DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis
Abstract
1. Introduction
2. Molecular Mechanism of RA Development
3. Epigenetic Predisposition for RA Development with Particular Emphasis on DNA Methylation
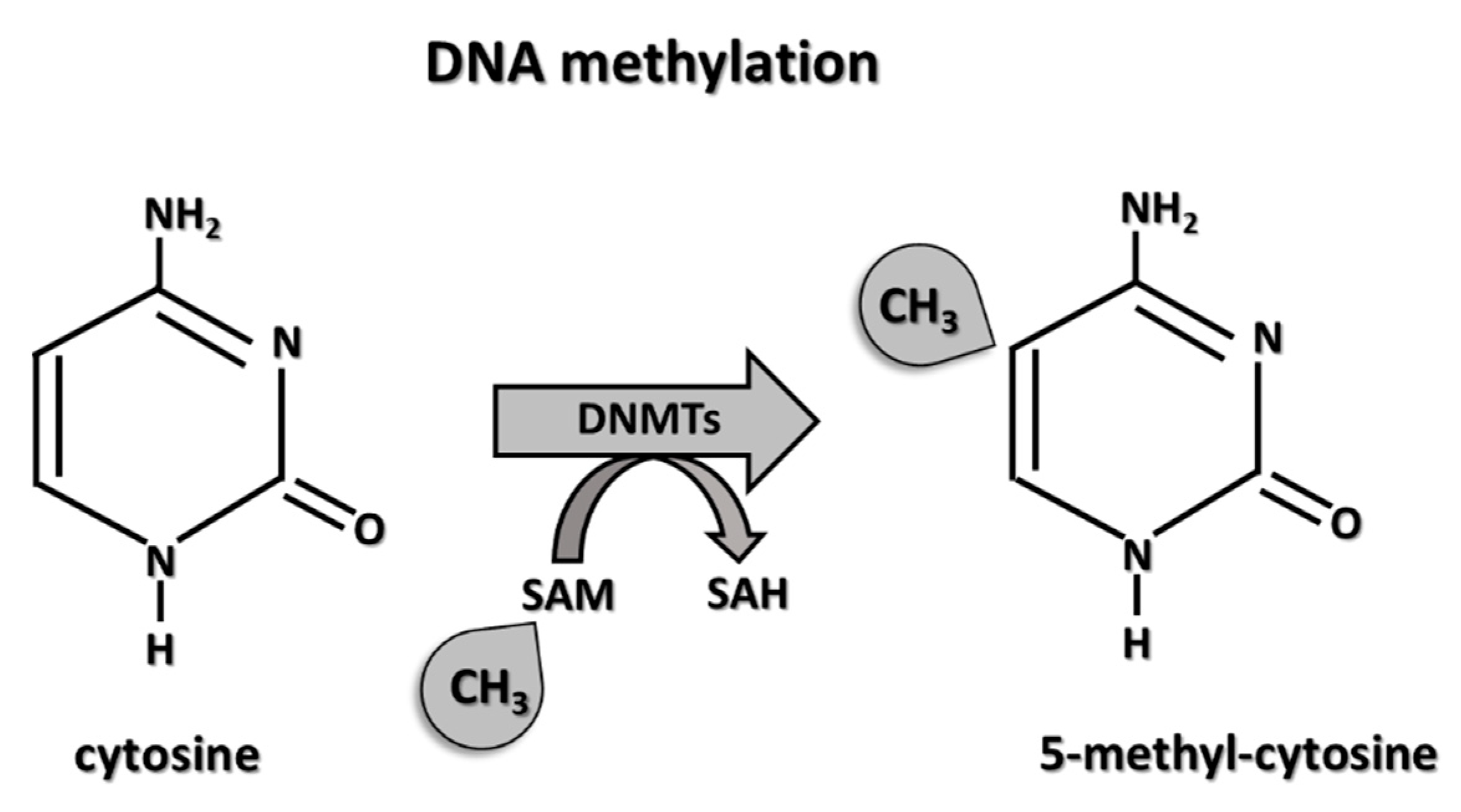
3.1. DNA Methylation
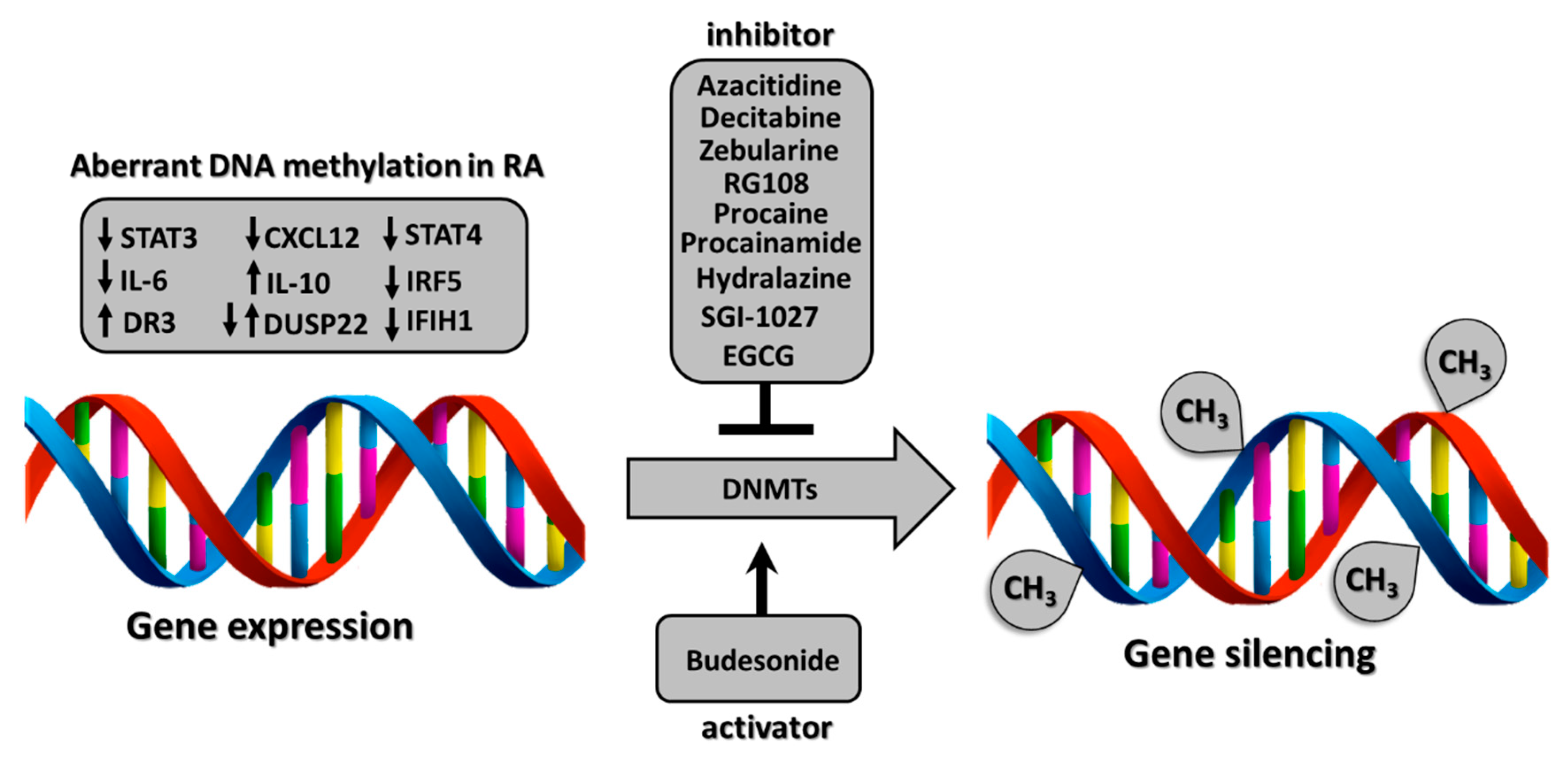
3.1.1. DNA Methylation-Targeted Drugs
3.1.2. DNA Methylation Pattern in RA
3.1.3. DNA Hydroxymethylation
3.2. Non-Coding RNA
3.3. Histone Modifications
4. Cell-Free Methylated DNA (ccfDNA) as Biomarkers
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Available online: http://www.eular.org/myUploadData/files/EU_Horizon_2020_EULAR_position_paper.pdf (accessed on 22 August 2011).
- Bogunia-Kubik, K.; Wysoczanska, B.; Piatek, D.; Iwaszko, M.; Ciechomska, M.; Swierkot, J. Significance of Polymorphism and Expression of miR-146a and NFkB1 Genetic Variants in Patients with Rheumatoid Arthritis. Arch. Immunol. Ther. Exp. 2016, 64, 131–136. [Google Scholar] [CrossRef]
- Symmons, D.P. Epidemiology of rheumatoid arthritis: Determinants of onset, persistence and outcome. Best Pract. Res. Clin. Rheumatol. 2002, 16, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid arthritis. Nat. Rev. Dis. Primers 2018, 4, 18001. [Google Scholar] [CrossRef] [PubMed]
- Aletaha, D.; Smolen, J.S. Diagnosis and Management of Rheumatoid Arthritis: A Review. JAMA 2018, 320, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Hitchon, C.A.; El-Gabalawy, H.S. The synovium in rheumatoid arthritis. Open Rheumatol. J. 2011, 5, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Laria, A.; Lurati, A.; Marrazza, M.; Mazzocchi, D.; Re, K.A.; Scarpellini, M. The macrophages in rheumatic diseases. J. Inflamm. Res. 2016, 9, 1–11. [Google Scholar] [PubMed]
- Ciechomska, M.; Wilson, C.L.; Floudas, A.; Hui, W.; Rowan, A.D.; van Eden, W.; Robinson, J.H.; Knight, A.M. Antigen-specific B lymphocytes acquire proteoglycan aggrecan from cartilage extracellular matrix resulting in antigen presentation and CD4+ T-cell activation. Immunology 2014, 141, 70–78. [Google Scholar] [CrossRef]
- Jarvinen, P.; Aho, K. Twin studies in rheumatic diseases. Semin. Arthritis Rheum. 1994, 24, 19–28. [Google Scholar] [CrossRef]
- Aho, K.; Koskenvuo, M.; Tuominen, J.; Kaprio, J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 1986, 13, 899–902. [Google Scholar]
- van der Woude, D.; Houwing-Duistermaat, J.J.; Toes, R.E.; Huizinga, T.W.; Thomson, W.; Worthington, J.; van der Helm-van Mil, A.H.; de Vries, R.R. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009, 60, 916–923. [Google Scholar] [CrossRef]
- Ciechomska, M.; O’Reilly, S. Epigenetic Modulation as a Therapeutic Prospect. for Treatment of Autoimmune Rheumatic Diseases. Mediators Inflamm. 2016, 2016, 9607946. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Wojdacz, T.K.; Thestrup, B.B.; Wiuf, C.; Hager, H.; Hansen, L.L. Quality assessment of DNA derived from up to 30 years old formalin fixed paraffin embedded (FFPE) tissue for PCR-based methylation analysis using SMART-MSP and MS-HRM. BMC Cancer 2009, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA methyltransferases: A novel target for prevention and therapy. Front. Oncol. 2014, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.M.; Wang, J.; Qian, D.M.; Song, J.Y.; Chen, H.; Zhu, X.L.; Zhou, R.; Zhao, Y.D.; Zhou, L.; Li, X.Z.; et al. DNA methylation level of promoter region of activating transcription factor 5 in glioma. J. Zhejiang Univ. Sci. B 2015, 16, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, C.S. Animal models to study environmental epigenetics. Biol. Reprod. 2010, 82, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.H.; Liu, Y.H.; Liu, J.D.; Xu, D.D.; Li, X.F.; Meng, X.M.; Ma, T.T.; Huang, C.; Li, J. MeCP2 Regulates PTCH1 Expression Through DNA Methylation in Rheumatoid Arthritis. Inflammation 2017, 40, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.H.; Ma, C.L.; Cong, B.; Li, S.J.; Chen, H.Y.; Zhang, J.G. Hypomethylation of proximal CpG motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis. Acta Pharmacol. Sin. 2011, 32, 1373–1380. [Google Scholar] [CrossRef]
