State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues
Abstract
:1. Introduction
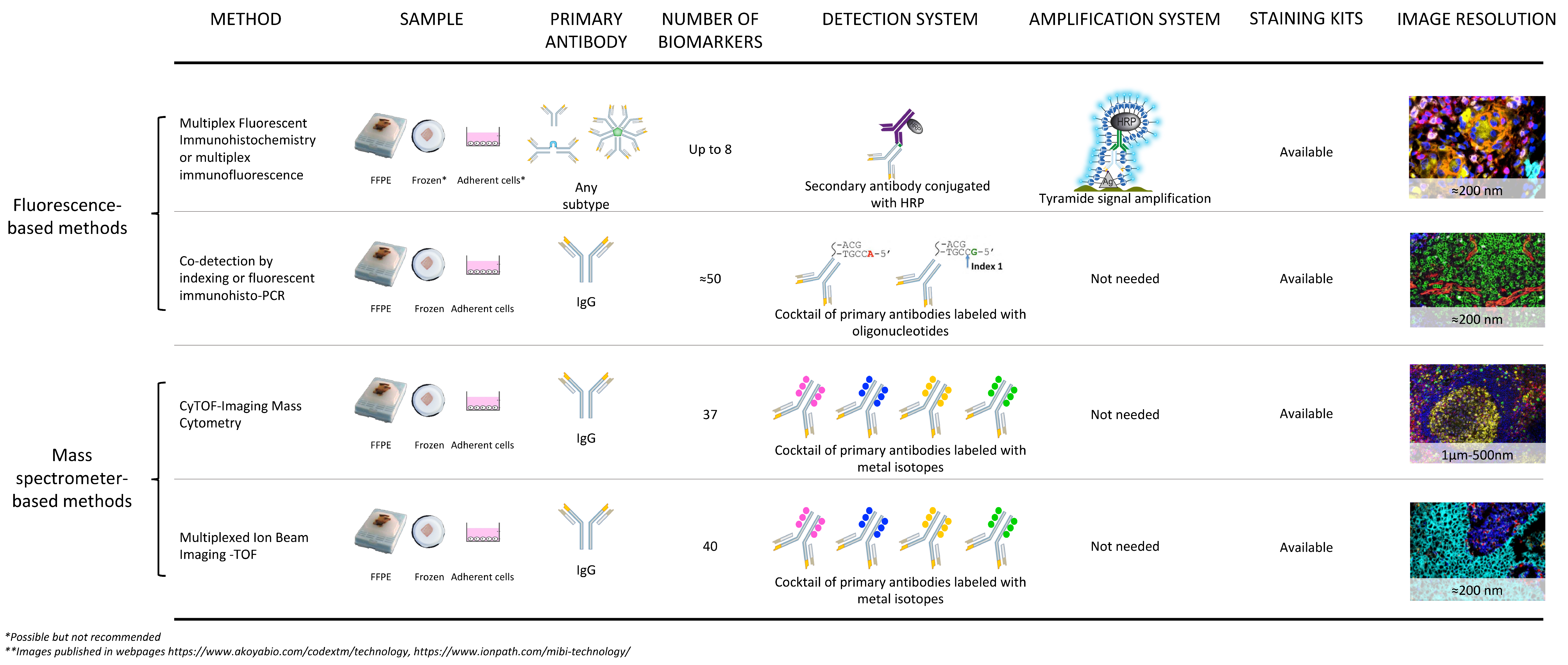
2. Non-Fluorescence-Based Platforms
2.1. Multiplexed Immunohistochemical Consecutive Staining on Single Slide
2.2. Sequential Immunoperoxidase Labeling and Erasing
3. Fluorescence-Based Platforms
3.1. Bleaching Techniques without Signal Amplification System
3.2. Multi-Epitope-Ligand Cartography
3.3. MultiOmyxTM Staining or Hyperplexed Immunofluorescence Assay
4. Tissue-Based Cyclic Immunofluorescence (t-CyCIF) Method
Co-Detection by Indexing or Fluorescent Immunohisto-PCR
5. Amplification of the Epitope Detection
5.1. Multiplex Modified Hapten-Based Technology
5.2. Tyramide Signal Amplification and Fluorescent Multiplex Immunohistochemistry
5.3. Nanocrystal Quantum Dots
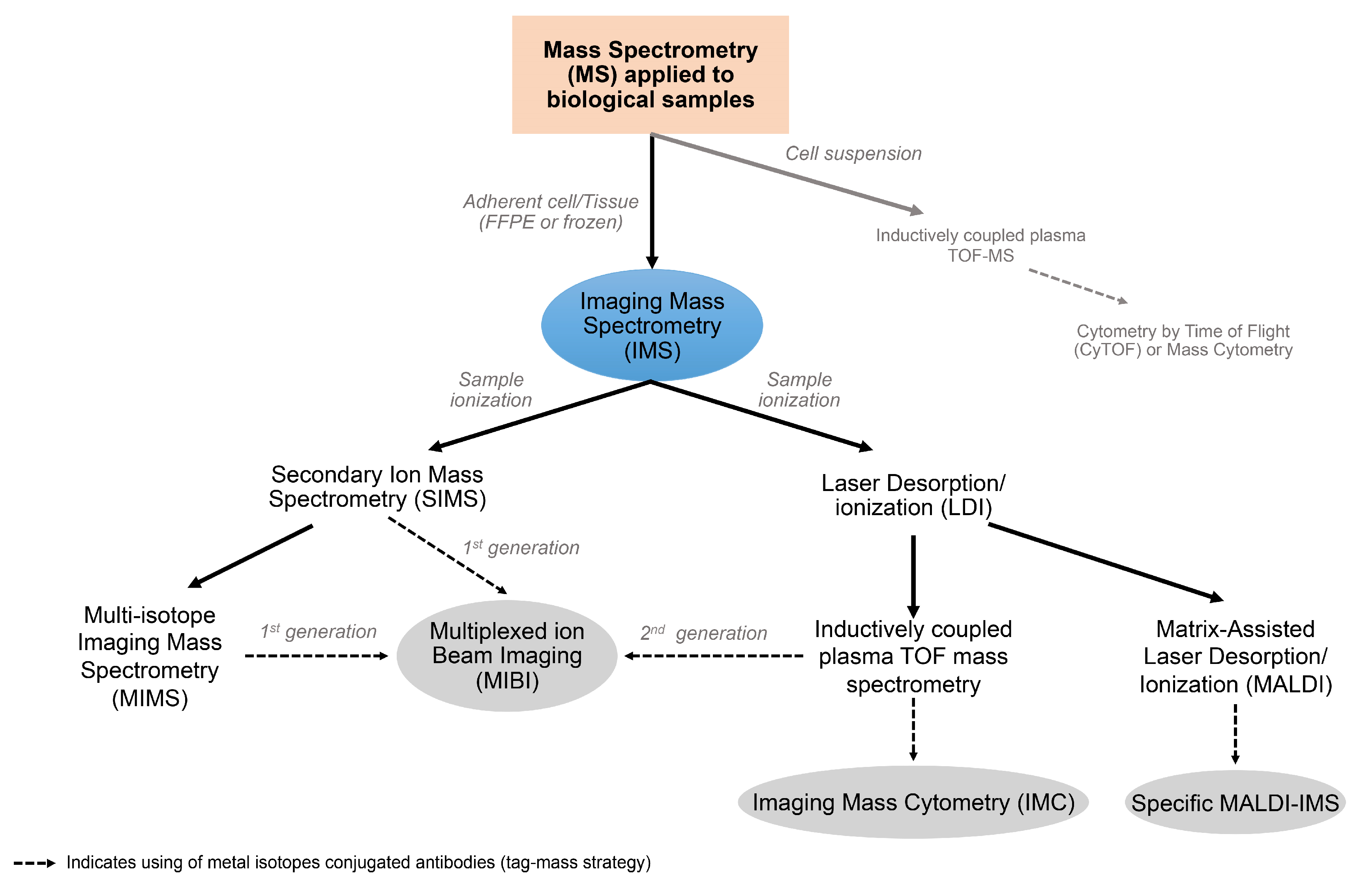
6. Fundamentals of Multiplexed Techniques Based on Mass Spectrometry
6.1. Imaging Mass Spectrometry
6.2. Secondary Ion Mass Spectrometry
6.3. Laser Desorption/Ionization
6.4. Matrix-Assisted Laser Desorption/Ionization
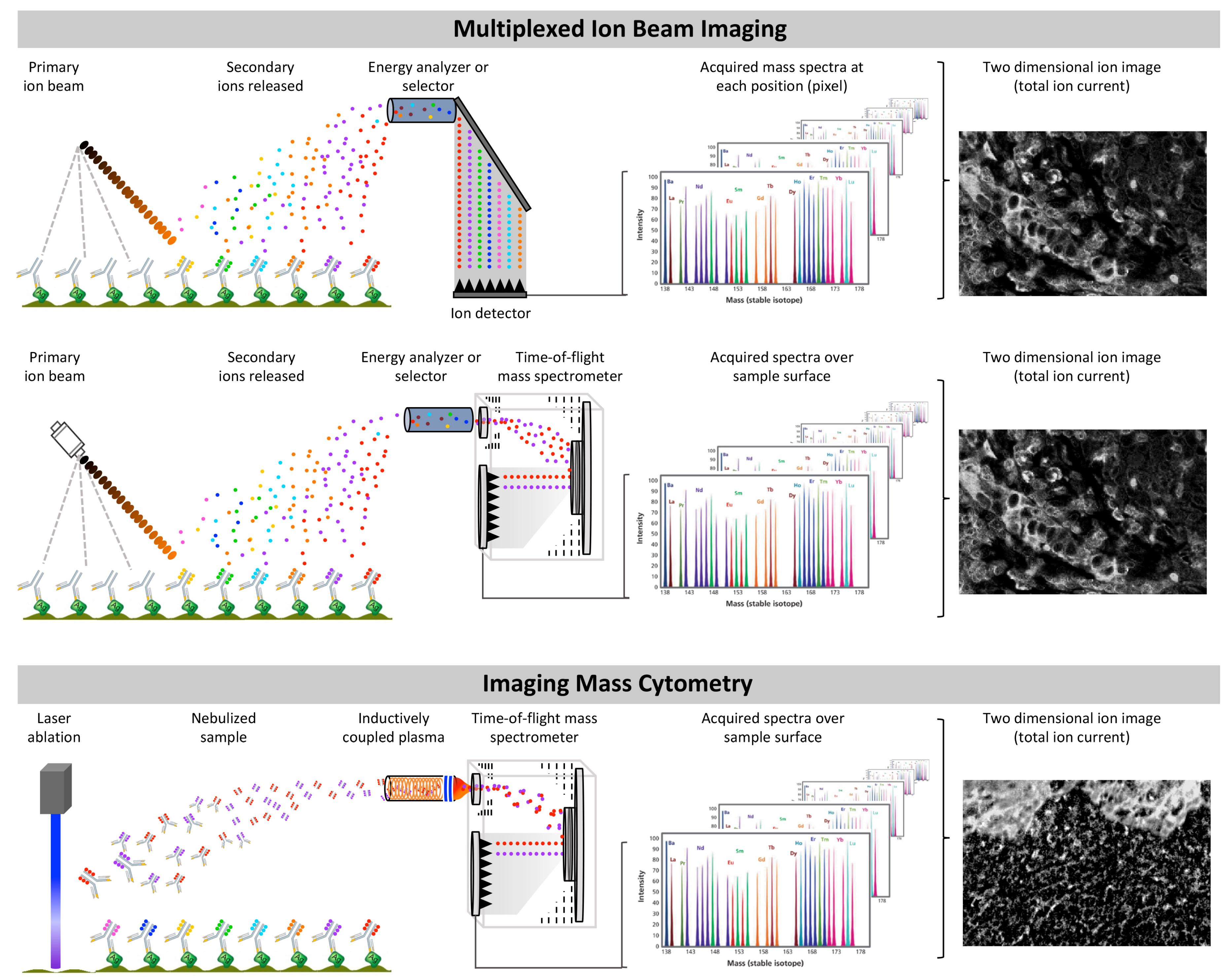
6.5. Multiplexed Ion Beam Imaging and Imaging Mass Cytometry: The Antibody-Based Tag-Mass IMS Strategy
6.6. Multiplexed Ion Beam Imaging
6.7. Imaging Mass Cytometry
7. Image Acquisition and Data Analysis
8. Clinical and Translational Use of Multiplexed Methodologies
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Steiner, C.; Ducret, A.; Tille, J.C.; Thomas, M.; McKee, T.A.; Rubbia-Brandt, L.; Scherl, A.; Lescuyer, P.; Cutler, P. Applications of mass spectrometry for quantitative protein analysis in formalin-fixed paraffin-embedded tissues. Proteomics 2014, 14, 441–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stauber, J.; MacAleese, L.; Franck, J.; Claude, E.; Snel, M.; Kaletas, B.K.; Wiel, I.M.; Wisztorski, M.; Fournier, I.; Heeren, R.M. On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Sood, A.; Miller, A.M.; Brogi, E.; Sui, Y.; Armenia, J.; McDonough, E.; Santamaria-Pang, A.; Carlin, S.; Stamper, A.; Campos, C.; et al. Multiplexed immunofluorescence delineates proteomic cancer cell states associated with metabolism. JCI Insight 2016, 1, e87030. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.A.J.; Halilovic, A.; Rabold, K.; van Duffelen, A.; Wickramasinghe, I.N.; Verweij, D.; Wortel, I.M.N.; Textor, J.C.; de Vries, I.J.M.; Figdor, C.G. Eight-Color Multiplex Immunohistochemistry for Simultaneous Detection of Multiple Immune Checkpoint Molecules within the Tumor Microenvironment. J. Immunol. 2017, 200, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Rost, S.; Giltnane, J.; Bordeaux, J.M.; Hitzman, C.; Koeppen, H.; Liu, S.D. Multiplexed ion beam imaging analysis for quantitation of protein expresssion in cancer tissue sections. Lab. Investig. 2017, 97, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- Stack, E.C.; Wang, C.; Roman, K.A.; Hoyt, C.C. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014, 70, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.R.; Bathany, C.; Tsuei, M.; White, J.; Barald, K.F.; Takayama, S. Recent developments in multiplexing techniques for immunohistochemistry. Expert Rev. Mol. Diagn. 2015, 15, 1171–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remark, R.; Merghoub, T.; Grabe, N.; Litjens, G.; Damotte, D.; Wolchok, J.D.; Merad, M.; Gnjatic, S. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci. Immunol. 2016, 1, aaf6925. [Google Scholar] [CrossRef] [PubMed]
