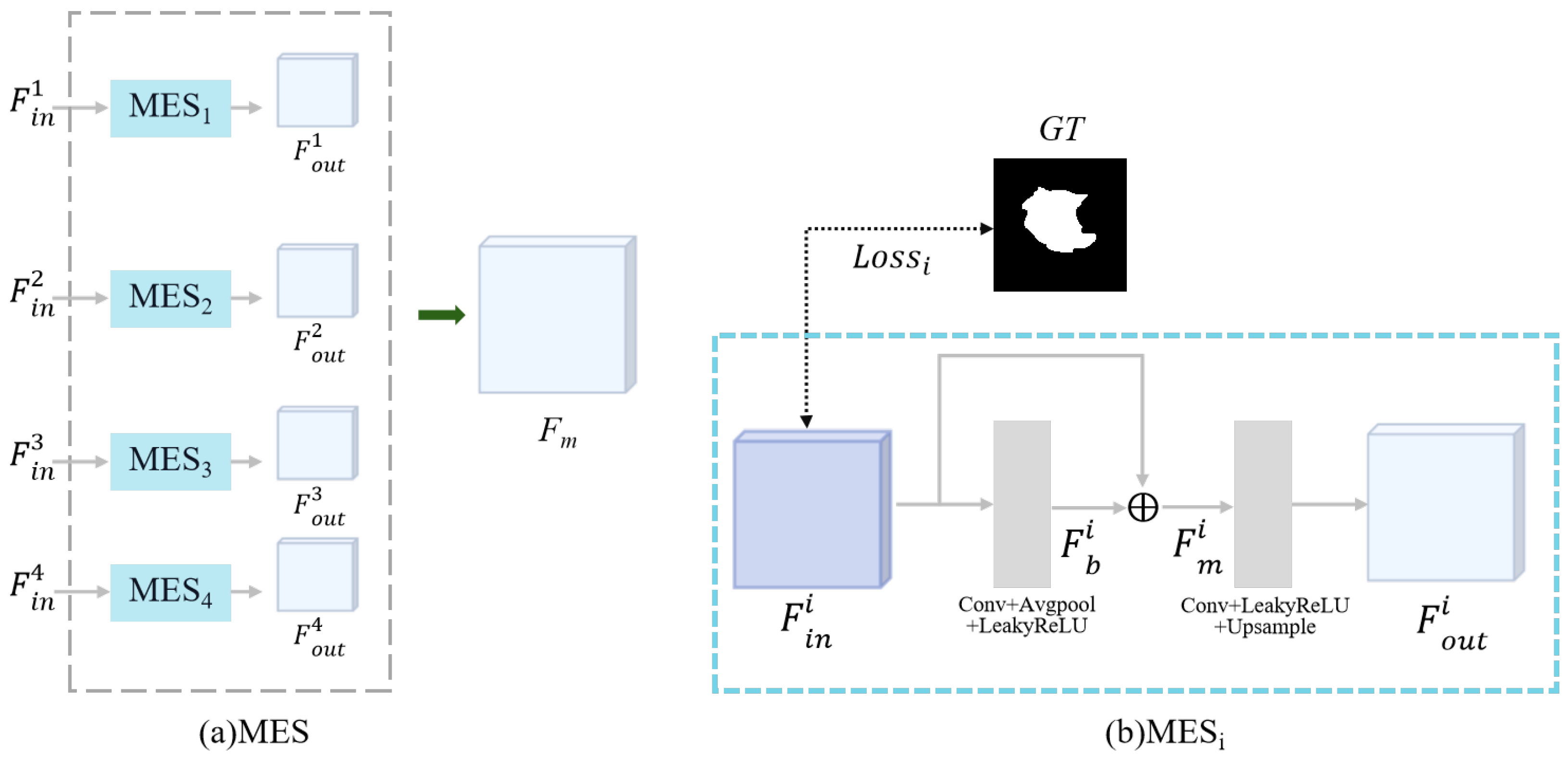
2.2.3. Multi-Input Edge Supplement Module
2.3. Loss Function
2.4. Evaluation Metrics
3. Results
3.1. Experiment Setting
3.2. Comparison Study
3.2.1. Comparison with Existing Methods
3.2.2. Comparison with Existing Multi-Scale Modules
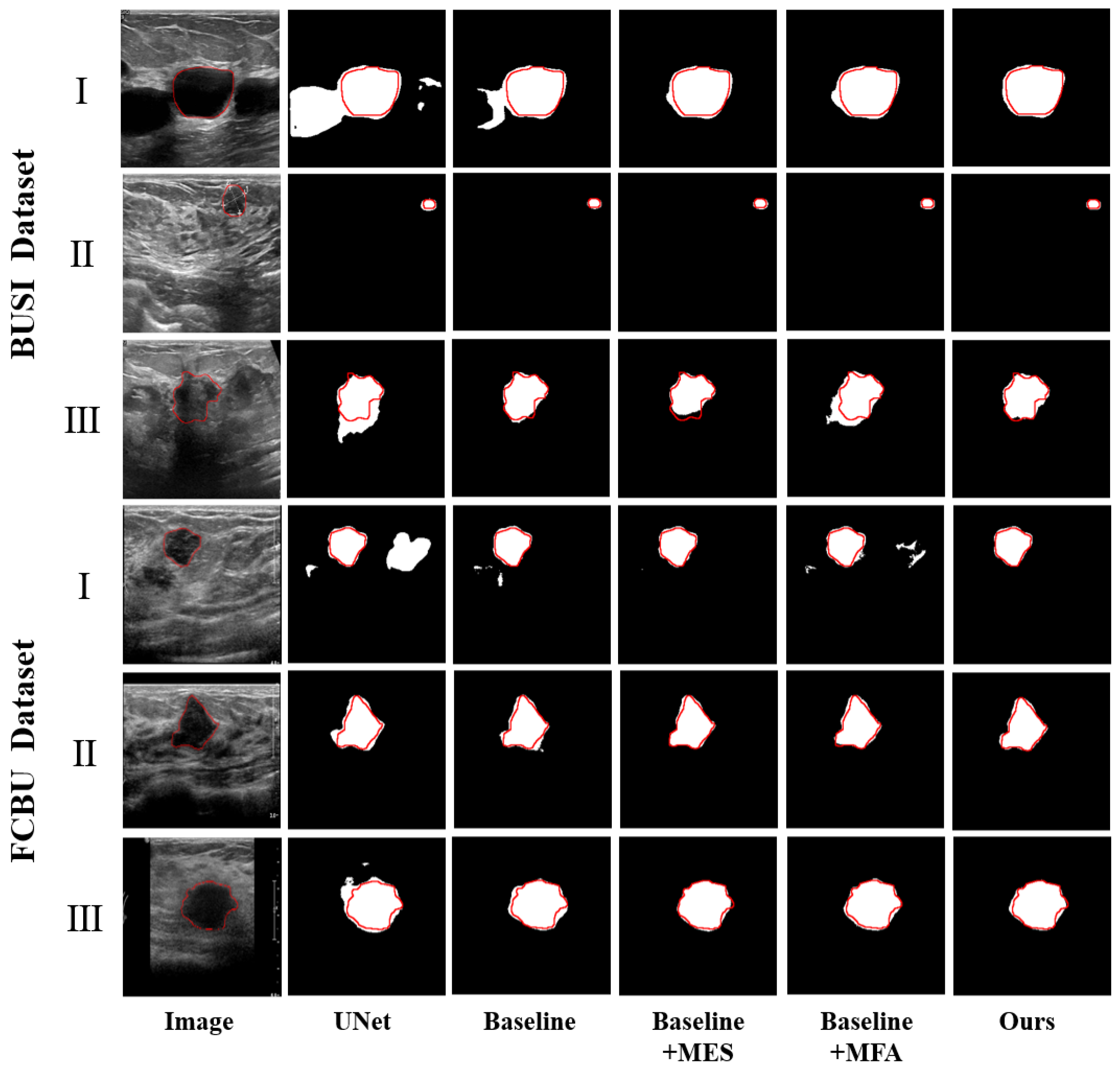
3.3. Ablation Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Yi, F.; Zhang, L.; Chen, Y.; Pi, Y.; Chen, Y.; Guo, J.; Wang, J.; Guo, Q.; Li, J.; et al. Computer-aided diagnosis of breast cancer in ultrasonography images by deep learning. Neurocomputing 2022, 472, 152–165. [Google Scholar] [CrossRef]
- AlZoubi, A.; Eskandari, A.; Yu, H.; Du, H. Explainable DCNN Decision Framework for Breast Lesion Classification from Ultrasound Images Based on Cancer Characteristics. Bioengineering 2024, 11, 453. [Google Scholar] [CrossRef]
- Arian, A.; Dinas, K.; Pratilas, G.C.; Alipour, S. The breast imaging-reporting and data system (BI-RADS) Made Easy. Iran. J. Radiol. 2022, 19, e121155. [Google Scholar] [CrossRef]
- Ning, Z.; Zhong, S.; Feng, Q.; Chen, W.; Zhang, Y. SMU-Net: Saliency-guided morphology-aware U-Net for breast lesion segmentation in ultrasound image. IEEE Trans. Med. Imaging 2021, 41, 476–490. [Google Scholar] [CrossRef] [PubMed]
- Hooley, R.J.; Scoutt, L.M.; Philpotts, L.E. Breast ultrasonography: State of the art. Radiology 2013, 268, 642–659. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.H.; Edirisinghe, E.; Bez, H. Processed images in human perception: A case study in ultrasound breast imaging. Eur. J. Radiol. 2010, 73, 682–687. [Google Scholar] [CrossRef]