- Konsta, O.D.; Le Dantec, C.; Charras, A.; Cornec, D.; Kapsogeorgou, E.K.; Tzioufas, A.G.; Pers, J.O.; Renaudineau, Y. Defective DNA methylation in salivary gland epithelial acini from patients with Sjogren’s syndrome is associated with SSB gene expression, anti-SSB/LA detection, and lymphocyte infiltration. J. Autoimmun. 2016, 68, 30–38. [Google Scholar] [CrossRef]
- O’Reilly, S.; Ciechomska, M.; Fullard, N.; Przyborski, S.; van Laar, J.M. IL-13 mediates collagen deposition via STAT6 and microRNA-135b: A role for epigenetics. Sci. Rep. 2016, 6, 25066. [Google Scholar] [CrossRef]
- Khan, C.; Pathe, N.; Fazal, S.; Lister, J.; Rossetti, J.M. Azacitidine in the management of patients with myelodysplastic syndromes. Ther. Adv. Hematol. 2012, 3, 355–373. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, Q.; Zhang, C.; Huang, C. Genome-wide DNA methylation analysis of articular chondrocytes identifies TRAF1, CTGF, and CX3CL1 genes as hypomethylated in osteoarthritis. Clin. Rheumatol. 2017, 36, 2335–2342. [Google Scholar] [CrossRef] [PubMed]
- Lubbert, M.; Suciu, S.; Hagemeijer, A.; Ruter, B.; Platzbecker, U.; Giagounidis, A.; Selleslag, D.; Labar, B.; Germing, U.; Salih, H.R.; et al. Decitabine improves progression-free survival in older high-risk MDS patients with multiple autosomal monosomies: Results of a subgroup analysis of the randomized phase III study 06011 of the EORTC Leukemia Cooperative Group and German MDS Study Group. Ann. Hematol. 2016, 95, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.T.; Sjogren, H.O.; Salford, L.G.; Widegren, B. An epigenetic mechanism for high, synergistic expression of indoleamine 2,3-dioxygenase 1 (IDO1) by combined treatment with zebularine and IFN-gamma: Potential therapeutic use in autoimmune diseases. Mol. Immunol. 2012, 51, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Vale, C.; Bhagat, T.; Verma, A. Role of DNA methylation in the pathogenesis and treatment of myelodysplastic syndromes. Semin. Hematol. 2013, 50, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Graca, I.; Sousa, E.J.; Baptista, T.; Almeida, M.; Ramalho-Carvalho, J.; Palmeira, C.; Henrique, R.; Jeronimo, C. Anti-tumoral effect of the non-nucleoside DNMT inhibitor RG108 in human prostate cancer cells. Curr. Pharm. Des. 2014, 20, 1803–1811. [Google Scholar] [CrossRef] [PubMed]
- Borges, S.; Doppler, H.R.; Storz, P. A combination treatment with DNA methyltransferase inhibitors and suramin decreases invasiveness of breast cancer cells. Breast Cancer Res. Treat. 2014, 144, 79–91. [Google Scholar] [CrossRef]
- Suzuki, T.; Tanaka, R.; Hamada, S.; Nakagawa, H.; Miyata, N. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 1124–1127. [Google Scholar] [CrossRef]
- Wu, F.; Wang, J.; Sun, J.; Shen, L.; Liu, M.; Zhao, E. Procaine stimulates aquaporin5 expression in human salivary gland ductal cells via the suppression of DNA methyltransferase1. Mol. Med. Rep. 2018, 17, 7996–8002. [Google Scholar]
- Medina-Franco, J.L.; Caulfield, T. Advances in the computational development of DNA methyltransferase inhibitors. Drug Discov. Today 2011, 16, 418–425. [Google Scholar] [CrossRef]