- Glass, G.; Papin, J.A.; Mandell, J.W. SIMPLE: A sequential immunoperoxidase labeling and erasing method. J. Histochem. Cytochem. 2009, 57, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, T.; Kumar, S.; Borkar, R.N.; Azimi, V.; Thibault, G.; Chang, Y.H.; Balter, A.; Kawashima, R.; Choe, G.; Sauer, D.; et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep. 2017, 19, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Schubert, W.; Bonnekoh, B.; Pommer, A.J.; Philipsen, L.; Bockelmann, R.; Malykh, Y.; Gollnick, H.; Friedenberger, M.; Bode, M.; Dress, A.W. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006, 24, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Friedenberger, M.; Bode, M.; Krusche, A.; Schubert, W. Fluorescence detection of protein clusters in individual cells and tissue sections by using toponome imaging system: Sample preparation and measuring procedures. Nat. Protoc. 2007, 2, 2285–2294. [Google Scholar] [CrossRef] [PubMed]
- Herman, B.; Krishnan, R.V.; Centonze, V.E. Microscopic analysis of fluorescence resonance energy transfer (FRET). Methods Mol. Biol. 2004, 261, 351–370. [Google Scholar] [CrossRef] [PubMed]
- Ostalecki, C.; Konrad, A.; Thurau, E.; Schuler, G.; Croner, R.S.; Pommer, A.J.; ael Sturzl, M. Combined multi-gene analysis at the RNA and protein levels in single FFPE tissue sections. Exp. Mol. Pathol. 2013, 95, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Philipsen, L.; Engels, T.; Schilling, K.; Gurbiel, S.; Fischer, K.D.; Tedford, K.; Schraven, B.; Gunzer, M.; Reichardt, P. Multimolecular analysis of stable immunological synapses reveals sustained recruitment and sequential assembly of signaling clusters. Mol. Cell. Proteom. 2013, 12, 2551–2567. [Google Scholar] [CrossRef] [PubMed]
- Ostalecki, C.; Wittki, S.; Lee, J.H.; Geist, M.M.; Tibroni, N.; Harrer, T.; Schuler, G.; Fackler, O.T.; Baur, A.S. HIV Nef- and Notch1-dependent Endocytosis of ADAM17 Induces Vesicular TNF Secretion in Chronic HIV Infection. EBioMedicine 2016, 13, 294–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berndt, U.; Philipsen, L.; Bartsch, S.; Wiedenmann, B.; Baumgart, D.C.; Hammerle, M.; Sturm, A. Systematic high-content proteomic analysis reveals substantial immunologic changes in colorectal cancer. Cancer Res. 2008, 68, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Berndt, U.; Philipsen, L.; Bartsch, S.; Hu, Y.; Rocken, C.; Bertram, W.; Hammerle, M.; Rosch, T.; Sturm, A. Comparative Multi-Epitope-Ligand-Cartography reveals essential immunological alterations in Barrett’s metaplasia and esophageal adenocarcinoma. Mol. Cancer 2010, 9, 177. [Google Scholar] [CrossRef]
- Hollman-Hewgley, D.; Lazare, M.; Bordwell, A.; Zebadua, E.; Tripathi, P.; Ross, A.S.; Fisher, D.; Adams, A.; Bouman, D.; O’Malley, D.P.; et al. A single slide multiplex assay for the evaluation of classical Hodgkin lymphoma. Am. J. Surg. Pathol. 2014, 38, 1193–1202. [Google Scholar] [CrossRef]
- Gerdes, M.J.; Sevinsky, C.J.; Sood, A.; Adak, S.; Bello, M.O.; Bordwell, A.; Can, A.; Corwin, A.; Dinn, S.; Filkins, R.J.; et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. USA 2013, 110, 11982–11987. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Ma, H.; Wang, Y.; Cao, Z.; Graves-Deal, R.; Powell, A.E.; Starchenko, A.; Ayers, G.D.; Washington, M.K.; Kamath, V.; et al. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J. Clin. Investig. 2014, 124, 2172–2187. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.R.; Izar, B.; Wang, S.; Yapp, C.; Mei, S.; Shah, P.M.; Santagata, S.; Sorger, P.K. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 2018, 7, e31657. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, M.M.; Manzoni, M.; Scalia, C.R.; Zannella, S.; Bosisio, F.M.; Faretta, M.; Cattoretti, G. Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. J. Histochem. Cytochem. 