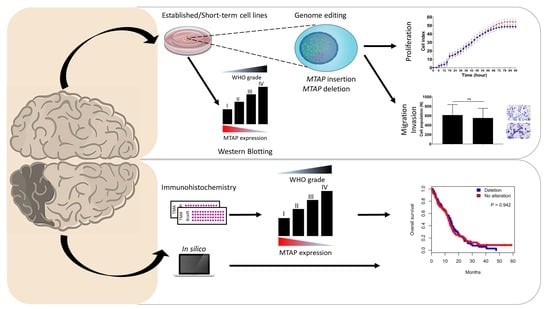
Loss of 5′-Methylthioadenosine Phosphorylase (MTAP) is Frequent in High-Grade Gliomas; Nevertheless, it is Not Associated with Higher Tumor Aggressiveness
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Gene Editing
2.2. Patients
2.3. DNA Isolation
2.4. RNA Extraction and RT-qPCR
2.5. IDH1 Mutation Analysis
2.6. TERT Mutation Analysis
2.7. MGMT Promoter Methylation
2.8. Western Blotting Analysis
2.9. Immunohistochemistry Analysis
2.10. In Silico Analysis
2.11. mRNA NanoStringTM Data Analysis
2.12. xCELLigence Proliferation Assay
2.13. Transwell Migration Assay
2.14. Cell Invasion Assay
2.15. Statistical Analysis
3. Results
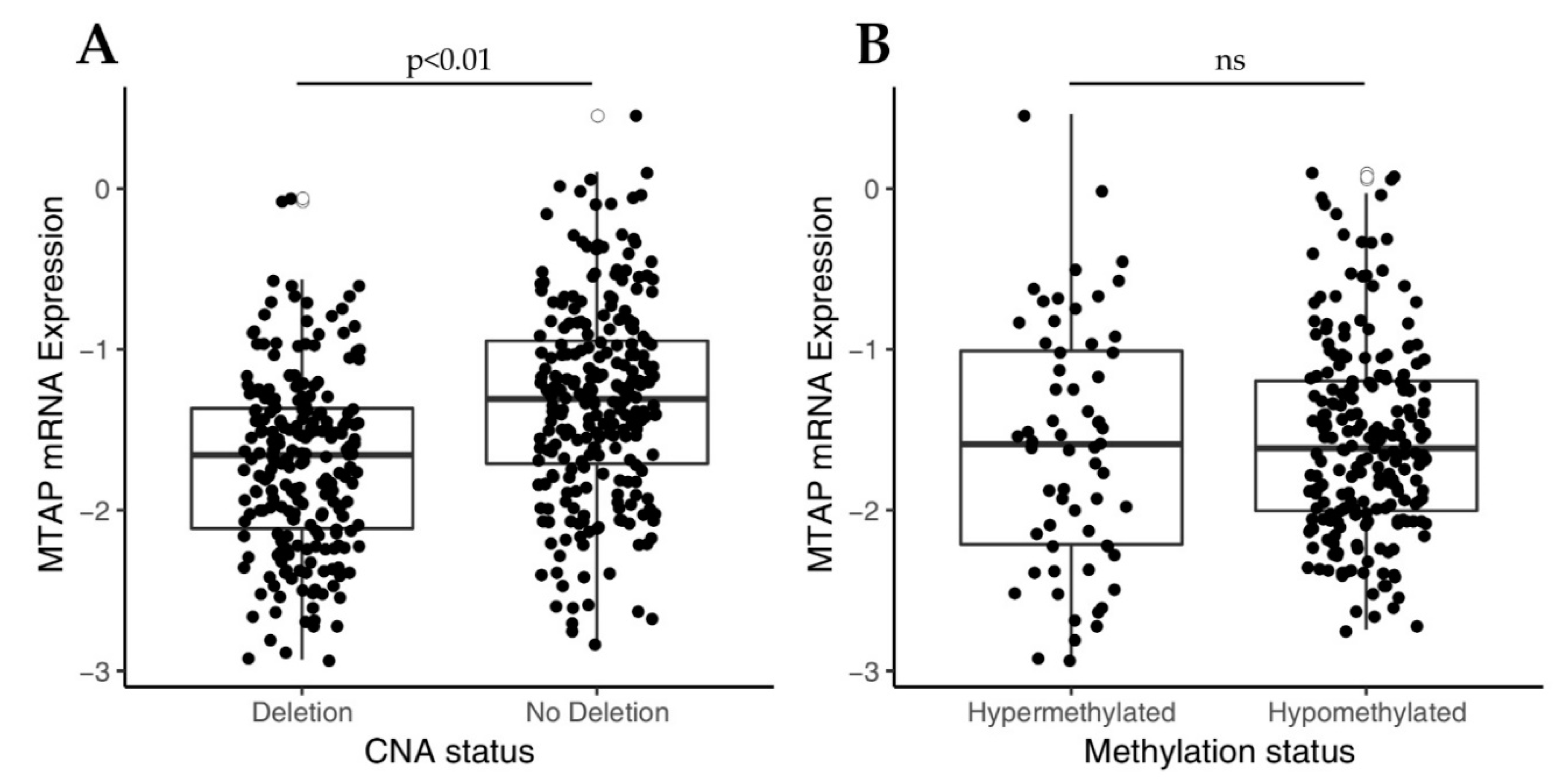
3.1. Loss of MTAP Expression is Associated with 9p21 Locus Deletion in Gliomas
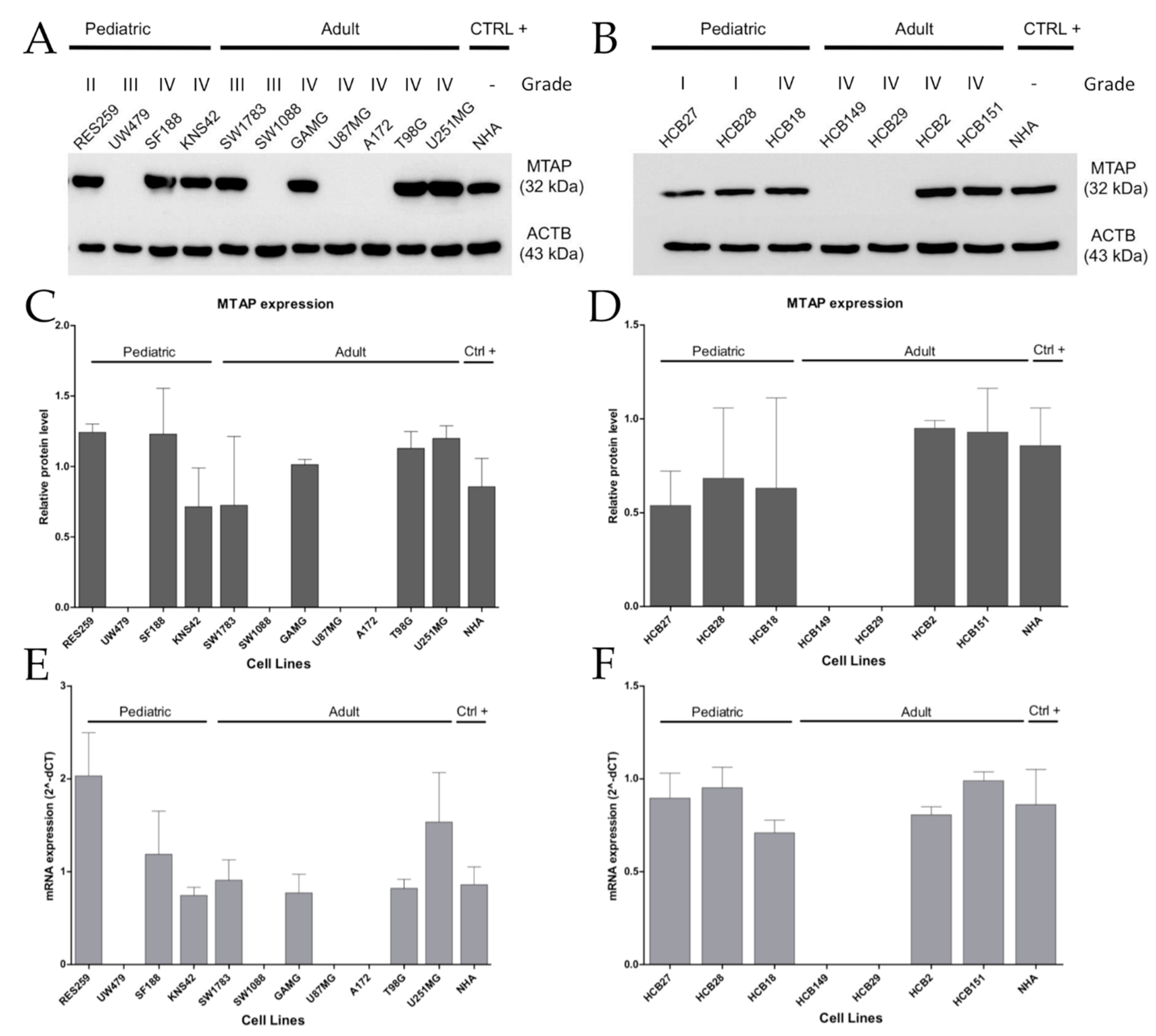
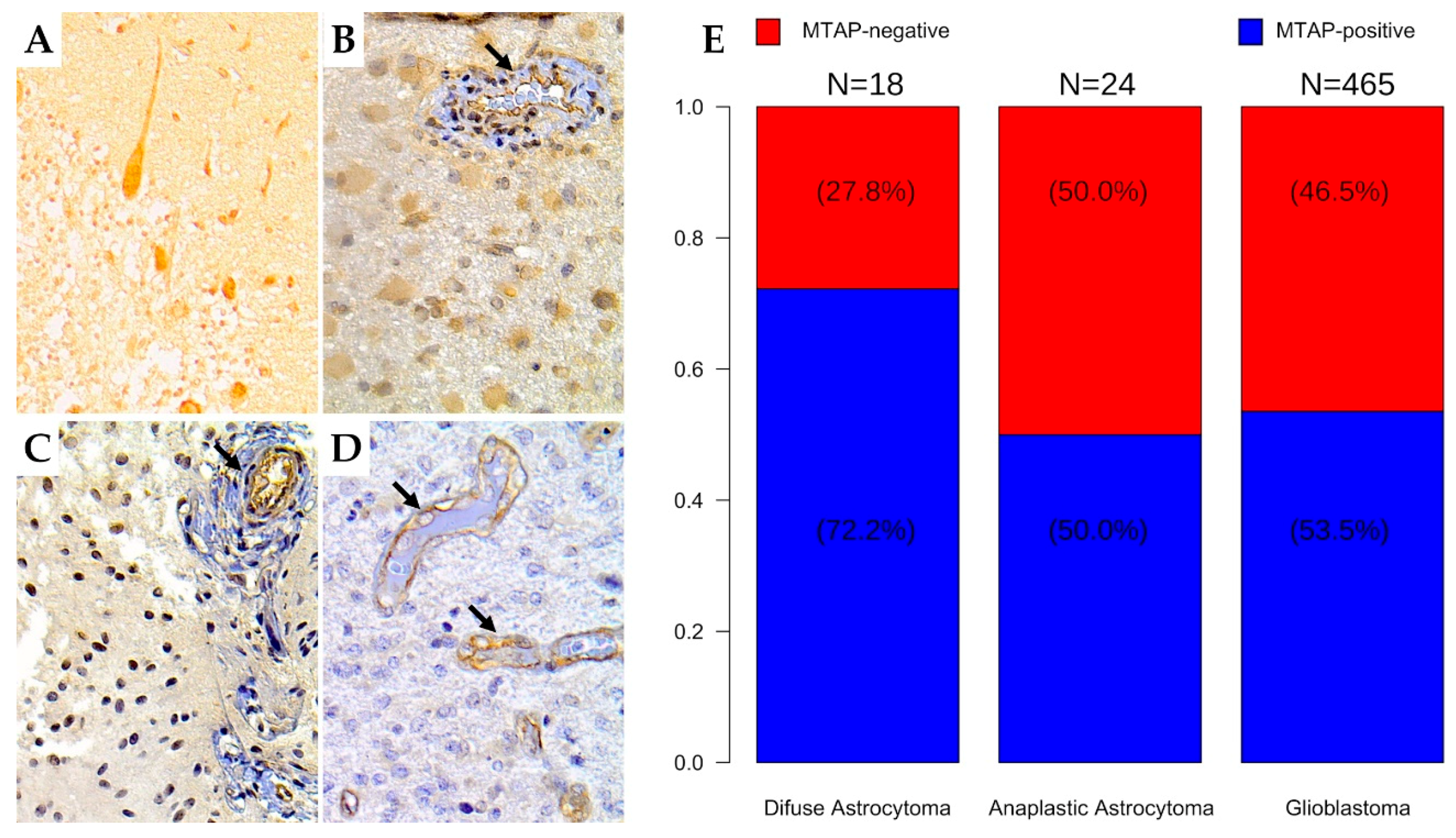
3.2. MTAP Expression Profile and Clinicopathological Association in Gliomas
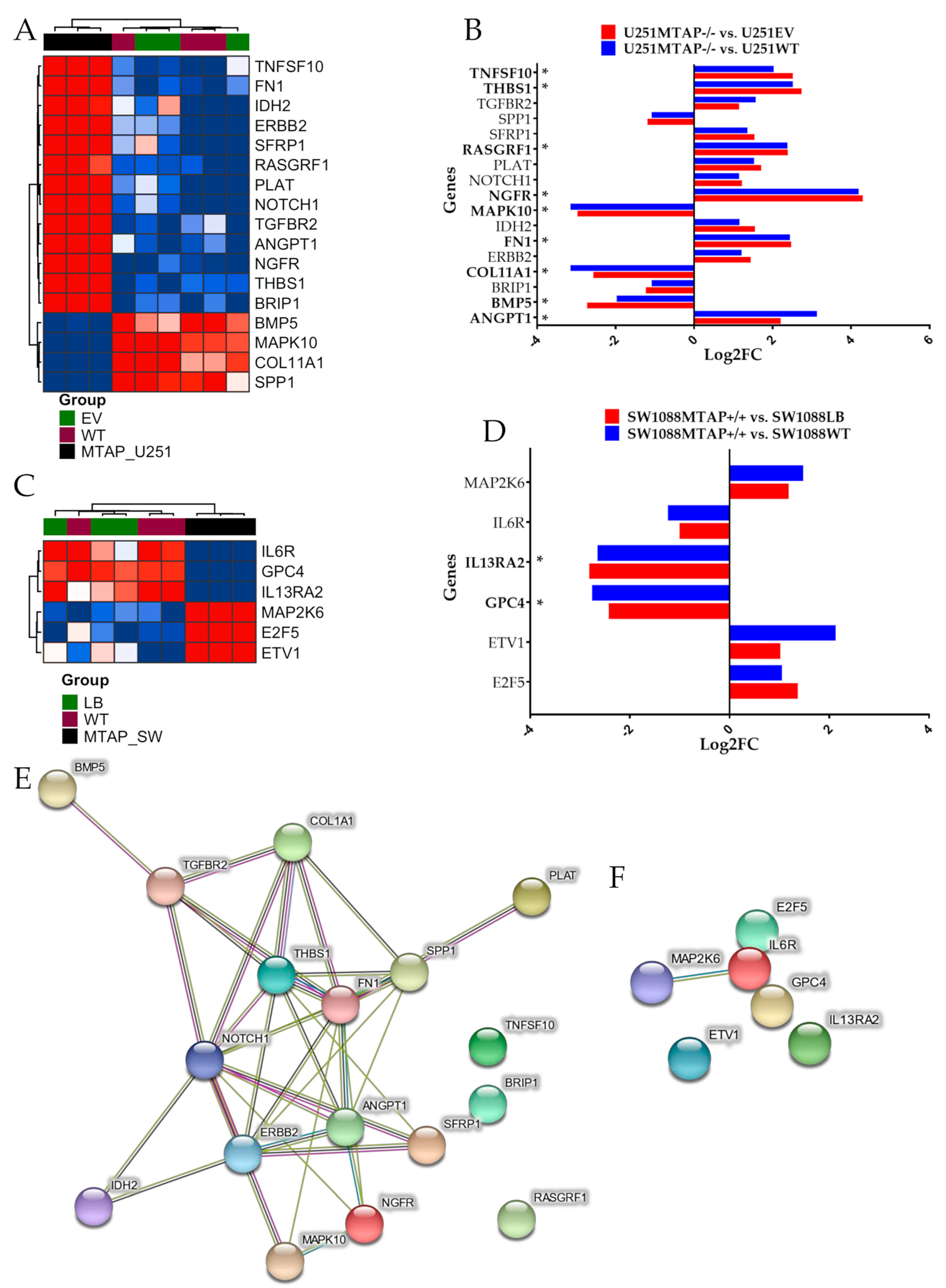
3.3. MTAP Cell Line Editing and Differential Gene Expression
3.4. MTAP Deletion Does Not Modulate Glioma Cell Proliferation
3.5. Loss of MTAP Gene is Not Associated with Cell Migration and Invasion
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
A.1. Cell Lines
A.2. Gene Editing
A.2.1. CRISPR/Cas9-Mediated MTAP Gene Editing
A.2.2. Cell Transduction and Generation of MTAP Overexpressing Cell Line
References
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro Oncol. 2018, 20, 86. [Google Scholar] [CrossRef] [PubMed]
- Ganau, L.; Paris, M.; Ligarotti, G.K.; Ganau, M. Management of Gliomas: Overview of the Latest Technological Advancements and Related Behavioral Drawbacks. Behav. Neurol. 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Appin, C.L.; Brat, D.J. Molecular genetics of gliomas. Cancer J. 2014, 20, 66–72. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef]
- Knizhnik, A.V.; Roos, W.P.; Nikolova, T.; Quiros, S.; Tomaszowski, K.H.; Christmann, M.; Kaina, B. Survival and death strategies in glioma cells: Autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS ONE 2013, 8, e55665. [Google Scholar] [CrossRef]
- Sturm, D.; Bender, S.; Jones, D.T.; Lichter, P.; Grill, J.; Becher, O.; Hawkins, C.; Majewski, J.; Jones, C.; Costello, J.F.; et al. Paediatric and adult glioblastoma: Multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 2014, 14, 92–107. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Burger, P.; Ellison, D.W.; Reifenberger, G.; von Deimling, A.; Aldape, K.; Brat, D.; Collins, V.P.; Eberhart, C.; et al. International Society of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014, 24, 429–435. [Google Scholar] [CrossRef]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A., Jr.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef]