- Yap, M.H.; Pons, G.; Marti, J.; Ganau, S.; Sentis, M.; Zwiggelaar, R.; Davison, A.K.; Marti, R. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J. Biomed. Health Inform. 2017, 22, 1218–1226. [Google Scholar] [CrossRef]
- Maolood, I.Y.; Al-Salhi, Y.E.A.; Lu, S. Thresholding for medical image segmentation for cancer using fuzzy entropy with level set algorithm. Open Med. 2018, 13, 374–383. [Google Scholar] [CrossRef]
- Daoud, M.I.; Atallah, A.A.; Awwad, F.; Al-Najjar, M.; Alazrai, R. Automatic superpixel-based segmentation method for breast ultrasound images. Expert Syst. Appl. 2019, 121, 78–96. [Google Scholar]
- Kozegar, E.; Soryani, M.; Behnam, H.; Salamati, M.; Tan, T. Mass segmentation in automated 3-D breast ultrasound using adaptive region growing and supervised edge-based deformable model. IEEE Trans. Med. Imaging 2017, 37, 918–928. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4. Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Ilesanmi, A.E.; Idowu, O.P.; Makhanov, S.S. Multiscale superpixel method for segmentation of breast ultrasound. Comput. Biol. Med. 2020, 125, 103879. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Chen, G.; Liu, Y.; Qian, J.; Zhang, J.; Yin, X.; Cui, L.; Dai, Y. DSEU-net: A novel deep supervision SEU-net for medical ultrasound image segmentation. Expert Syst. Appl. 2023, 223, 119939. [Google Scholar] [CrossRef]
- Byra, M.; Jarosik, P.; Szubert, A.; Galperin, M.; Ojeda-Fournier, H.; Olson, L.; O’Boyle, M.; Comstock, C.; Andre, M. Breast mass segmentation in ultrasound with selective kernel U-Net convolutional neural network. Biomed. Signal Process. Control 2020, 61, 102027. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, Q.; Chen, J.; Chen, L.; Chen, Z. Effective hybrid attention network based on pseudo-color enhancement in ultrasound image segmentation. Image Vis. Comput. 2023, 137, 104742. [Google Scholar]