2017, 65, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Goltsev, Y.; Samusik, N.; Kennedy-Darling, J.; Bhate, S.; Hale, M.; Vasquez, G.; Nolan, G. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 2018, 174, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Woehrstein, J.B.; Donoghue, N.; Dai, M.; Avendano, M.S.; Schackmann, R.C.J.; Zoeller, J.J.; Wang, S.S.H.; Tillberg, P.W.; Park, D.; et al. Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues. Nano Lett. 2017, 17, 6131–6139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jungmann, R.; Avendano, M.S.; Woehrstein, J.B.; Dai, M.; Shih, W.M.; Yin, P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11, 313–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, M.; Kron, S.J.; Schwartz, D.; Snyder, H. Rapid 5-marker multiplex phenotyping of breast cancer subtypes & tumor-infiltrating leukocytes “in situ” in FFPE sections. Cancer Res. 2016, 76. [Google Scholar] [CrossRef]
- Bobrow, M.N.; Harris, T.D.; Shaughnessy, K.J.; Litt, G.J. Catalyzed reporter deposition, a novel method of signal amplification. Application to immunoassays. J. Immunol. Methods 1989, 125, 279–285. [Google Scholar] [CrossRef]
- Bobrow, M.N.; Shaughnessy, K.J.; Litt, G.J. Catalyzed reporter deposition, a novel method of signal amplification. II. Application to membrane immunoassays. J. Immunol. Methods 1991, 137, 103–112. [Google Scholar] [CrossRef]
- Parra, E.R.; Uraoka, N.; Jiang, M.; Cook, P.; Gibbons, D.; Forget, M.A.; Bernatchez, C.; Haymaker, C.; Wistuba, I.I.; Rodriguez-Canales, J. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci. Rep. 2017, 7, 13380. [Google Scholar] [CrossRef] [Green Version]
- Faget, L.; Hnasko, T.S. Tyramide Signal Amplification for Immunofluorescent Enhancement. Methods Mol. Biol. 2015, 1318, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Shen, R.; Huang, P.; Zhai, J.; Qian, X.; Wang, Q.; Chen, M. Predictive relevance of PD-L1 expression with pre-existing TILs in gastric cancer. Oncotarget 2017, 8, 99372–99381. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.D.; Gusenleitner, D.; Lipschitz, M.; Roemer, M.G.M.; Stack, E.C.; Gjini, E.; Hu, X.; Redd, R.; Freeman, G.J.; Neuberg, D.; et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017, 130, 2420–2430. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Wilmott, J.S.; Madore, J.; Gide, T.N.; Quek, C.; Tasker, A.; Ferguson, A.L.; Chen, J.; Hewavisenti, R.; Hersey, P.; et al. CD103+ tumor-resident CD8+ T cells are associated with improved survival in immunotherapy naive melanoma patients and expand significantly during anti-PD1 treatment. Clin. Cancer Res. 2018, 24, 3036–3045. [Google Scholar] [CrossRef] [PubMed]
- Buisseret, L.; Pommey, S.; Allard, B.; Garaud, S.; Bergeron, M.; Cousineau, I.; Ameye, L.; Bareche, Y.; Paesmans, M.; Crown, J.P.A.; et al. Clinical significance of CD73 in triple-negative breast cancer: Multiplex analysis of a phase III clinical trial. Ann. Oncol. 2018, 29, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Peng, C.W.; Pang, D.W.; Li, Y. Quantum dots for cancer research: Current status, remaining issues, and future perspectives. Cancer Biol. Med. 2012, 9, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Bostick, R.M.; Kong, K.Y.; Ahearn, T.U.; Chaudry, Q.; Cohen, V.; Wang, M.D. Detecting and quantifying biomarkers of risk for colorectal cancer using quantum dots and novel image analysis algorithms. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006, 1, 3313–3316. [Google Scholar] [CrossRef] [PubMed]