- Richardson, B.; Scheinbart, L.; Strahler, J.; Gross, L.; Hanash, S.; Johnson, M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990, 33, 1665–1673. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lin, Z.; Huo, Y.; Geng, X.; Li, M.; Yang, Y.; Li, B. Procainamide-induced autoimmunity: Relationship to T-helper 2-type T-cell activation. Hum. Exp. Toxicol. 2018, 37, 647–662. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Yegnasubramanian, S.; Lin, X.; Nelson, W.G. Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 2005, 280, 40749–40756. [Google Scholar] [CrossRef] [PubMed]
- Riegsecker, S.; Wiczynski, D.; Kaplan, M.J.; Ahmed, S. Potential benefits of green tea polyphenol EGCG in the prevention and treatment of vascular inflammation in rheumatoid arthritis. Life Sci. 2013, 93, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Umar, S.; Riegsecker, S.; Chourasia, M.; Ahmed, S. Regulation of Transforming Growth Factor beta-Activated Kinase Activation by Epigallocatechin-3-Gallate in Rheumatoid Arthritis Synovial Fibroblasts: Suppression of K(63)-Linked Autoubiquitination of Tumor Necrosis Factor Receptor-Associated Factor 6. Arthritis Rheumatol. 2016, 68, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, V.; Vaid, M.; Katiyar, S.K. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 2011, 32, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, C. Hydralazine inhibits human cervical cancer cell growth in vitro in association with APC demethylation and re-expression. Cancer Chemother. Pharmacol. 2009, 63, 605–613. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, J.; Zhang, C.; Zhao, B.; Jiao, A. DNMTs inhibitor SGI-1027 induces apoptosis in Huh7 human hepatocellular carcinoma cells. Oncol. Lett. 2018, 16, 5799–5806. [Google Scholar] [CrossRef]
- Pereira, M.A.; Tao, L.; Liu, Y.; Li, L.; Steele, V.E.; Lubet, R.A. Modulation by budesonide of DNA methylation and mRNA expression in mouse lung tumors. Int. J. Cancer 2007, 120, 1150–1153. [Google Scholar] [CrossRef]
- Alyaqoub, F.S.; Tao, L.; Kramer, P.M.; Steele, V.E.; Lubet, R.A.; Gunning, W.T.; Pereira, M.A. Prevention of mouse lung tumors and modulation of DNA methylation by combined treatment with budesonide and R115777 (Zarnestra MT). Carcinogenesis 2007, 28, 124–129. [Google Scholar] [CrossRef]
- Rohle, D.; Popovici-Muller, J.; Palaskas, N.; Turcan, S.; Grommes, C.; Campos, C.; Tsoi, J.; Clark, O.; Oldrini, B.; Komisopoulou, E.; et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013, 340, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.; Araujo Cruz, M.M.; Jyotsana, N.; Sharma, A.; Yun, H.; Gorlich, K.; Wichmann, M.; Schwarzer, A.; Preller, M.; Thol, F.; et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood 2013, 122, 2877–2887. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; Issa, J.P.; Garcia-Manero, G.; Kantarjian, H. Evolution of decitabine development: Accomplishments, ongoing investigations, and future strategies. Cancer 2008, 112, 2341–2351. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.C.; Matsen, C.B.; Gonzales, F.A.; Ye, W.; Greer, S.; Marquez, V.E.; Jones, P.A.; Selker, E.U. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J. Natl. Cancer Inst. 2003, 95, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Oakes, C.C.; Kelly, T.L.; Robaire, B.; Trasler, J.M. Adverse effects of 5-aza-2’-deoxycytidine on spermatogenesis include reduced sperm function and selective inhibition of de novo DNA methylation. J. Pharmacol. Exp. Ther. 2007, 322, 1171–1180. [Google Scholar] [CrossRef]
- Stresemann, C.; Brueckner, B.; Musch, T.; Stopper, H.; Lyko, F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006, 66, 2794–2800. [Google Scholar] [CrossRef]
- Segura-Pacheco, B.; Perez-Cardenas, E.; Taja-Chayeb, L.; Chavez-Blanco, A.; Revilla-Vazquez, A.; Benitez-Bribiesca, L.; Duenas-Gonzalez, A. Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J. Transl. Med. 2006, 4, 32. [Google Scholar] [CrossRef]
