Antibiofilm Effect of Curcumin on Saccharomyces boulardii during Beer Fermentation and Bottle Aging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Beer Brewing Process
- i.
- Malt crushing: the malt was mixed and ground with a two-roller mill at a setting of 0.5 mm.
- ii.
- Malt mashing: mashing was carried out in a 27 L capacity Brewferm® (Sunderland, UK) stainless steel boiler equipped with a mash agitator with a water ratio of 1:4 for one hour at 66 °C.
- iii.
- Lautering: a mash filter was used for lautering, and the mash was washed with 70 °C water until the Brix value reached 5° Bx.
- iv.
- Wort boiling: the wort was boiled for one hour; bittering hops (10 g Columbus hops) were added at the beginning of the boil, and aroma hops (30 g Hüll Melon) were added 10 min before the end.
- v.
- Beer fermentation: the wort was pitched with S. boulardii (6.2 × 105 cells/mL) in a total volume of 10 L in each flask. Prior to treatment, curcumin (Sigma cat # C1386) was dissolved in alcohol (99.8%) to obtain a stock solution (3 mg/mL) and used for treatment at a concentration of 25 μg/mL. Fermentation was also conducted in 250 mL bottles to investigate the impact of curcumin on fermentation performance and biofilm development. A control sample was prepared without curcumin addition. All samples were fermented in the dark at 20 °C for two weeks. During this period, they were regularly tested every two days to monitor fermentation performance and biofilm development.
- vi.
- Beer aging: Beer samples were supplemented with d-glucose (5 g/L) and subsequently aged for a duration of four weeks in 250 mL bottles. The bottles were stored in a dark environment at a temperature of 20 °C.
2.2. Biofilm Surface and Experiment Time Points
2.3. Quantification of Biofilm Level