- Kairdolf, B.A.; Smith, A.M.; Stokes, T.H.; Wang, M.D.; Young, A.N.; Nie, S. Semiconductor quantum dots for bioimaging and biodiagnostic applications. Annu. Rev. Anal. Chem. 2013, 6, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.W.; Liu, X.L.; Chen, C.; Liu, X.; Yang, X.Q.; Pang, D.W.; Zhu, X.B.; Li, Y. Patterns of cancer invasion revealed by QDs-based quantitative multiplexed imaging of tumor microenvironment. Biomaterials 2011, 32, 2907–2917. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lau, S.K.; Varma, V.A.; Moffitt, R.A.; Caldwell, M.; Liu, T.; Young, A.N.; Petros, J.A.; Osunkoya, A.O.; Krogstad, T.; et al. Molecular mapping of tumor heterogeneity on clinical tissue specimens with multiplexed quantum dots. ACS Nano 2010, 4, 2755–2765. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.R.; Gossage, K.W.; Hoyt, C.C.; Levenson, R.M. Autofluorescence removal, multiplexing, and automated analysis methods for in-vivo fluorescence imaging. J. Biomed. Opt. 2005, 10, 41207. [Google Scholar] [CrossRef] [PubMed]
- Zrazhevskiy, P.; True, L.D.; Gao, X. Multicolor multicycle molecular profiling with quantum dots for single-cell analysis. Nat. Protoc. 2013, 8, 1852–1869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, T.H. Quantum dot enabled molecular sensing and diagnostics. Theranostics 2012, 2, 631–654. [Google Scholar] [CrossRef] [PubMed]
- Perez-Trevino, P.; la Cerda, H.H.; Perez-Trevino, J.; Fajardo-Ramirez, O.R.; Garcia, N.; Altamirano, J. 3D Imaging Detection of HER2 Based in the Use of Novel Affibody-Quantum Dots Probes and Ratiometric Analysis. Transl. Oncol. 2018, 11, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Matros, A.; Mock, H.P. Mass spectrometry based imaging techniques for spatially resolved analysis of molecules. Front. Plant Sci. 2013, 4, 89. [Google Scholar] [CrossRef] [PubMed]
- Reyzer, M.L.; Caprioli, R.M. The Development of Imaging Mass Spectrometry. Encycl. Mass Spectrom. 2016, 9, 285–304. [Google Scholar]
- Dong, Y.; Li, B.; Malitsky, S.; Rogachev, I.; Aharoni, A.; Kaftan, F.; Svatos, A.; Franceschi, P. Sample Preparation for Mass Spectrometry Imaging of Plant Tissues: A Review. Front. Plant Sci. 2016, 7, 60. [Google Scholar] [CrossRef]
- Pohl, L.; Kolbl, A.; Werner, F.; Mueller, C.W.; Hoschen, C.; Hausler, W.; Kogel-Knabner, I. Imaging of Al/Fe ratios in synthetic Al-goethite revealed by nanoscale secondary ion mass spectrometry. Rapid Commun. Mass Spectrom. 2018, 32, 619–628. [Google Scholar] [CrossRef]
- Andersen, C.A.; Hinthorne, J.R. Ion microprobe mass analyzer. Science 1972, 175, 853–860. [Google Scholar] [CrossRef]
- Liebl, H. SIMS Instrumentation and Imaging Techniques. Scanning 1980, 3, 79–89. [Google Scholar] [CrossRef]
- Morrison, G.H.; Slodzian, G. Ion Microscopy. Anal. Chem. 1975, 47, 932A–943A. [Google Scholar] [CrossRef]
- Liebl, H. Ion Microprobe Mass Analyzer. J. Appl. Phys. 1967, 38, 5277–5283. [Google Scholar] [CrossRef]
- Baker, M.J.; Brown, M.D.; Gazi, E.; Clarke, N.W.; Vickerman, J.C.; Lockyer, N.P. Discrimination of prostate cancer cells and non-malignant cells using secondary ion mass spectrometry. Analyst 2008, 133, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Gostek, J.; Awsiuk, K.; Pabijan, J.; Rysz, J.; Budkowski, A.; Lekka, M. Differentiation between single bladder cancer cells using principal component analysis of time-of-flight secondary ion mass spectrometry. Anal. Chem. 2015, 87, 3195–3201. [Google Scholar] [CrossRef] [PubMed]
- Kulp, K.S.; Berman, E.S.; Knize, M.G.; Shattuck, D.L.; Nelson, E.J.; Wu, L.; Montgomery, J.L.; Felton, J.S.; Wu, K.J. Chemical and biological differentiation of three human breast cancer cell types using time-of-flight secondary ion mass spectrometry. Anal. Chem. 2006, 78, 3651–3658. [Google Scholar] [CrossRef] [PubMed]