- Bidinotto, L.T.; Torrieri, R.; Mackay, A.; Almeida, G.C.; Viana-Pereira, M.; Cruvinel-Carloni, A.; Spina, M.L.; Campanella, N.C.; Pereira de Menezes, W.; Clara, C.A.; et al. Copy Number Profiling of Brazilian Astrocytomas. G3 2016, 6, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Foote, M.B.; Papadopoulos, N.; Diaz, L.A., Jr. Genetic Classification of Gliomas: Refining Histopathology. Cancer Cell 2015, 28, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Lie, A.; Li, T.; Chowdhury, R.; Liu, F.; Ozer, B.; Wei, B.; Green, R.M.; Ellingson, B.M.; Wang, H.J.; et al. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro-Oncol. 2017, 19, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Yamasaki, K.; Matsushita, Y.; Nakamura, T.; Shimokawa, A.; Takami, H.; Tanaka, S.; Mukasa, A.; Shirahata, M.; Shimizu, S.; et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol. Commun. 2016, 4, 79. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef]
- Savarese, T.M.; Crabtree, G.W.; Parks, R.E. 5′-methylthioadenosine phosphorylase—I. Biochem. Pharmacol. 1981, 30, 189–199. [Google Scholar] [CrossRef]
- Traweek, S.T.; Riscoe, M.K.; Ferro, A.J.; Braziel, R.M.; Magenis, R.E.; Fitchen, J.H. Methylthioadenosine phosphorylase deficiency in acute leukemia: Pathologic, cytogenetic, and clinical features. Blood 1988, 71, 1568–1573. [Google Scholar] [CrossRef]
- Tang, B.; Lee, H.O.; An, S.S.; Cai, K.Q.; Kruger, W.D. Specific Targeting of MTAP-Deleted Tumors with a Combination of 2’-Fluoroadenine and 5’-Methylthioadenosine. Cancer Res. 2018, 78, 4386–4395. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Hoffman, R.M.; Bertino, J.R. Exploiting methionine restriction for cancer treatment. Biochem. Pharmacol. 2018, 154, 170–173. [Google Scholar] [CrossRef]
- Batova, A.; Diccianni, M.B.; Nobori, T.; Vu, T.; Yu, J.; Bridgeman, L.; Yu, A.L. Frequent deletion in the methylthioadenosine phosphorylase gene in T-cell acute lymphoblastic leukemia: Strategies for enzyme-targeted therapy. Blood 1996, 88, 3083–3090. [Google Scholar] [CrossRef] [PubMed]
- Mirebeau, D.; Acquaviva, C.; Suciu, S.; Bertin, R.; Dastugue, N.; Robert, A.; Boutard, P.; Mechinaud, F.; Plouvier, E.; Otten, J.; et al. The prognostic significance of CDKN2A, CDKN2B and MTAP inactivation in B-lineage acute lymphoblastic leukemia of childhood. Results of the EORTC studies 58881 and 58951. Haematologica 2006, 91, 881–885. [Google Scholar]
- Illei, P.B.; Rusch, V.W.; Zakowski, M.F.; Ladanyi, M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin. Cancer Res. 2003, 9, 2108–2113. [Google Scholar] [PubMed]