- Singh, N.; Duenas-Gonzalez, A.; Lyko, F.; Medina-Franco, J.L. Molecular modeling and molecular dynamics studies of hydralazine with human DNA methyltransferase 1. ChemMedChem 2009, 4, 792–799. [Google Scholar] [CrossRef]
- Datta, J.; Ghoshal, K.; Denny, W.A.; Gamage, S.A.; Brooke, D.G.; Phiasivongsa, P.; Redkar, S.; Jacob, S.T. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009, 69, 4277–4285. [Google Scholar] [CrossRef]
- Amato, R.J.; Stephenson, J.; Hotte, S.; Nemunaitis, J.; Belanger, K.; Reid, G.; Martell, R.E. MG98, a second-generation DNMT1 inhibitor, in the treatment of advanced renal cell carcinoma. Cancer Investig. 2012, 30, 415–421. [Google Scholar] [CrossRef]
- Li, Y.C.; Wang, Y.; Li, D.D.; Zhang, Y.; Zhao, T.C.; Li, C.F. Procaine is a specific DNA methylation inhibitor with anti-tumor effect for human gastric cancer. J. Cell Biochem. 2018, 119, 2440–2449. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Fang, T.J.; Ou, T.T.; Wu, C.C.; Li, R.N.; Lin, Y.C.; Lin, C.H.; Tsai, W.C.; Liu, H.W.; Yen, J.H. Global DNA methylation, DNMT1, and MBD2 in patients with rheumatoid arthritis. Immunol. Lett. 2011, 135, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Whitaker, J.W.; Boyle, D.L.; Wang, W.; Firestein, G.S. DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis. 2013, 72, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. Interleukin 6 in autoimmune and inflammatory diseases: A personal memoir. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Ai, R.; Hammaker, D.; Boyle, D.L.; Morgan, R.; Walsh, A.M.; Fan, S.; Firestein, G.S.; Wang, W. Joint-specific DNA methylation and transcriptome signatures in rheumatoid arthritis identify distinct pathogenic processes. Nat. Commun. 2016, 7, 11849. [Google Scholar] [CrossRef] [PubMed]
- Rhead, B.; Holingue, C.; Cole, M.; Shao, X.; Quach, H.L.; Quach, D.; Shah, K.; Sinclair, E.; Graf, J.; Link, T.; et al. Rheumatoid Arthritis Naive T Cells Share Hypermethylation Sites With Synoviocytes. Arthritis Rheumatol. 2017, 69, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Glossop, J.R.; Emes, R.D.; Nixon, N.B.; Haworth, K.E.; Packham, J.C.; Dawes, P.T.; Fryer, A.A.; Mattey, D.L.; Farrell, W.E. Genome-wide DNA methylation profiling in rheumatoid arthritis identifies disease-associated methylation changes that are distinct to individual T- and B-lymphocyte populations. Epigenetics 2014, 9, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Nile, C.J.; Read, R.C.; Akil, M.; Duff, G.W.; Wilson, A.G. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008, 58, 2686–2693. [Google Scholar] [CrossRef]
- Karouzakis, E.; Rengel, Y.; Jungel, A.; Kolling, C.; Gay, R.E.; Michel, B.A.; Tak, P.P.; Gay, S.; Neidhart, M.; Ospelt, C. DNA methylation regulates the expression of CXCL12 in rheumatoid arthritis synovial fibroblasts. Genes Immun. 2011, 12, 643–652. [Google Scholar] [CrossRef]
- Mok, A.; Rhead, B.; Holingue, C.; Shao, X.; Quach, H.L.; Quach, D.; Sinclair, E.; Graf, J.; Imboden, J.; Link, T.; et al. Hypomethylation of CYP2E1 and DUSP22 Promoters Associated With Disease Activity and Erosive Disease Among Rheumatoid Arthritis Patients. Arthritis Rheumatol. 2018, 70, 528–536. [Google Scholar] [CrossRef]
- Takami, N.; Osawa, K.; Miura, Y.; Komai, K.; Taniguchi, M.; Shiraishi, M.; Sato, K.; Iguchi, T.; Shiozawa, K.; Hashiramoto, A.; et al. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis Rheum. 2006, 54, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Boyle, D.L.; Firestein, G.S. Regulation of DNA methylation in rheumatoid arthritis synoviocytes. J. Immunol. 2013, 190, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Y.; Beier, U.H.; Han, R.; Bhatti, T.R.; Akimova, T.; Hancock, W.W. Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood 2013, 121, 3631–3639. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xiong, L.; Zeng, T.; Xiao, K.; Huang, Y.; Guo, H.; Ren, J. Hypermethylated ERG as a cell-free fetal DNA biomarker for non-invasive prenatal testing of Down syndrome. Clin. Chim. Acta 2015, 444, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Stefan, M.; Wei, C.; Lombardi, A.; Li, C.W.; Concepcion, E.S.; Inabnet, W.B., III; Owen, R.; Zhang, W.; Tomer, Y. Genetic-epigenetic dysregulation of thymic TSH receptor gene expression triggers thyroid autoimmunity. Proc. Natl. Acad. Sci. USA 2014, 111, 12562–12567. [Google Scholar] [CrossRef] [PubMed]