- Wu, X.; Jiang, Y.; Xing, H.; Song, W.; Wu, P.; Cui, X.w.; Xu, G. ULS4US: Universal lesion segmentation framework for 2D ultrasound images. Phys. Med. Biol. 2023, 68, 165009. [Google Scholar]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. Resunet++: An advanced architecture for medical image segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 225–2255. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Sharif, M. PDF-UNet: A semi-supervised method for segmentation of breast tumor images using a U-shaped pyramid-dilated network. Expert Syst. Appl. 2023, 221, 119718. [Google Scholar] [CrossRef]
- Liu, S.; Huang, D.; Wang, H. Receptive field block net for accurate and fast object detection. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 385–400. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar]
- Zhu, C.; Chai, X.; Xiao, Y.; Liu, X.; Zhang, R.; Yang, Z.; Wang, Z. Swin-Net: A swin-transformer-based network combing with multi-scale features for segmentation of breast tumor ultrasound images. Diagnostics 2024, 14, 269. [Google Scholar] [CrossRef]
- Tagnamas, J.; Ramadan, H.; Yahyaouy, A.; Tairi, H. Multi-task approach based on combined CNN-transformer for efficient segmentation and classification of breast tumors in ultrasound images. Vis. Comput. Ind. Biomed. Art 2024, 7, 2. [Google Scholar]

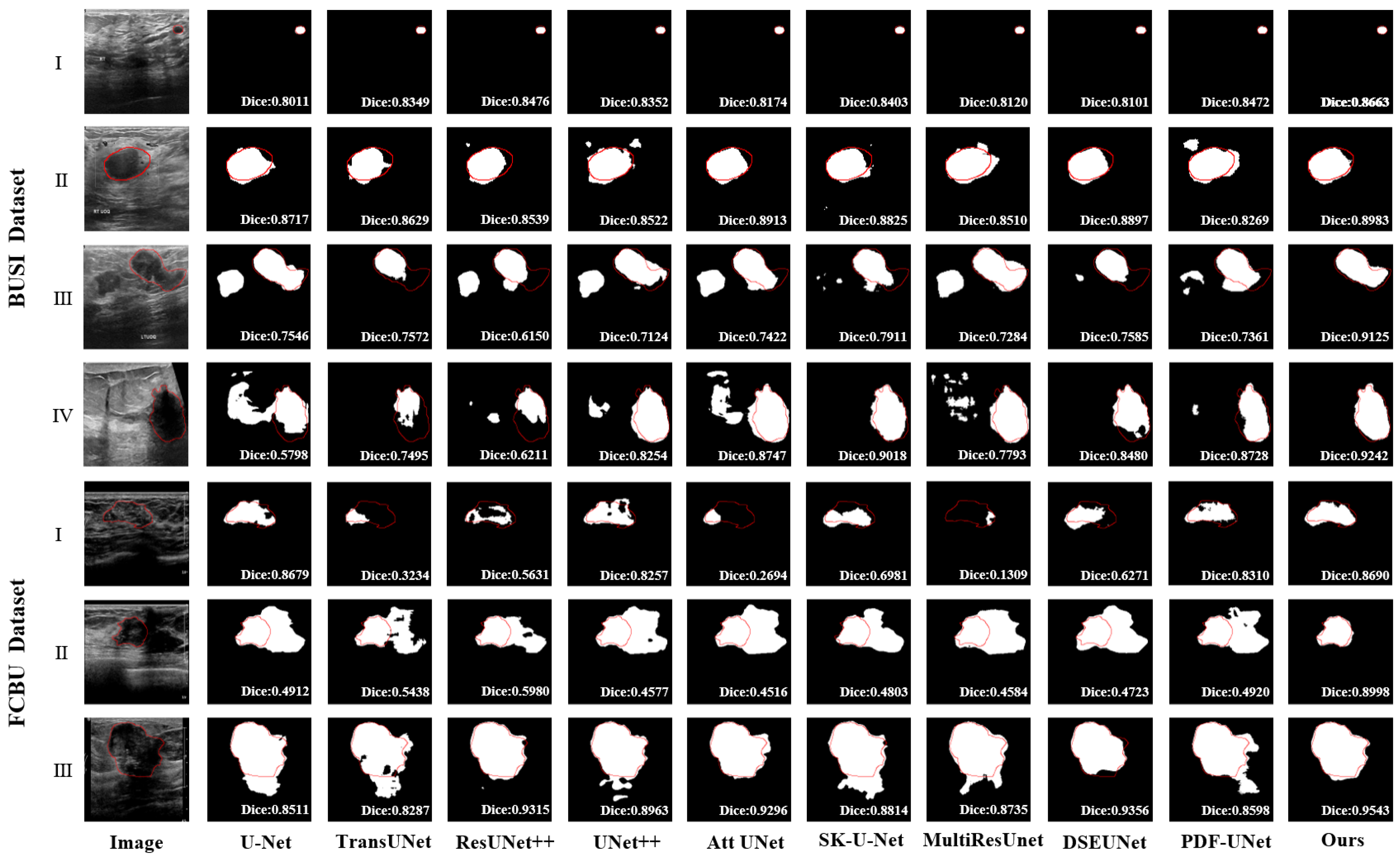
| Method | Dataset | Dice % | PA % | IoU % | Recall % | HD (mm) |
|---|---|---|---|---|---|---|