- Theiner, S.; Van Malderen, S.J.M.; Van Acker, T.; Legin, A.; Keppler, B.K.; Vanhaecke, F.; Koellensperger, G. Fast High-Resolution Laser Ablation-Inductively Coupled Plasma Mass Spectrometry Imaging of the Distribution of Platinum-Based Anticancer Compounds in Multicellular Tumor Spheroids. Anal. Chem. 2017, 89, 12641–12645. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, E.B.; de la Monte, S.M. Review of matrix-assisted laser desorption ionization-imaging mass spectrometry for lipid biochemical histopathology. J. Histochem. Cytochem. 2015, 63, 762–771. [Google Scholar] [CrossRef]
- Di Girolamo, F.; Lante, I.; Muraca, M.; Putignani, L. The Role of Mass Spectrometry in the “Omics” Era. Curr. Org. Chem. 2013, 17, 2891–2905. [Google Scholar] [CrossRef]
- Chan, T.W.; Duan, L.; Sze, T.P. Accurate mass measurements for peptide and protein mixtures by using matrix-assisted laser desorption/ionization Fourier transform mass spectrometry. Anal. Chem. 2002, 74, 5282–5289. [Google Scholar] [CrossRef]
- Sauer, S.; Kliem, M. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 2010, 8, 74–82. [Google Scholar] [CrossRef]
- Gao, X.; Tan, B.H.; Sugrue, R.J.; Tang, K. MALDI mass spectrometry for nucleic acid analysis. Top. Curr. Chem. 2013, 331, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Vogel, N.; Schiebel, K.; Humeny, A. Technologies in the Whole-Genome Age: MALDI-TOF-Based Genotyping. Transfus. Med. Hemother. 2009, 36, 253–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanagisawa, K.; Shyr, Y.; Xu, B.J.; Massion, P.P.; Larsen, P.H.; White, B.C.; Roberts, J.R.; Edgerton, M.; Gonzalez, A.; Nadaf, S.; et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 2003, 362, 433–439. [Google Scholar] [CrossRef]
- Steinhauser, M.L.; Bailey, A.P.; Senyo, S.E.; Guillermier, C.; Perlstein, T.S.; Gould, A.P.; Lee, R.T.; Lechene, C.P. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 2012, 481, 516–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelo, M.; Bendall, S.C.; Finck, R.; Hale, M.B.; Hitzman, C.; Borowsky, A.D.; Levenson, R.M.; Lowe, J.B.; Liu, S.D.; Zhao, S.; et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 2014, 20, 436–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandura, D.R.; Baranov, V.I.; Ornatsky, O.I.; Antonov, A.; Kinach, R.; Lou, X.; Pavlov, S.; Vorobiev, S.; Dick, J.E.; Tanner, S.D. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009, 81, 6813–6822. [Google Scholar] [CrossRef] [PubMed]
- Rimm, D.L. What brown cannot do for you. Nat. Biotechnol. 2006, 24, 914–916. [Google Scholar] [CrossRef] [PubMed]
- Keren, L.; Bosse, M.; Marquez, D.; Angoshtari, R.; Jain, S.; Varma, S.; Yang, S.R.; Kurian, A.; Van Valen, D.; West, R.; et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, 174, 1373–1387. [Google Scholar] [CrossRef]
- Di Palma, S.; Bodenmiller, B. Unraveling cell populations in tumors by single-cell mass cytometry. Curr. Opin. Biotechnol. 2015, 31, 122–129. [Google Scholar] [CrossRef]
- Dempsey, L.A. CyTOF analysis of anti-tumor responses. Nat. Immunol. 2017, 18, 254. [Google Scholar] [CrossRef]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; Amir el, A.D.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Giesen, C.; Wang, H.A.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schuffler, P.J.; Grolimund, D.; Buhmann, J.M.; Brandt, S.; et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.; Zanotelli, V.R.T.; Fischer, J.R.; Schapiro, D.; Engler, S.; Lun, X.K.; Jackson, H.W.; Bodenmiller, B. Simultaneous Multiplexed Imaging of mRNA and Proteins with Subcellular Resolution in Breast Cancer Tissue Samples by Mass Cytometry. Cell Syst. 2018, 6, 531. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Dummer, R.; Puzanov, I.; VanderWalde, A.; Andtbacka, R.H.I.; Michielin, O.; Olszanski, A.J.; Malvehy, J.; Cebon, J.; Fernandez, E.; et al. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell 2017, 170, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.P.; van der Weiden, M.; Kros, J.M. Fast tracking of co-localization of multiple markers by using the nanozoomer slide scanner and NDPViewer. J. Cell. Physiol. 2014, 229, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Saylor, J.; Ma, Z.; Goodridge, H.S.; Huang, F.; Cress, A.E.; Pandol, S.J.; Shiao, S.L.; Vidal, A.C.; Wu, L.; Nickols, N.G.; et al. Spatial Mapping of Myeloid Cells and Macrophages by Multiplexed Tissue Staining. Front. Immunol. 2018, 9, 2925. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Smith, I.; Parker, I.; Bootman, M.D. Fluorescence microscopy. Cold Spring Harb. Protoc. 2014, 2014, pdb.top071795. [Google Scholar] [CrossRef]
- Spindel, S.; Sapsford, K.E. Evaluation of optical detection platforms for multiplexed detection of proteins and the need for point-of-care biosensors for clinical use. Sensors 2014, 14, 22313–22341. [Google Scholar] [CrossRef]
- Blom, S.; Paavolainen, L.; Bychkov, D.; Turkki, R.; Maki-Teeri, P.; Hemmes, A.; Valimaki, K.; Lundin, J.; Kallioniemi, O.; Pellinen, T. Systems pathology by multiplexed immunohistochemistry and whole-slide digital image analysis. Sci. Rep. 2017, 7, 15580. [Google Scholar] [CrossRef] [Green Version]
- Isse, K.; Lesniak, A.; Grama, K.; Roysam, B.; Minervini, M.I.; Demetris, A.J. Digital transplantation pathology: Combining whole slide imaging, multiplex staining and automated image analysis. Am. J. Transplant. 2012, 12, 27–37. [Google Scholar] [CrossRef]
- Bodenmiller, B. Multiplexed Epitope-Based Tissue Imaging for Discovery and Healthcare Applications. Cell Syst. 2016, 2, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Lyons, C.; Lawler, D. Aperio Cellular IF Algorithm Validation. Pathologist 2016. [Google Scholar] [CrossRef]
- Harder, N.; Athelogou, M.; Hessel, H.; Brieu, N.; Yigitsoy, M.; Zimmermann, J.; Baatz, M.; Buchner, A.; Stief, C.G.; Kirchner, T.; et al. Tissue Phenomics for prognostic biomarker discovery in low- and intermediate-risk prostate cancer. Sci. Rep. 2018, 8, 4470. [Google Scholar] [CrossRef] [PubMed]
- Klimowicz, A.C.; Bose, P.; Petrillo, S.K.; Magliocco, A.M.; Dort, J.C.; Brockton, N.T. The prognostic impact of a combined carbonic anhydrase IX and Ki67 signature in oral squamous cell carcinoma. Br. J. Cancer 2013, 109, 1859–1866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojo, M.G.; Bueno, G.; Slodkowska, J. Review of imaging solutions for integrated quantitative immunohistochemistry in the Pathology daily practice. Folia Histochem. Cytobiol. 2009, 47, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Shiao, S.L.; Yoshida, E.J.; Swartwood, S.; Huang, F.; Doche, M.E.; Chung, A.P.; Knudsen, B.S.; Gertych, A. Data integration from pathology slides for quantitative imaging of multiple cell types within the tumor immune cell infiltrate. Diagn. Pathol. 2017, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Sideras, K.; Galjart, B.; Vasaturo, A.; Pedroza-Gonzalez, A.; Biermann, K.; Mancham, S.; Nigg, A.L.; Hansen, B.E.; Stoop, H.A.; Zhou, G.; et al. Prognostic value of intra-tumoral CD8(+) /FoxP3(+) lymphocyte ratio in patients with resected colorectal cancer liver metastasis. J. Surg. Oncol. 2018, 118, 68–76. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [Green Version]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- De Chaumont, F.; Dallongeville, S.; Chenouard, N.; Herve, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef]