- Ghodke-Puranik, Y.; Niewold, T.B. Immunogenetics of systemic lupus erythematosus: A comprehensive review. J. Autoimmun. 2015, 64, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Kochi, Y. Genetics of autoimmune diseases: Perspectives from genome-wide association studies. Int. Immunol. 2016, 28, 155–161. [Google Scholar] [CrossRef]
- Chen, S.; Pu, W.; Guo, S.; Jin, L.; He, D.; Wang, J. Genome-Wide DNA Methylation Profiles Reveal Common Epigenetic Patterns of Interferon-Related Genes in Multiple Autoimmune Diseases. Front. Genet. 2019, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Karouzakis, E.; Gay, R.E.; Michel, B.A.; Gay, S.; Neidhart, M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009, 60, 3613–3622. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Zhang, Q.H.; Tang, Q.; Jiang, Z.; Xing, S.; Li, J.; Pang, Y. Comprehensive analysis of gene expression and DNA methylation datasets identify valuable biomarkers for rheumatoid arthritis progression. Oncotarget 2018, 9, 2977–2983. [Google Scholar] [CrossRef]
- Svendsen, A.J.; Gervin, K.; Lyle, R.; Christiansen, L.; Kyvik, K.; Junker, P.; Nielsen, C.; Houen, G.; Tan, Q. Differentially Methylated DNA Regions in Monozygotic Twin Pairs Discordant for Rheumatoid Arthritis: An. Epigenome-Wide Study. Front. Immunol. 2016, 7, 510. [Google Scholar] [CrossRef] [PubMed]
- Plant, D.; Webster, A.; Nair, N.; Oliver, J.; Smith, S.L.; Eyre, S.; Hyrich, K.L.; Wilson, A.G.; Morgan, A.W.; Isaacs, J.D.; et al. Differential Methylation as a Biomarker of Response to Etanercept in Patients With Rheumatoid Arthritis. Arthritis Rheumatol. 2016, 68, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, X.; Clark, E.; Mulcahey, M.; Huang, S.; Shi, Y.G. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010, 20, 1390–1393. [Google Scholar] [CrossRef] [PubMed]
- de Andres, M.C.; Perez-Pampin, E.; Calaza, M.; Santaclara, F.J.; Ortea, I.; Gomez-Reino, J.J.; Gonzalez, A. Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis Res. Ther. 2015, 17, 233. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Luo, M.; Pan, K.; Ahmad, T.; Zhou, T.; Miao, Z.; Zhou, H.; Sun, H.; Xu, X.; Namaka, M.; et al. DNA hydroxymethylation changes in response to spinal cord damage in a multiple sclerosis mouse model. Epigenomics 2019, 11, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Moran-Moguel, M.C.; Petarra-Del Rio, S.; Mayorquin-Galvan, E.E.; Zavala-Cerna, M.G. Rheumatoid Arthritis and miRNAs: A Critical Review through a Functional View. J. Immunol. Res. 2018, 2018, 2474529. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; Zarecki, P.; Merdas, M.; Swierkot, J.; Morgiel, E.; Wiland, P.; Maslinski, W.; Bogunia-Kubik, K. The role of microRNA-5196 in the pathogenesis of systemic sclerosis. Eur. J. Clin. Investig. 2017, 47, 555–564. [Google Scholar] [CrossRef]
- Ciechomska, M.; O’Reilly, S.; Suwara, M.; Bogunia-Kubik, K.; van Laar, J.M. MiR-29a reduces TIMP-1 production by dermal fibroblasts via targeting TGF-beta activated kinase 1 binding protein 1, implications for systemic sclerosis. PLoS ONE 2014, 9, e115596. [Google Scholar] [CrossRef]
- Li, J.; Wan, Y.; Guo, Q.; Zou, L.; Zhang, J.; Fang, Y.; Zhang, J.; Zhang, J.; Fu, X.; Liu, H.; et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res. Ther. 2010, 12, R81. [Google Scholar] [CrossRef]