| UNet | BUSI | 69.56 ± 2.57 Δ | 95.90 ± 0.43 * | 77.15 ± 1.31 Δ | 83.14 ± 2.24 * | 32.30 ± 3.43 * |
| TransUNet | 62.96 ± 3.07 Δ | 94.72 ± 0.73 Δ | 73.88 ± 1.62 Δ | 82.58 ± 2.52 Δ | 38.02 ± 4.11 Δ | |
| ResUNet++ | 70.83 ± 2.44 Δ | 95.91 ± 0.46 * | 77.73 ± 1.30 Δ | 85.35 ± 2.02 Δ | 30.71 ± 3.21 * | |
| UNet++ | 73.06 ± 2.23 Δ | 96.21 ± 0.45 * | 78.92 ± 1.23 Δ | 84.93 ± 2.04 * | 32.50 ± 3.40 * | |
| Att UNet | 74.64 ± 2.14 Δ | 96.27 ± 0.45 * | 79.76 ± 1.17 Δ | 84.26 ± 2.06 | 32.37 ± 3.63 * | |
| SK UNet | 73.19 ± 2.32 Δ | 96.01 ± 0.55 * | 79.12 ± 1.32 Δ | 85.23 ± 1.99 Δ | 30.08 ± 3.31 * | |
| MultiResUNet | 73.91 ± 2.20 Δ | 96.12 ± 0.48 * | 79.36 ± 1.22 Δ | 84.72 ± 1.99 | 26.85 ± 2.93 * | |
| DSEUNet | 77.06 ± 2.16 | 96.57 ± 0.42 | 81.52 ± 1.16 * | 85.53 ± 1.97 | 22.98 ± 3.02 * | |
| PDF-UNet | 78.29 ± 1.92 | 96.56 ± 0.42 | 82.02 ± 1.13 | 86.58 ± 1.79 | 22.10 ± 2.55 | |
| Ours | 79.43 ± 1.97 | 96.84 ± 0.45 | 83.00 ± 1.12 | 87.17 ± 1.80 | 19.71 ± 2.67 | |
| UNet | FCBU | 86.74 ± 1.39 Δ | 96.95 ± 0.28 Δ | 87.37 ± 0.92 Δ | 90.84 ± 1.46 * | 18.63 ± 2.18 * |
| TransUNet | 84.08 ± 1.73 Δ | 96.29 ± 0.36 Δ | 85.33 ± 1.10 Δ | 90.38 ± 1.51 * | 22.37 ± 2.09 Δ | |
| ResUNet++ | 87.16 ± 1.13 Δ | 96.98 ± 0.29 Δ | 87.53 ± 0.89 Δ | 92.06 ± 1.20 | 18.35 ± 2.06 Δ | |
| UNet++ | 87.45 ± 1.07 Δ | 96.91 ± 0.32 Δ | 87.64 ± 0.86 Δ | 91.77 ± 1.20 | 19.50 ± 2.31 Δ | |
| Att UNet | 88.02 ± 1.18 * | 97.14 ± 0.31 * | 88.33 ± 0.90 * | 92.91 ± 1.07 | 16.79 ± 2.00 * | |
| SK UNet | 88.05 ± 0.87 Δ | 97.02 ± 0.28 Δ | 87.98 ± 0.74 Δ | 91.94 ± 1.22 | 15.39 ± 1.41 Δ | |
| MultiResUNet | 87.71 ± 1.18 * | 96.98 ± 0.30 Δ | 87.91 ± 0.83 Δ | 92.27 ± 1.10 | 15.46 ± 1.70 * | |