- Huang, H.Y.; Li, S.H.; Yu, S.C.; Chou, F.F.; Tzeng, C.C.; Hu, T.H.; Uen, Y.H.; Tian, Y.F.; Wang, Y.H.; Fang, F.M.; et al. Homozygous deletion of MTAP gene as a poor prognosticator in gastrointestinal stromal tumors. Clin. Cancer Res. 2009, 15, 6963–6972. [Google Scholar] [CrossRef] [PubMed]
- Nobori, T.; Szinai, I.; Amox, D.; Parker, B.; Olopade, O.I.; Buchhagen, D.L.; Carson, D.A. Methylthioadenosine phosphorylase deficiency in human non-small cell lung cancers. Cancer Res. 1993, 53, 1098–1101. [Google Scholar] [PubMed]
- Christopher, S.A.; Diegelman, P.; Porter, C.W.; Kruger, W.D. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 2002, 62, 6639–6644. [Google Scholar]
- Behrmann, I.; Wallner, S.; Komyod, W.; Heinrich, P.C.; Schuierer, M.; Buettner, R.; Bosserhoff, A.K. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am. J. Pathol. 2003, 163, 683–690. [Google Scholar] [CrossRef]
- Hellerbrand, C.; Muhlbauer, M.; Wallner, S.; Schuierer, M.; Behrmann, I.; Bataille, F.; Weiss, T.; Scholmerich, J.; Bosserhoff, A.K. Promoter-hypermethylation is causing functional relevant downregulation of methylthioadenosine phosphorylase (MTAP) expression in hepatocellular carcinoma. Carcinogenesis 2006, 27, 64–72. [Google Scholar] [CrossRef]
- Kirovski, G.; Stevens, A.P.; Czech, B.; Dettmer, K.; Weiss, T.S.; Wild, P.; Hartmann, A.; Bosserhoff, A.K.; Oefner, P.J.; Hellerbrand, C. Down-regulation of methylthioadenosine phosphorylase (MTAP) induces progression of hepatocellular carcinoma via accumulation of 5’-deoxy-5’-methylthioadenosine (MTA). Am J. Pathol. 2011, 178, 1145–1152. [Google Scholar] [CrossRef]
- Tang, B.; Kadariya, Y.; Chen, Y.; Slifker, M.; Kruger, W.D. Expression of MTAP inhibits tumor-related phenotypes in HT1080 cells via a mechanism unrelated to its enzymatic function. G3 2014, 5, 35–44. [Google Scholar] [CrossRef]
- Basu, I.; Cordovano, G.; Das, I.; Belbin, T.J.; Guha, C.; Schramm, V.L. A transition state analogue of 5’-methylthioadenosine phosphorylase induces apoptosis in head and neck cancers. J. Biol. Chem. 2007, 282, 21477–21486. [Google Scholar] [CrossRef] [PubMed]
- Basu, I.; Locker, J.; Cassera, M.B.; Belbin, T.J.; Merino, E.F.; Dong, X.; Hemeon, I.; Evans, G.B.; Guha, C.; Schramm, V.L. Growth and metastases of human lung cancer are inhibited in mouse xenografts by a transition state analogue of 5′-methylthioadenosine phosphorylase. J. Biol. Chem. 2011, 286, 4902–4911. [Google Scholar] [CrossRef] [PubMed]
- Bistulfi, G.; Affronti, H.C.; Foster, B.A.; Karasik, E.; Gillard, B.; Morrison, C.; Mohler, J.; Phillips, J.G.; Smiraglia, D.J. The essential role of methylthioadenosine phosphorylase in prostate cancer. Oncotarget 2016, 7, 14380–14393. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.; Kim, J.H.; Lee, E.; Jang, Y.J.; Son, J.E.; Kwon, J.Y.; Lim, T.G.; Kim, S.; Park, J.H.; Kim, J.E.; et al. Methionine deprivation suppresses triple-negative breast cancer metastasis in vitro and in vivo. Oncotarget 2016, 7, 67223–67234. [Google Scholar] [CrossRef]