- Jones, T.R.; Kang, I.H.; Wheeler, D.B.; Lindquist, R.A.; Papallo, A.; Sabatini, D.M.; Golland, P.; Carpenter, A.E. CellProfiler Analyst: Data exploration and analysis software for complex image-based screens. BMC Bioinform. 2008, 9, 482. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, M.R.; Sabatini, D.M.; Carpenter, A.E. CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques 2007, 42, 71–75. [Google Scholar] [CrossRef] [PubMed]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [PubMed]
- Wiesmann, V.; Franz, D.; Held, C.; Munzenmayer, C.; Palmisano, R.; Wittenberg, T. Review of free software tools for image analysis of fluorescence cell micrographs. J. Microsc. 2015, 257, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Fang, P.; Sharma, A.; Fujimoto, J.; Wistuba, I.; Rao, A.U.K.; Lin, S.H. Spatial interaction of tumor cells and regulatory T cells correlates with survival in non-small cell lung cancer. Lung Cancer 2018, 117, 73–79. [Google Scholar] [CrossRef]
- Stack, E.C.; Foukas, P.G.; Lee, P.P. Multiplexed tissue biomarker imaging. J. Immunother. Cancer 2016, 4, 9. [Google Scholar] [CrossRef]
- Johansson, A.C.; Visse, E.; Widegren, B.; Sjogren, H.O.; Siesjo, P. Computerized image analysis as a tool to quantify infiltrating leukocytes: A comparison between high- and low-magnification images. J. Histochem. Cytochem. 2001, 49, 1073–1079. [Google Scholar] [CrossRef]
- Henriksen, M. Quantitative imaging cytometry: Instrumentation of choice for automated cellular and tissue analysis. Nat. Methods 2010, 7, 330. [Google Scholar] [CrossRef]
- Miller, A.; Nagy, C.; Knapp, B.; Laengle, J.; Ponweiser, E.; Groeger, M.; Starkl, P.; Bergmann, M.; Wagner, O.; Haschemi, A. Exploring Metabolic Configurations of Single Cells within Complex Tissue Microenvironments. Cell Metab. 2017, 26, 788–800. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef]
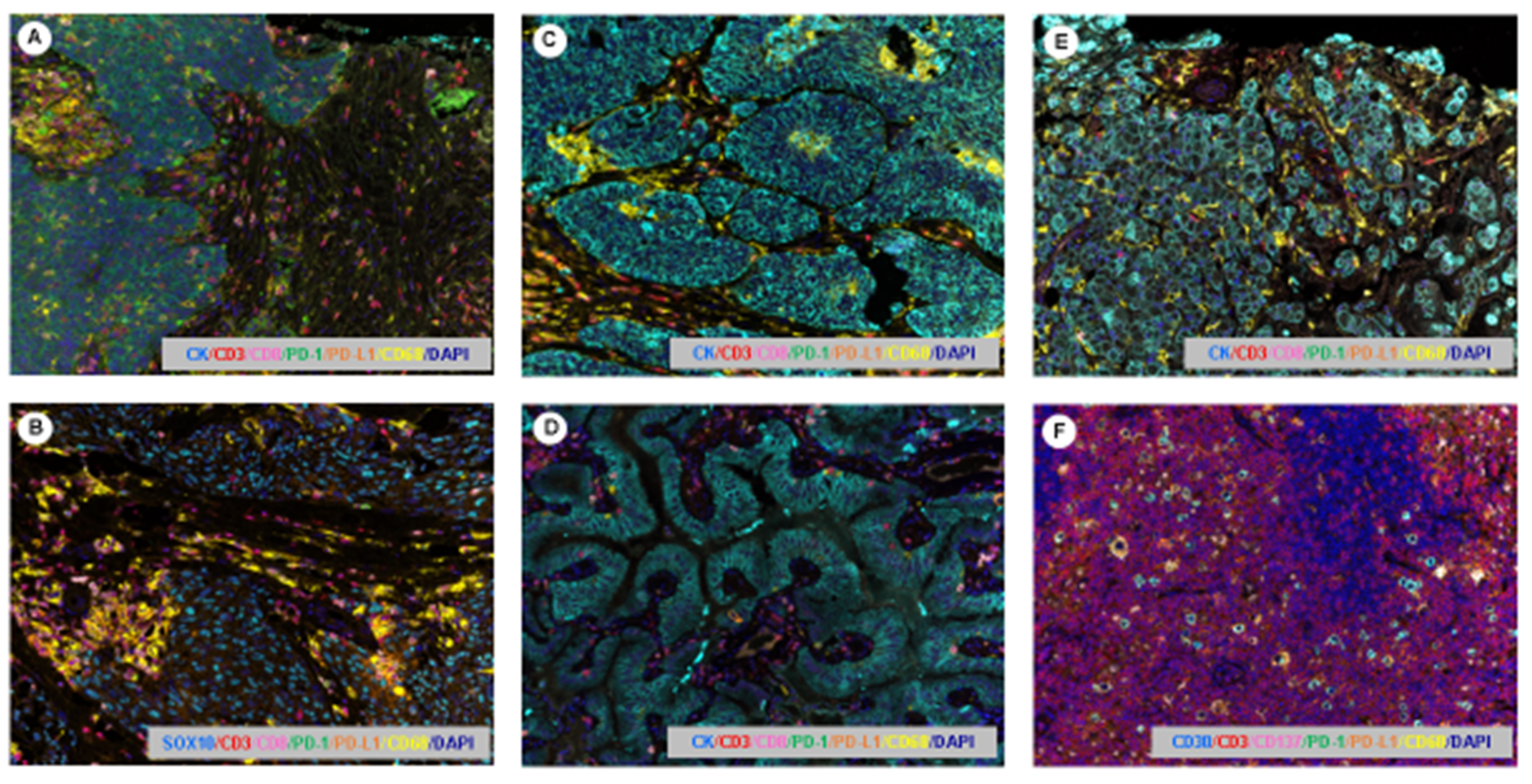
- Parra, E.R.; Villalobos, P.; Behrens, C.; Jiang, M.; Pataer, A.; Swisher, S.G.; William, W.N., Jr.; Zhang, J.; Lee, J.; Cascone, T.; et al. Effect of neoadjuvant chemotherapy on the immune microenvironment in non-small cell lung carcinomas as determined by multiplex immunofluorescence and image analysis approaches. J. Immunother. Cancer 2018, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R. Novel Technology to Assess Programmed Death-Ligand 1 Expression by Multiplex Immunofluorescence and Image Analysis. Appl. Immunohistochem. Mol. Morphol. 2018, 26, e22–e24. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R. Novel Platforms of Multiplexed Immunofluorescence for Study of Paraffin Tumor Tissues. J. Cancer Treat. Diagn. 2018, 2, 43–53. [Google Scholar] [CrossRef]
| Multiplex Staining Method | Advantage | Disadvantage |
|---|---|---|
| Non-fluorescence based platform | ||
| Multiplexed immunohistochemical consecutive staining on a single slide |
|
|
| Sequential immunoperoxidase labeling and erasing |
|
|
| Fluorescence based platform | ||
| Bleaching techniques without signal amplification system | ||
| Multi-epitope-ligand cartography |
|
|
| MultiOmyxTM staining or hyperplexed Immunofluorescence Assay |
|
|
| Tissue-based cyclic immunofluorescence method |
|
|
| Co-detection by indexing or fluorescent immunohisto-PCR |
|
|
| DNA exchange imaging |
|
|
| Amplification of the epitope detection | ||
| Hapten-based modified multiplex |
|
|
| Tyramide signal amplification |
|
|
| Nanocrystal quantum dots |
|
|
| Mass Spectrometry Imaging | ||
| Secondary Ion Mass Spectrometry |
|
|
| Laser Desorption/Ionization |
|
|
| Matrix-assisted laser desorption/ionization |
|
|
| Multiplexed ion beam imaging |
|
|