- Churov, A.V.; Oleinik, E.K.; Knip, M. MicroRNAs in rheumatoid arthritis: Altered expression and diagnostic potential. Autoimmun. Rev. 2015, 14, 1029–1037. [Google Scholar] [CrossRef]
- Niimoto, T.; Nakasa, T.; Ishikawa, M.; Okuhara, A.; Izumi, B.; Deie, M.; Suzuki, O.; Adachi, N.; Ochi, M. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet. Disord. 2010, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; Bonek, K.; Merdas, M.; Zarecki, P.; Swierkot, J.; Gluszko, P.; Bogunia-Kubik, K.; Maslinski, W. Changes in MiRNA-5196 Expression as a Potential Biomarker of Anti-TNF-alpha Therapy in Rheumatoid Arthritis and Ankylosing Spondylitis Patients. Arch. Immunol. Ther. Exp. 2018, 66, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Wohlers, I.; Bertram, L.; Lill, C.M. Evidence for a potential role of miR-1908-5p and miR-3614-5p in autoimmune disease risk using integrative bioinformatics. J. Autoimmun. 2018, 94, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Stanczyk, J.; Ospelt, C.; Karouzakis, E.; Filer, A.; Raza, K.; Kolling, C.; Gay, R.; Buckley, C.D.; Tak, P.P.; Gay, S.; et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011, 63, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Nemtsova, M.V.; Zaletaev, D.V.; Bure, I.V.; Mikhaylenko, D.S.; Kuznetsova, B.; Alekseeva, E.A.; Beloukhova, M.I.; Deviatkin, A.A.; Lukashev, A.N.; Zamyatnin, A.A., Jr. Epigenetic Changes in the Pathogenesis of Rheumatoid Arthritis. Front. Genet. 2019, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Huber, L.C.; Brock, M.; Hemmatazad, H.; Giger, O.T.; Moritz, F.; Trenkmann, M.; Distler, J.H.; Gay, R.E.; Kolling, C.; Moch, H.; et al. Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthritis Rheum. 2007, 56, 1087–1093. [Google Scholar] [CrossRef]
- Wada, T.T.; Araki, Y.; Sato, K.; Aizaki, Y.; Yokota, K.; Kim, Y.T.; Oda, H.; Kurokawa, R.; Mimura, T. Aberrant histone acetylation contributes to elevated interleukin-6 production in rheumatoid arthritis synovial fibroblasts. Biochem. Biophys. Res. Commun. 2014, 444, 682–686. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Kijima, M.; Akita, M.; Beppu, T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990, 265, 17174–17179. [Google Scholar]
- Jungel, A.; Baresova, V.; Ospelt, C.; Simmen, B.R.; Michel, B.A.; Gay, R.E.; Gay, S.; Seemayer, C.A.; Neidhart, M. Trichostatin A sensitises rheumatoid arthritis synovial fibroblasts for TRAIL-induced apoptosis. Ann. Rheum. Dis. 2006, 65, 910–912. [Google Scholar] [CrossRef]
- Morinobu, A.; Wang, B.; Liu, J.; Yoshiya, S.; Kurosaka, M.; Kumagai, S. Trichostatin A cooperates with Fas-mediated signal to induce apoptosis in rheumatoid arthritis synovial fibroblasts. J. Rheumatol. 2006, 33, 1052–1060. [Google Scholar]
- Nakamura, C.; Matsushita, I.; Kosaka, E.; Kondo, T.; Kimura, T. Anti-arthritic effects of combined treatment with histone deacetylase inhibitor and low-intensity ultrasound in the presence of microbubbles in human rheumatoid synovial cells. Rheumatology 2008, 47, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, B. Trichostatin A, an Inhibitor of Histone Deacetylase, Inhibits the Viability and Invasiveness of Hypoxic Rheumatoid Arthritis Fibroblast-Like Synoviocytes via PI3K/Akt Signaling. J. Biochem. Mol. Toxicol. 2016, 30, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Grabiec, A.M.; Korchynskyi, O.; Tak, P.P.; Reedquist, K.A. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann. Rheum. Dis. 2012, 71, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; O’Reilly, S.; Przyborski, S.; Oakley, F.; Bogunia-Kubik, K.; van Laar, J.M. Histone Demethylation and Toll-like Receptor 8-Dependent Cross-Talk in Monocytes Promotes Transdifferentiation of Fibroblasts in Systemic Sclerosis Via Fra-2. Arthritis Rheumatol. 