- Casero, R.A., Jr.; Murray Stewart, T.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Zhong, Y.; Lu, K.; Zhu, S.; Li, W.; Sun, S. Characterization of methylthioadenosin phosphorylase (MTAP) expression in colorectal cancer. Artif. Cellsnanomedicineand Biotechnol. 2018, 46, 2082–2087. [Google Scholar] [CrossRef]
- Nobori, T.; Karras, J.G.; Della Ragione, F.; Waltz, T.A.; Chen, P.P.; Carson, D.A. Absence of methylthioadenosine phosphorylase in human gliomas. Cancer Res. 1991, 51, 3193–3197. [Google Scholar]
- Olopade, O.I.; Jenkins, R.B.; Ransom, D.T.; Malik, K.; Pomykala, H.; Nobori, T.; Cowan, J.M.; Rowley, J.D.; Diaz, M.O. Molecular analysis of deletions of the short arm of chromosome 9 in human gliomas. Cancer Res. 1992, 52, 2523–2529. [Google Scholar]
- Frazao, L.; do Carmo Martins, M.; Nunes, V.M.; Pimentel, J.; Faria, C.; Miguens, J.; Sagarribay, A.; Matos, M.; Salgado, D.; Nunes, S.; et al. BRAF V600E mutation and 9p21: CDKN2A/B and MTAP co-deletions—Markers in the clinical stratification of pediatric gliomas. Bmc Cancer 2018, 18, 1259. [Google Scholar] [CrossRef]
- Hansen, L.J.; Sun, R.; Yang, R.; Singh, S.X.; Chen, L.H.; Pirozzi, C.J.; Moure, C.J.; Hemphill, C.; Carpenter, A.B.; Healy, P.; et al. MTAP Loss Promotes Stemness in Glioblastoma and Confers Unique Susceptibility to Purine Starvation. Cancer Res. 2019, 79, 3383–3394. [Google Scholar] [CrossRef]
- Huang, T.; Li, S.; Yang, Z.; Liu, J.; Han, Y. Loss of Heterozygosity of 9p Is Associated with Poorer Survival in Patients with Gliomas. Mol. Neurobiol. 2016, 53, 6407–6412. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.P.; Scapulatempo-Neto, C.; Menezes, W.P.; Clara, C.; Machado, H.R.; Oliveira, R.S.; Neder, L.; Reis, R.M. Expression of Methylthioadenosine Phosphorylase (MTAP) in Pilocytic Astrocytomas. Pathobiology 2015, 82, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Simms, D.; Paul, E.; Cizdziel, P.E.; Piotr, C.N. Trizol: A new reagent for optimal single step of RNA. Focus 1993, 15, 532–535. [Google Scholar]
- Batista, R.; Cruvinel-Carloni, A.; Vinagre, J.; Peixoto, J.; Catarino, T.A.; Campanella, N.C.; Menezes, W.; Becker, A.P.; de Almeida, G.C.; Matsushita, M.M.; et al. The prognostic impact of TERT promoter mutations in glioblastomas is modified by the rs2853669 single nucleotide polymorphism. Int. J. Cancer 2016, 139, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, N.; Marshall, L.; Perryman, L.; Bax, D.A.; Little, S.E.; Viana-Pereira, M.; Sharp, S.Y.; Vassal, G.; Pearson, A.D.; Reis, R.M.; et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010, 70, 9243–9252. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 5. [Google Scholar] [CrossRef]