| DSEUNet | 88.71 ± 0.95 * | 97.10 ± 0.33 * | 88.63 ± 0.82 * | 92.46 ± 1.15 | 14.71 ± 1.48 * | |
| PDF-UNet | 89.12 ± 1.27 * | 97.47 ± 0.26 | 89.40 ± 0.83 * | 93.79 ± 1.15 * | 14.85 ± 1.58 * | |
| Ours | 90.97 ± 0.62 | 97.77 ± 0.23 | 90.62 ± 0.59 | 94.63 ± 0.77 | 11.02 ± 1.00 |
| Methods | Benign | Malignant | ||
|---|---|---|---|---|
| Dice | HD | Dice | HD | |
| UNet | 72.47 ± 2.99 | 25.58 ± 4.07 | 63.80 ± 4.79 | 44.72 ± 5.79 |
| TransUNet | 65.19 ± 3.80 | 33.31 ± 5.00 | 58.55 ± 5.22 | 47.34 ± 7.04 |
| ResUNet++ | 74.45 ± 2.82 | 25.26 ± 3.90 | 63.66 ± 4.50 | 41.74 ± 5.31 |
| UNet++ | 75.14 ± 2.70 | 27.55 ± 4.34 | 68.95 ± 3.90 | 41.79 ± 5.19 |
| Att UNet | 74.39 ± 2.71 | 29.97 ± 4.79 | 75.13 ± 3.50 | 36.87 ± 5.30 |
| SK UNet | 75.98 ± 2.82 | 25.51 ± 4.34 | 67.68 ± 4.00 | 38.88 ± 4.68 |
| MultiResUNet | 76.10 ± 2.47 | 20.18 ± 3.18 | 69.59 ± 4.35 | 39.87 ± 5.54 |
| DSEUNet | 78.60 ± 2.54 | 19.11 ± 3.82 | 74.02 ± 4.00 | 30.80 ± 4.68 |
| PDF-UNet | 79.50 ± 2.32 | 16.75 ± 2.80 | 75.90 ± 3.41 | 32.66 ± 4.84 |
| Ours | 79.96 ± 2.53 | 15.68 ± 3.26 | 78.39 ± 3.09 | 27.59 ± 4.43 |
| Methods | Dataset | Dice % | PA % | IoU % | Recall % | HD (mm) |
|---|---|---|---|---|---|---|
| Baseline + RFB | BUSI | 75.65 ± 2.22 | 96.40 ± 0.44 | 80.69 ± 1.23 | 85.59 ± 1.97 | 23.10 ± 2.52 |
| Baseline + SPP | 75.79 ± 2.14 | 96.16 ± 0.50 | 80.58 ± 1.26 | 85.42 ± 1.99 | 29.17 ± 3.31 | |
| Baseline + ASPP | 75.52 ± 2.24 | 96.38 ± 0.45 | 80.61 ± 1.23 | 85.04 ± 2.04 | 24.26 ± 2.88 | |
| Baseline + SPPCSPC | 76.61 ± 2.12 | 96.55 ± 0.44 | 81.23 ± 1.19 | 86.17 ± 1.88 | 27.92 ± 3.45 | |
| Baseline + MFA | 77.83 ± 1.89 | 96.31 ± 0.50 | 81.51 ± 1.14 | 85.86 ± 1.84 | 22.20 ± 2.53 | |