2016, 68, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Duraisamy, A.J.; Mishra, M.; Kowluru, R.A. Crosstalk Between Histone and DNA Methylation in Regulation of Retinal Matrix Metalloproteinase-9 in Diabetes. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6440–6448. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.G.; Yang, Y.Y.; He, X.; Li, J. New advances of DNA methylation and histone modifications in rheumatoid arthritis, with special emphasis on MeCP2. Cell Signal. 2013, 25, 875–882. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tsou, P.S.; Khanna, D.; Sawalha, A.H. Methyl-CpG-binding protein 2 mediates antifibrotic effects in scleroderma fibroblasts. Ann. Rheum. Dis. 2018, 77, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Hamzeh, E.; Aslani, S.; Ziaee, V.; Poursani, S.; Rezaei, N. Single nucleotide polymorphism of Methyl-CpG-binding protein 2 gene associates with juvenile idiopathic arthritis. Clin. Rheumatol. 2018, 37, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Sawalha, A.H.; Webb, R.; Han, S.; Kelly, J.A.; Kaufman, K.M.; Kimberly, R.P.; Alarcon-Riquelme, M.E.; James, J.A.; Vyse, T.J.; Gilkeson, G.S.; et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS ONE 2008, 3, e1727. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, Y.; Tong, D.; Qin, Y.; Yang, J.; Xue, M.; Du, N.; Liu, L.; Guo, B.; Hou, N.; et al. MeCP2 Promotes Gastric Cancer Progression Through Regulating FOXF1/Wnt5a/beta-Catenin and MYOD1/Caspase-3 Signaling Pathways. EBioMedicine 2017, 16, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Neupane, M.; Clark, A.P.; Landini, S.; Birkbak, N.J.; Eklund, A.C.; Lim, E.; Culhane, A.C.; Barry, W.T.; Schumacher, S.E.; Beroukhim, R.; et al. MECP2 Is a Frequently Amplified Oncogene with a Novel Epigenetic Mechanism That Mimics the Role of Activated RAS in Malignancy. Cancer Discov. 2016, 6, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, J.; Gao, N.; Wang, Y.; Chen, Y.; Han, J. MECP2 expression in gastric cancer and its correlation with clinical pathological parameters. Medicine 2017, 96, e7691. [Google Scholar] [CrossRef] [PubMed]
- Raza, K.; Saber, T.P.; Kvien, T.K.; Tak, P.P.; Gerlag, D.M. Timing the therapeutic window of opportunity in early rheumatoid arthritis: Proposal for definitions of disease duration in clinical trials. Ann. Rheum. Dis. 2012, 71, 1921–1923. [Google Scholar] [CrossRef] [PubMed]
- Raza, K.; Buckley, C.E.; Salmon, M.; Buckley, C.D. Treating very early rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 2006, 20, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Laird, P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer 2003, 3, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Mahon, K.L.; Qu, W.; Devaney, J.; Paul, C.; Castillo, L.; Wykes, R.J.; Chatfield, M.D.; Boyer, M.J.; Stockler, M.R.; Marx, G.; et al. Methylated Glutathione S-transferase 1 (mGSTP1) is a potential plasma free DNA epigenetic marker of prognosis and response to chemotherapy in castrate-resistant prostate cancer. Br. J. Cancer 2014, 111, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Zurita, M.; Lara, P.C.; del Moral, R.; Torres, B.; Linares-Fernandez, J.L.; Arrabal, S.R.; Martinez-Galan, J.; Oliver, F.J.; Ruiz de Almodovar, J.M. Hypermethylated 14-3-3-sigma and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer 2010, 10, 217. [Google Scholar] [CrossRef]
- Montavon, C.; Gloss, B.S.; Warton, K.; Barton, C.A.; Statham, A.L.; Scurry, J.P.; Tabor, B.; Nguyen, T.V.; Qu, W.; Samimi, G.; et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol. Oncol. 2012, 124, 582–588. [Google Scholar] [CrossRef]
- Warton, K.; Mahon, K.L.; Samimi, G. Methylated circulating tumor DNA in blood: Power in cancer prognosis and response. Endocr. Relat. Cancer 2016, 23, R157–R171. [Google Scholar] [CrossRef]