- Wan, Y.-W.; Allen, G.I.; Liu, Z. TCGA2STAT: Simple TCGA data access for integrated statistical analysis in R. Bioinformatics 2016, 32, 952–954. [Google Scholar] [CrossRef]
- Waggott, D.; Chu, K.; Yin, S.; Wouters, B.G.; Liu, F.F.; Boutros, P.C. NanoStringNorm: An extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics 2012, 28, 1546–1548. [Google Scholar] [CrossRef]
- Zhao, M.; Zhao, Z. Concordance of copy number loss and down-regulation of tumor suppressor genes: A pan-cancer study. Bmc Genom. 2016, 17 (Suppl. 7), 532. [Google Scholar] [CrossRef]
- Crespo, I.; Tao, H.; Nieto, A.B.; Rebelo, O.; Domingues, P.; Vital, A.L.; Patino Mdel, C.; Barbosa, M.; Lopes, M.C.; Oliveira, C.R.; et al. Amplified and homozygously deleted genes in glioblastoma: Impact on gene expression levels. PLoS ONE 2012, 7, e46088. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Z.H.; Savarese, T.M. Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines. Cancer Genet. Cytogenet. 1996, 86, 22–28. [Google Scholar] [CrossRef]
- Su, C.Y.; Chang, Y.C.; Chan, Y.C.; Lin, T.C.; Huang, M.S.; Yang, C.J.; Hsiao, M. MTAP is an independent prognosis marker and the concordant loss of MTAP and p16 expression predicts short survival in non-small cell lung cancer patients. Eur. J. Surg. Oncol. 2014, 40, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Ueki, K.; Ono, Y.; Henson, J.W.; Efird, J.T.; von Deimling, A.; Louis, D.N. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996, 56, 150–153. [Google Scholar] [PubMed]
- Lopez, F.; Sampedro, T.; Llorente, J.L.; Hermsen, M.; Alvarez-Marcos, C. Alterations of p14 (ARF), p15 (INK4b), and p16 (INK4a) Genes in Primary Laryngeal Squamous Cell Carcinoma. Pathol. Oncol. Res. 2017, 23, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Krasinskas, A.M.; Bartlett, D.L.; Cieply, K.; Dacic, S. CDKN2A and MTAP deletions in peritoneal mesotheliomas are correlated with loss of p16 protein expression and poor survival. Mod Pathol 2010, 23, 531–538. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Malta, T.M.; de Souza, C.F.; Sabedot, T.S.; Silva, T.C.; Mosella, M.S.; Kalkanis, S.N.; Snyder, J.; Castro, A.V.B.; Noushmehr, H. Glioma CpG island methylator phenotype (G-CIMP): Biological and clinical implications. Neuro Oncol. 2018, 20, 608–620. [Google Scholar] [CrossRef]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef]
- Velázquez Vega, J.E.; Brat, D.J. Molecular-Genetic Classification of Gliomas and Its Practical Application to Diagnostic Neuropathology. In Diffuse Low-Grade Gliomas in Adults; Springer: Berlin/Heidelberg, Germany, 2017; pp. 73–100. [Google Scholar] [CrossRef]