| Baseline + RFB | FCBU | 88.58 ± 1.34 | 97.43 ± 0.25 | 89.01 ± 0.86 | 93.30 ± 1.19 | 15.00 ± 1.57 |
| Baseline + SPP | 88.34 ± 0.93 | 97.02 ± 0.31 | 88.26 ± 0.79 | 93.30 ± 1.04 | 16.73 ± 1.57 | |
| Baseline + ASPP | 88.64 ± 0.82 | 97.19 ± 0.27 | 88.50 ± 0.71 | 92.49 ± 1.10 | 16.82 ± 1.68 | |
| Baseline + SPPCSPC | 89.51 ± 0.90 | 97.26 ± 0.32 | 89.33 ± 0.79 | 93.41 ± 1.11 | 14.35 ± 1.54 | |
| Baseline + MFA | 89.74 ± 0.75 | 97.52 ± 0.23 | 89.54 ± 0.64 | 93.26 ± 0.94 | 14.00 ± 1.33 |
| Methods | Dataset | Dice % | PA % | IoU % | Recall % | HD (mm) |
|---|---|---|---|---|---|---|
| UNet | BUSI | 69.56 ± 2.57 | 95.90 ± 0.43 | 77.15 ± 1.31 | 83.14 ± 2.24 | 32.30 ± 3.43 |
| Baseline | 71.47 ± 2.44 | 96.01 ± 0.48 | 78.24 ± 1.32 | 83.77 ± 2.18 | 29.42 ± 3.09 | |
| Baseline + MES | 74.17 ± 2.51 | 96.56 ± 0.47 | 80.36 ± 1.33 | 86.71 ± 2.01 | 20.52 ± 2.47 | |
| Baseline + MFA | 77.83 ± 1.89 | 96.31 ± 0.50 | 81.51 ± 1.14 | 85.86 ± 1.84 | 22.20 ± 2.53 | |
| Baseline + MES + MFA | 79.43 ± 1.97 | 96.84 ± 0.45 | 83.00 ± 1.12 | 87.17 ± 1.80 | 19.71 ± 2.67 | |
| UNet | FCBU | 86.74 ± 1.39 | 96.95 ± 0.28 | 87.37 ± 0.92 | 90.84 ± 1.46 | 18.63 ± 2.18 |
| Baseline | 87.85 ± 1.08 | 97.09 ± 0.28 | 88.08 ± 0.85 | 92.26 ± 1.21 | 17.71 ± 1.89 | |
| Baseline + MES | 88.75 ± 0.97 | 97.21 ± 0.28 | 88.76 ± 0.81 | 93.02 ± 1.14 | 15.91 ± 1.77 | |
| Baseline + MFA | 88.89 ± 0.90 | 97.36 ± 0.25 | 88.86 ± 0.71 | 93.09 ± 0.98 | 15.97 ± 1.74 | |
| Baseline + MES + MFA | 90.97 ± 0.62 | 97.77 ± 0.23 | 90.62 ± 0.59 | 94.63 ± 0.77 | 11.02 ± 1.00 |
| Methods | Benign | Malignant | ||
|---|---|---|---|---|
| Dice | HD | Dice | HD | |