| Imaging Mass Cytometry |
|
|
| Vendor | Software Package | Capabilities | Data Visualization | Availability | Reference |
|---|---|---|---|---|---|
| Akoya/PerkinElmer | InForm | Color-Based Co-localization, Tissue Segmentation, Cell/Object Segmentation, Cell Phenotyping, Scoring and Automated Quantitation using Batch Analysis | Density Raw Data | Licensed | [6,96] |
| Neo Genomics | MultiOmyx Quantification Program | Epithelial tissue reconstruction, Cellular and Subcellular Segmentation, Cell Phenotyping, Quantification Algorithms | Density Raw Data | Licensed | [3,20] |
| Leica Biosystems | Aperio eSlide Manager Analysis | Pixel-Based Analysis, Cellular identification, Area Quantification and Positive Pixel Count IF Algorithm | Density Raw Data | Licensed | [82] |
| Definiens | Tissue Studio/Image Developer | Imaging Segmentation, Marker Intensity Measurement, Cell Quantification, Batch Analysis, Statistical Analysis, and Algorithm Creator. | Histograms and Profile Plots | Licensed | [83] |
| HistoRx | AQUAnalysis | Signal Intensity Quantification Per Unit Area and Per Layer | Density Raw Data | Licensed | [84] |
| SlidePath | SlidePath’s Tissue Image Analysis | Membrane, Nuclear and Positive Pixel Quantification | Density Raw Data | Licensed | [85] |
| Indica Labs | HALO | Membrane, Co-localization, Immune Cell Proximity, Spatial Analysis, Batch Analysis | Spatial Plot, Histogram | Licensed | [86] |
| VISIOPHARM | Visimoph Tissuemorph | Signal Intensity, Area, Counting Objects, Spatial Analysis, Clustering Statistical Analysis, Batch Analysis and Algorithm Creator. | Phenotypic Matrix, t-SNE Plots | Licensed | [87] |
| Media Cybernetics | Image-Pro | Color-Based, Nuclear segmentation, Cell quantification, Macro-enabled Advanced Image Processing Solution | Density Raw Data | Licensed | [97] |
| CompuCyte | iCyte/iBroser/iNovator | Nucleus Segmentation or Phantom Contouring, Measures Associated Signals | Density Raw Data | Licensed | [98] |
| TissueGnostics | HistoQuest/TissueQuest/StrataQuest | Nuclei-Based Segmentation of Tissues, Cell Phenotyping | Density Raw Data | Licensed | [99] |
| NIH | Image J | Color-Based, User Interactive Segmentation | Histograms and Profile Plots | Open | [88] |
| https://qupath.github.io | QuPath | View Measurements in Context by Color Coding Objects According to Their Features, Flexible Object Classification, Trainable Cell Classification and Quantification | Density Raw Data | Open | [89] |
| http://icy.bioimageanalysis.org | Icy | Based and Color Object Identification, Size, Shape, Color Intensity, Texture, Spatial Analysis. | Plots, Histogram | Open | [90] |
| https://cellprofiler.org/ | Cell Profiler/Cell Analyst | Based and Color Object Identification, Size, Shape, Color Intensity, Texture, and Number Neighbor Quantification. | Density Plot, Histogram | Open | [91,92,93] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra, E.R.; Francisco-Cruz, A.; Wistuba, I.I. State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues. Cancers 2019, 11, 247. https://doi.org/10.3390/cancers11020247
Parra ER, Francisco-Cruz A, Wistuba II. State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues. Cancers. 2019; 11(2):247. https://doi.org/10.3390/cancers11020247
Chicago/Turabian StyleParra, Edwin Roger, Alejandro Francisco-Cruz, and Ignacio Ivan Wistuba. 2019. "State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues" Cancers 11, no. 2: 247. https://doi.org/10.3390/cancers11020247
APA StyleParra, E. R., Francisco-Cruz, A., & Wistuba, I. I. (2019). State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues. Cancers, 11(2), 247. https://doi.org/10.3390/cancers11020247