- Dou, C.Y.; Fan, Y.C.; Cao, C.J.; Yang, Y.; Wang, K. Sera DNA Methylation of CDH1, DNMT3b and ESR1 Promoters as Biomarker for the Early Diagnosis of Hepatitis B Virus-Related Hepatocellular Carcinoma. Dig. Dis. Sci. 2016, 61, 1130–1138. [Google Scholar] [CrossRef]
- van den Oever, J.M.; Balkassmi, S.; Segboer, T.; Verweij, E.J.; van der Velden, P.A.; Oepkes, D.; Bakker, E.; Boon, E.M. Mrassf1a-pap, a novel methylation-based assay for the detection of cell-free fetal DNA in maternal plasma. PLoS ONE 2013, 8, e84051. [Google Scholar] [CrossRef] [PubMed]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., III; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 rheumatoid arthritis classification criteria: An. American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann. Rheum. Dis. 2010, 69, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Suresh, E. Diagnosis of early rheumatoid arthritis: What the non-specialist needs to know. J. R. Soc. Med. 2004, 97, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Rantapaa-Dahlqvist, S.; de Jong, B.A.; Berglin, E.; Hallmans, G.; Wadell, G.; Stenlund, H.; Sundin, U.; van Venrooij, W.J. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003, 48, 2741–2749. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T. Biomarkers as a treatment guide in rheumatoid arthritis. Clin. Immunol. 2018, 186, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Filkova, M.; Jungel, A.; Gay, R.E.; Gay, S. MicroRNAs in rheumatoid arthritis: Potential role in diagnosis and therapy. BioDrugs 2012, 26, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.epigenomics.com/wp-content/uploads/2016/06/approval_pm_eng.pdf (accessed on 22 August 2016).
| Drug | Epigenetic Effect | Disease |
|---|---|---|
| Azacitidine (5′-AZA) | DNA methyltransferase inhibitor | RA [18,19], primary Sjogren’s syndrome (pSS) [20], systemic sclerosis (SSc) [21], myelodysplastic syndromes (MDS) [22] |
| Decitabine | DNA methyltransferase inhibitor | RA [23], OA [23], chronic myelomonocytic leukemia [24] |
| Zebularine | DNA methyltransferase inhibitor | autoimmunity/chronic inflammation [25], MDS [26] |
| RG108 | DNA methyltransferase inhibitor | prostate cancer [27], breast cancer [28], colon cancer [29] |
| Procaine | DNA methyltransferase inhibitor | SS [30], gastric carcinoma [31] |
| Procainamide | DNA methyltransferase inhibitor | RA [32], SLE [32], drug-induced autoimmunity [33], solid tumors [34] |
| Epigallocatechin-3-gallate (EGCG) | DNA methyltransferase inhibitor | RA [35,36], skin cancer [37] |
| Hydralazine | DNA methyltransferase inhibitor | RA [32], SLE [32], cervical cancer [38] |
| SGI-1027 | DNA methyltransferase inhibitor | hepatoma [39] |
| Budesonide | DNA methyltransferase activator | lung cancer [40,41] |
| AGI-5198 | DNA hydroxymethylation inhibitor | brain tumor [42] |
| HMS-101 | DNA hydroxymethylation inhibitor | acute myeloid leukemia (AML) [43] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciechomska, M.; Roszkowski, L.; Maslinski, W. DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis. Cells 2019, 8, 953. https://doi.org/10.3390/cells8090953
Ciechomska M, Roszkowski L, Maslinski W. DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis. Cells. 2019; 8(9):953. https://doi.org/10.3390/cells8090953
Chicago/Turabian StyleCiechomska, Marzena, Leszek Roszkowski, and Wlodzimierz Maslinski. 2019. "DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis" Cells 8, no. 9: 953. https://doi.org/10.3390/cells8090953
APA StyleCiechomska, M., Roszkowski, L., & Maslinski, W. (2019). DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis. Cells, 8(9), 953. https://doi.org/10.3390/cells8090953