| UNet | 72.47 ± 2.99 | 25.58 ± 4.07 | 63.80 ± 4.79 | 44.72 ± 5.79 |
| Baseline | 74.25 ± 2.88 | 24.52 ± 3.66 | 65.96 ± 4.43 | 39.33 ± 5.40 |
| Baseline + MES | 75.79 ± 3.08 | 14.53 ± 2.50 | 70.96 ± 4.32 | 32.06 ± 4.92 |
| Baseline + MFA | 79.21 ± 2.29 | 16.70 ± 2.78 | 75.10 ± 3.33 | 32.94 ± 4.75 |
| Baseline + MES + MFA | 79.96 ± 2.53 | 15.68 ± 3.26 | 78.39 ± 3.09 | 27.59 ± 4.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, L.; Tang, M.; Chen, X.; Du, Z.; Huang, D.; Yang, M.; Chen, Y. M2UNet: Multi-Scale Feature Acquisition and Multi-Input Edge Supplement Based on UNet for Efficient Segmentation of Breast Tumor in Ultrasound Images. Diagnostics 2025, 15, 944. https://doi.org/10.3390/diagnostics15080944
Pan L, Tang M, Chen X, Du Z, Huang D, Yang M, Chen Y. M2UNet: Multi-Scale Feature Acquisition and Multi-Input Edge Supplement Based on UNet for Efficient Segmentation of Breast Tumor in Ultrasound Images. Diagnostics. 2025; 15(8):944. https://doi.org/10.3390/diagnostics15080944
Chicago/Turabian StylePan, Lin, Mengshi Tang, Xin Chen, Zhongshi Du, Danfeng Huang, Mingjing Yang, and Yijie Chen. 2025. "M2UNet: Multi-Scale Feature Acquisition and Multi-Input Edge Supplement Based on UNet for Efficient Segmentation of Breast Tumor in Ultrasound Images" Diagnostics 15, no. 8: 944. https://doi.org/10.3390/diagnostics15080944
APA StylePan, L., Tang, M., Chen, X., Du, Z., Huang, D., Yang, M., & Chen, Y. (2025). M2UNet: Multi-Scale Feature Acquisition and Multi-Input Edge Supplement Based on UNet for Efficient Segmentation of Breast Tumor in Ultrasound Images. Diagnostics, 15(8), 944. https://doi.org/10.3390/diagnostics15080944