- Suzuki, T.; Maruno, M.; Wada, K.; Kagawa, N.; Fujimoto, Y.; Hashimoto, N.; Izumoto, S.; Yoshimine, T. Genetic analysis of human glioblastomas using a genomic microarray system. Brain Tumor Pathol. 2004, 21, 27–34. [Google Scholar] [CrossRef]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.T.; Konermann, C.; Pfaff, E.; Tonjes, M.; Sill, M.; Bender, S.; et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar] [CrossRef] [PubMed]
- Rubinfeld, H.; Seger, R. The ERK Cascade: A Prototype of MAPK Signaling. Mol. Biotechnol. 2005, 31, 151–174. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, C.; Ye, Y.; Wang, Z.; He, Y.; Li, Y.; Mao, H. High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncol. Lett. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hao, Q.; Liao, P.; Luo, S.; Zhang, M.; Hu, G.; Liu, H.; Zhang, Y.; Cao, B.; Baddoo, M.; et al. Nerve growth factor receptor negates the tumor suppressor p53 as a feedback regulator. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Daubon, T.; Leon, C.; Clarke, K.; Andrique, L.; Salabert, L.; Darbo, E.; Pineau, R.; Guerit, S.; Maitre, M.; Dedieu, S.; et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat. Commun. 2019, 10, 1146. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Deng, L.; Huang, J.; Cai, R.; Zhu, X.; Liu, F.; Wang, Q.; Zhang, J.; Zheng, Y. High expression of Fibronectin 1 suppresses apoptosis through the NF-κB pathway and is associated with migration in nasopharyngeal carcinoma. Am. J. Transl. Res. 2017, 9, 4502–4511. [Google Scholar]
- Zhang, Z.; Fang, C.; Wang, Y.; Zhang, J.; Yu, J.; Zhang, Y.; Wang, X.; Zhong, J. COL1A1: A potential therapeutic target for colorectal cancer expressing wild-type or mutant KRAS. Int. J. Oncol. 2018, 53, 1869–1880. [Google Scholar] [CrossRef]
- De Miguel, D.; Lemke, J.; Anel, A.; Walczak, H.; Martinez-Lostao, L. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016, 23, 733–747. [Google Scholar] [CrossRef]
- Knight, M.J.; Riffkin, C.D.; Muscat, A.M.; Ashley, D.M.; Hawkins, C.J. Analysis of FasL and TRAIL induced apoptosis pathways in glioma cells. Oncogene 2001, 20, 5789–5798. [Google Scholar] [CrossRef]
- Kirkpatrick, C.A.; Selleck, S.B. Heparan sulfate proteoglycans at a glance. J. Cell Sci. 2007, 120, 1829–1832. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.P.; Cummings, B.S. Role of glypicans in regulation of the tumor microenvironment and cancer progression. Biochem. Pharmacol. 2019, 168, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K.; Kawakami, M.; Snoy, P.J.; Husain, S.R.; Puri, R.K. In vivo overexpression of IL-13 receptor alpha2 chain inhibits tumorigenicity of human breast and pancreatic tumors in immunodeficient mice. J. Exp. Med. 2001, 194, 1743–1754. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Joshi, B.H.; Puri, R.K. IL-13 regulates cancer invasion and metastasis through IL-13Ralpha2 via ERK/AP-1 pathway in mouse model of human ovarian cancer. Int. J. Cancer 2012, 131, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Caragher, S.; Chalmers, A.J.; Gomez-Roman, N. Glioblastoma’s Next Top Model: Novel Culture Systems for Brain Cancer Radiotherapy Research. Cancers 2019, 11. [Google Scholar] [CrossRef]
- Bertino, J.R.; Waud, W.R.; Parker, W.B.; Lubin, M. Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity: Current strategies. Cancer Biol. 2011, 11, 627–632. [Google Scholar] [CrossRef]
- Bax, D.A.; Little, S.E.; Gaspar, N.; Perryman, L.; Marshall, L.; Viana-Pereira, M.; Jones, T.A.; Williams, R.D.; Grigoriadis, A.; Vassal, G.; et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PLoS ONE 2009, 4, e5209. [Google Scholar] [CrossRef]
- Martinho, O.O.R.; Miranda-Gonçalves, V.; Clara, C.; Almeida, J.R.; Carvalho, A.L.; Barata, J.T.; Reis, R.M. In Vitro and In Vivo Analysis of RTK Inhibitor Efficacy and Identification of Its Novel Targets in Glioblastomas. Transl. Oncol. 2013, 6, 187–196. [Google Scholar] [CrossRef]
- Cruvinel-Carloni, A.; Silva-Oliveira, R.; Torrieri, R.; Bidinotto, L.T.; Berardinelli, G.N.; Oliveira-Silva, V.A.; Clara, C.A.; de Almeida, G.C.; Martinho, O.; Squire, J.A.; et al. Molecular characterization of short-term primary cultures and comparison with corresponding tumor tissue of Brazilian glioblastoma patients. Transl. Cancer Res. 2017, 6, 332–345. [Google Scholar] [CrossRef]



| Subtype | MTAP Positive | MTAP Negative | Total | p-Value |
|---|---|---|---|---|
| Classical | 23 (34.8%) | 43 (65.2%) | 66 (100%) | <0.001 |
| Mesenchymal | 36 (41.9%) | 50 (58.1%) | 86 (100%) | |
| Neural | 19 (37.3%) | 32 (62.7%) | 51 (100%) | |
| Proneural | 30 (49.2%) | 31 (50.8%) | 61 (100%) | |
| G-CIMP+ | 25 (92.6%) | 2 (7.4%) | 27 (100%) |
| MTAP Expression | |||||
|---|---|---|---|---|---|
| Total | (%) | Negative (%) | Positive (%) | p-Value | |
| Gender (N = 491) Male | 300 | (61.1) | 141 (47.0) | 159 (53.0) | 0.503 |
| Female | 191 | (38.9) | 83 (43.5) | 108 (56.5) | |
| Age group (N = 504) | |||||
| 0–19 | 49 | (9.7) | 26 (53.1) | 23 (46.9) | 0.479 |
| 20–59 | 256 | (50.8) | 112 (43.8) | 144 (56.2) | |
| >59 | 199 | (39.5) | 92 (46.2) | 107 (53.8) | |
| Location (N = 210) | |||||
| Frontal Lobe | 88 | (41.9) | 24 (27.3) | 64 (72.7) | 0.013 |
| Parietal Lobe | 41 | (19.5) | 9 (22.0) | 32 (78.0) | |
| Temporal Lobe | 66 | (31.4) | 28 (42.4) | 38 (57.6) | |
| Occipital Lobe | 10 | (4.8) | 5 (50.0) | 5 (50.0) | |
| Cerebellum | 5 | (2.4) | 4 (80.0) | 1 (20.0) | |
| KPS (N = 168) | |||||
| <70 | 75 | (44.6) | 11 (14.7) | 64 (85.3) | 0.539 |
| >70 | 93 | (55.4) | 18 (19.4) | 75 (80.6) | |
| Grade (N = 507) | |||||
| Low Grade | 18 | (3.6) | 5 (27.8) | 13 (72.2) | 0.149 |
| High Grade | 489 | (96.4) | 228 (46.6) | 261 (53.4) | |
| Histologic Subtype (N = 507) | |||||
| DA (WHO Grade II, NOS) | 18 | (3.6) | 5 (27.8) | 13 (72.2) | 0.272 |
| AA (WHO Grade III, NOS) | 24 | (4.7) | 12 (50.0) | 12 (50.0) | |
| Ped. GBM (WHO Grade IV, NOS) | 42 | (8.3) | 23 (54.8) | 19 (45.2) | |
| Ad. GBM (WHO Grade IV, NOS) | 423 | (83.4) | 193 (45.6) | 230 (54.4) | |
| GBM status mutation (N = 460) | |||||
| IDH1 mutation (N = 229) | |||||
| No | 220 | (96.1) | 117 (53.2) | 103 (46.8) | 0.185 |
| Yes | 9 | (3.9) | 7 (77.8) | 2 (22.2) | |
| TERT promoter mutation (N = 45) | |||||
| No | 11 | (24.4) | 3 (27.3) | 8 (72.7) | 0.687 |
| Yes | 34 | (75.6) | 7 (20.6) | 27 (79.4) | |
| MGMT promoter methylation (N = 186) | |||||
| No | 120 | (64.5) | 75 (62.5) | 45 (37.5) | 0.753 |
| Yes | 66 | (35.5) | 39 (59.1) | 27 (40.9) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Menezes, W.P.; Silva, V.A.O.; Gomes, I.N.F.; Rosa, M.N.; Spina, M.L.C.; Carloni, A.C.; Alves, A.L.V.; Melendez, M.; Almeida, G.C.; da Silva, L.S.; et al. Loss of 5′-Methylthioadenosine Phosphorylase (MTAP) is Frequent in High-Grade Gliomas; Nevertheless, it is Not Associated with Higher Tumor Aggressiveness. Cells 2020, 9, 492. https://doi.org/10.3390/cells9020492
de Menezes WP, Silva VAO, Gomes INF, Rosa MN, Spina MLC, Carloni AC, Alves ALV, Melendez M, Almeida GC, da Silva LS, et al. Loss of 5′-Methylthioadenosine Phosphorylase (MTAP) is Frequent in High-Grade Gliomas; Nevertheless, it is Not Associated with Higher Tumor Aggressiveness. Cells. 2020; 9(2):492. https://doi.org/10.3390/cells9020492
Chicago/Turabian Stylede Menezes, Weder Pereira, Viviane Aline Oliveira Silva, Izabela Natália Faria Gomes, Marcela Nunes Rosa, Maria Luisa Corcoll Spina, Adriana Cruvinel Carloni, Ana Laura Vieira Alves, Matias Melendez, Gisele Caravina Almeida, Luciane Sussuchi da Silva, and et al. 2020. "Loss of 5′-Methylthioadenosine Phosphorylase (MTAP) is Frequent in High-Grade Gliomas; Nevertheless, it is Not Associated with Higher Tumor Aggressiveness" Cells 9, no. 2: 492. https://doi.org/10.3390/cells9020492
APA Stylede Menezes, W. P., Silva, V. A. O., Gomes, I. N. F., Rosa, M. N., Spina, M. L. C., Carloni, A. C., Alves, A. L. V., Melendez, M., Almeida, G. C., da Silva, L. S., Clara, C., Cunha, I. W. d., Hajj, G. N. M., Jones, C., Bidinotto, L. T., & Reis, R. M. (2020). Loss of 5′-Methylthioadenosine Phosphorylase (MTAP) is Frequent in High-Grade Gliomas; Nevertheless, it is Not Associated with Higher Tumor Aggressiveness. Cells, 9(2), 492. https://doi.org/10.3390/cells9020492