2.3.1. Crystal Violet Staining Assay
2.3.2. Biofilm XTT Reduction Assay
2.4. Cell Count
2.5. Physicochemical Analysis of Wort and Beer
2.5.1. HPLC Analysis
2.5.2. pH Measurement
2.6. Reverse Transcription Quantitative PCR (RT-qPCR) Analysis of FLO Genes
2.7. Microscopic Techniques
2.7.1. Confocal Laser Scanning Microscopy (CLSM) Observation
2.7.2. Transmission Electron Microscopy (TEM) Analysis
2.8. Determination of Total Phenolics
2.9. Sensorial Analysis
2.10. Statistical Analysis
3. Results
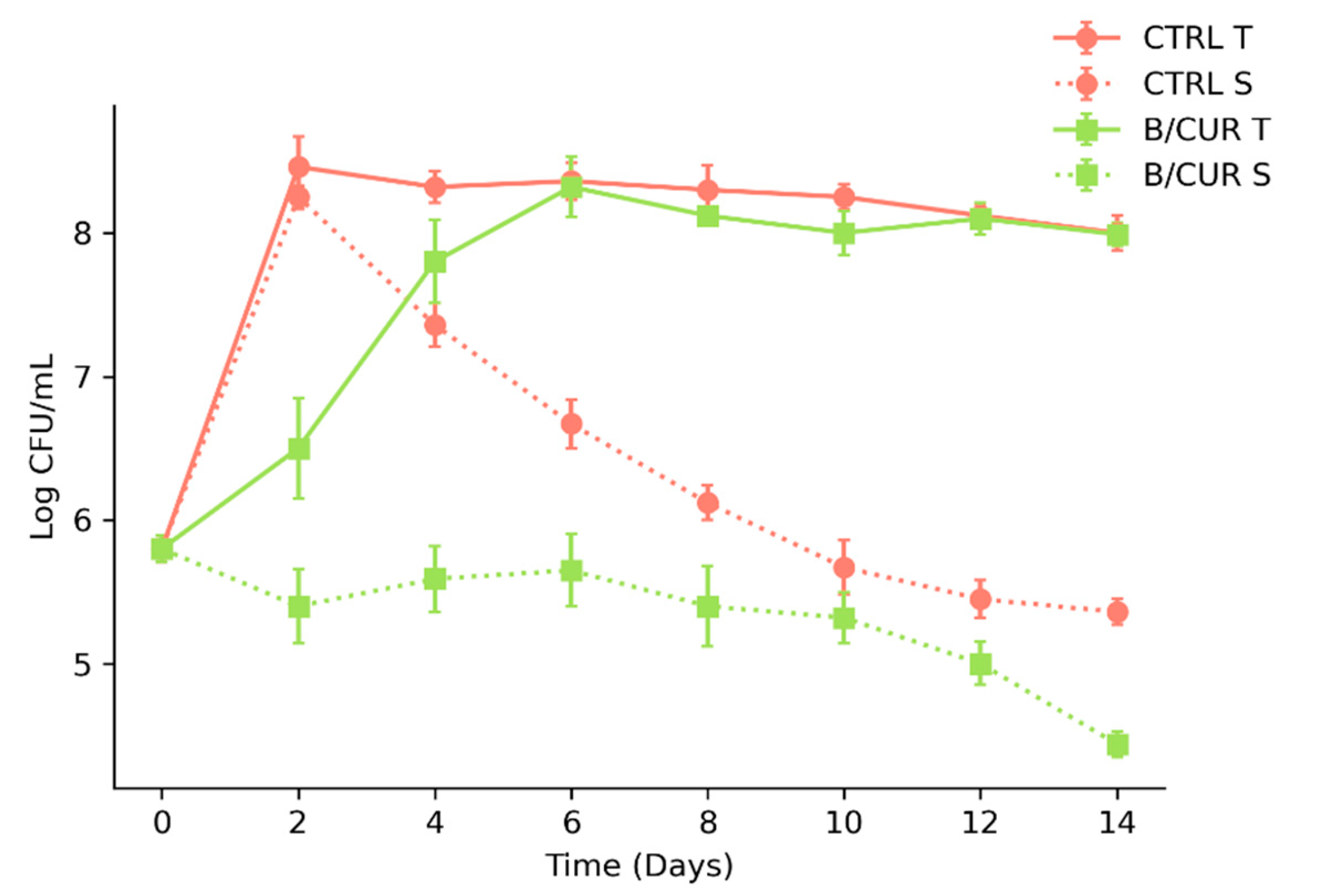
3.1. Curcumin as a Potential Inhibitor of Biofilm Formation in S. boulardii
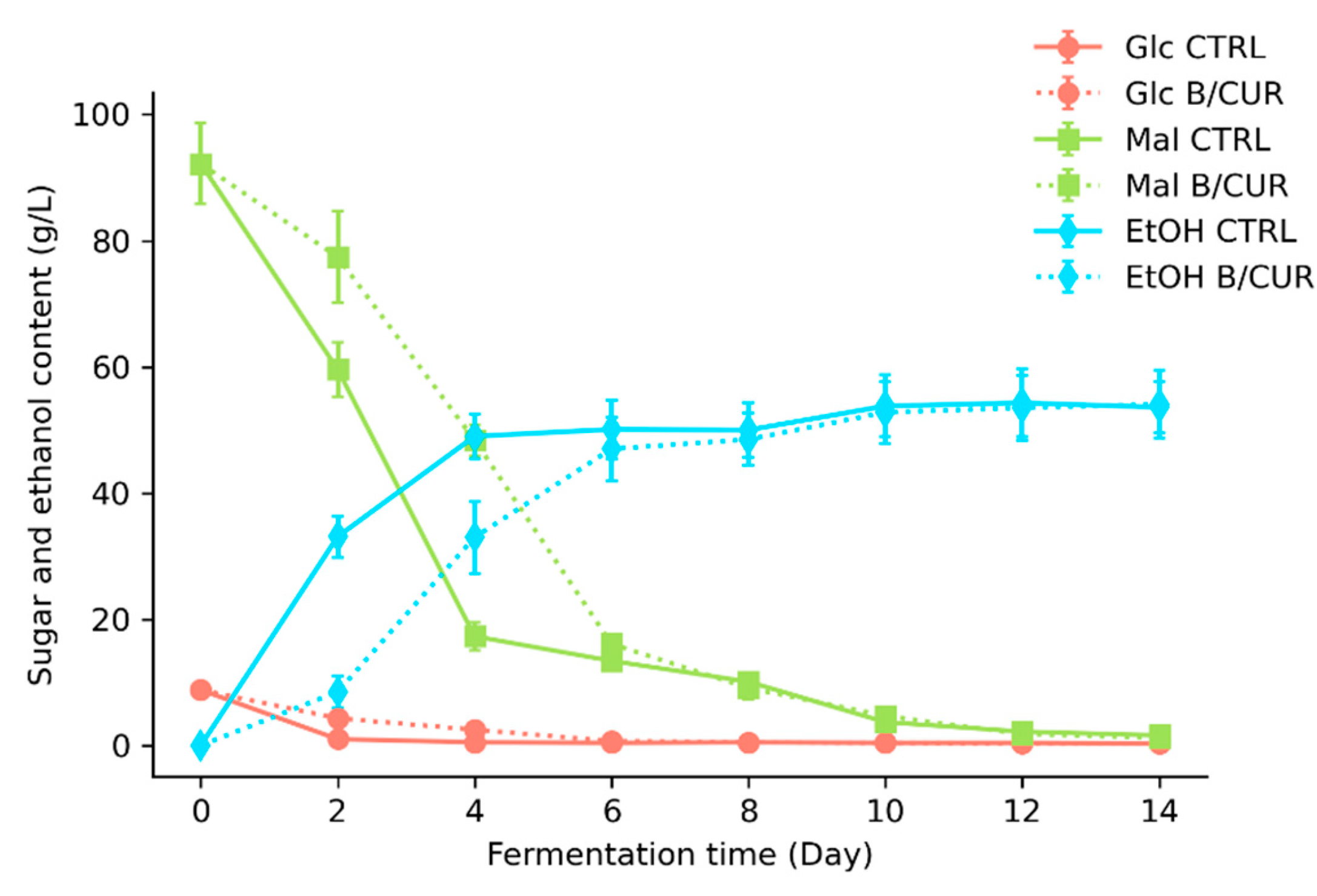
3.2. Influence of Curcumin on Fermentation Performance
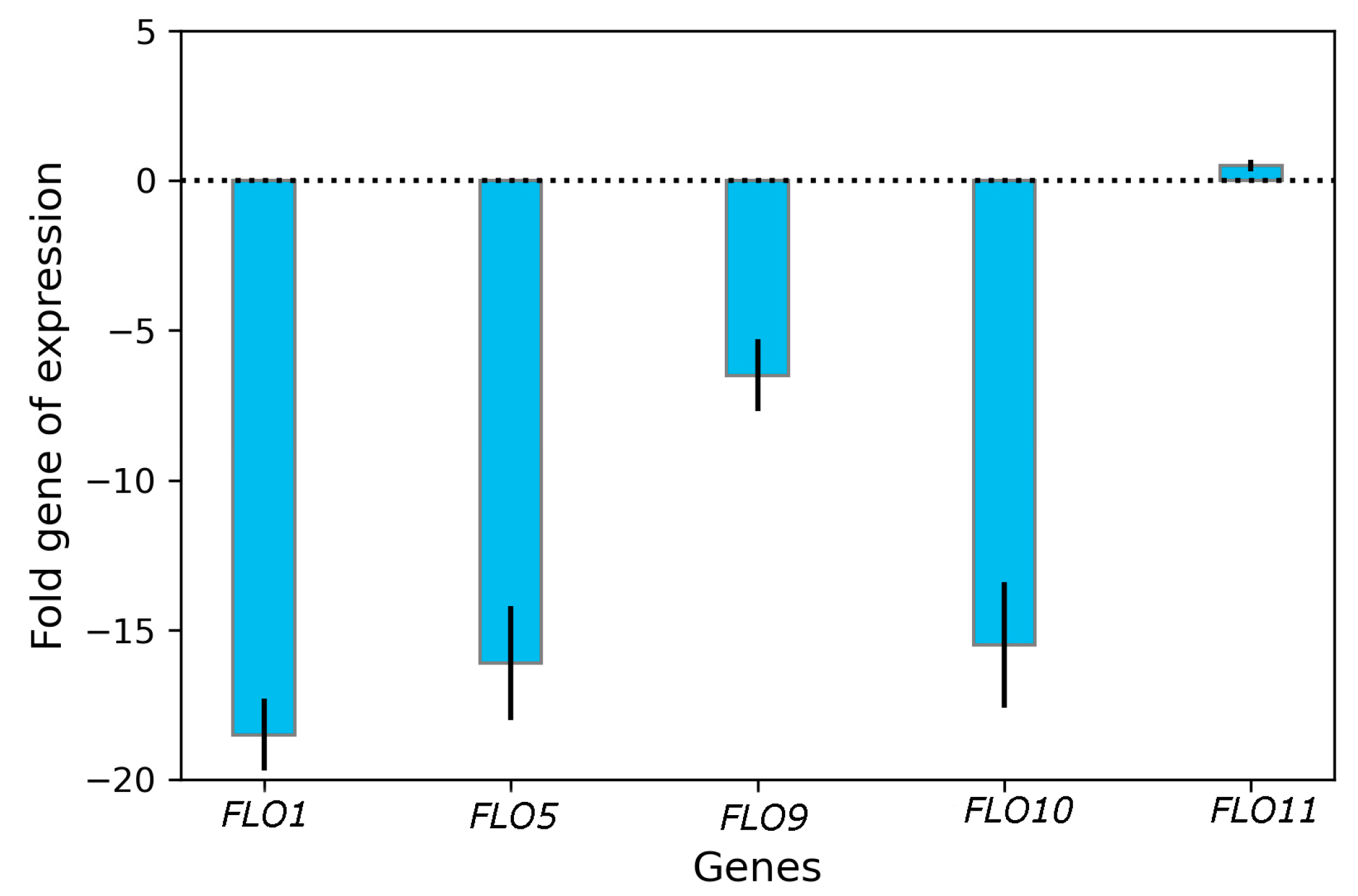
3.3. Impact of Curcumin on Transcriptional Changes in FLO Genes
3.4. Prevention of Biofilm Formation on Glass Microscope Slide Covers
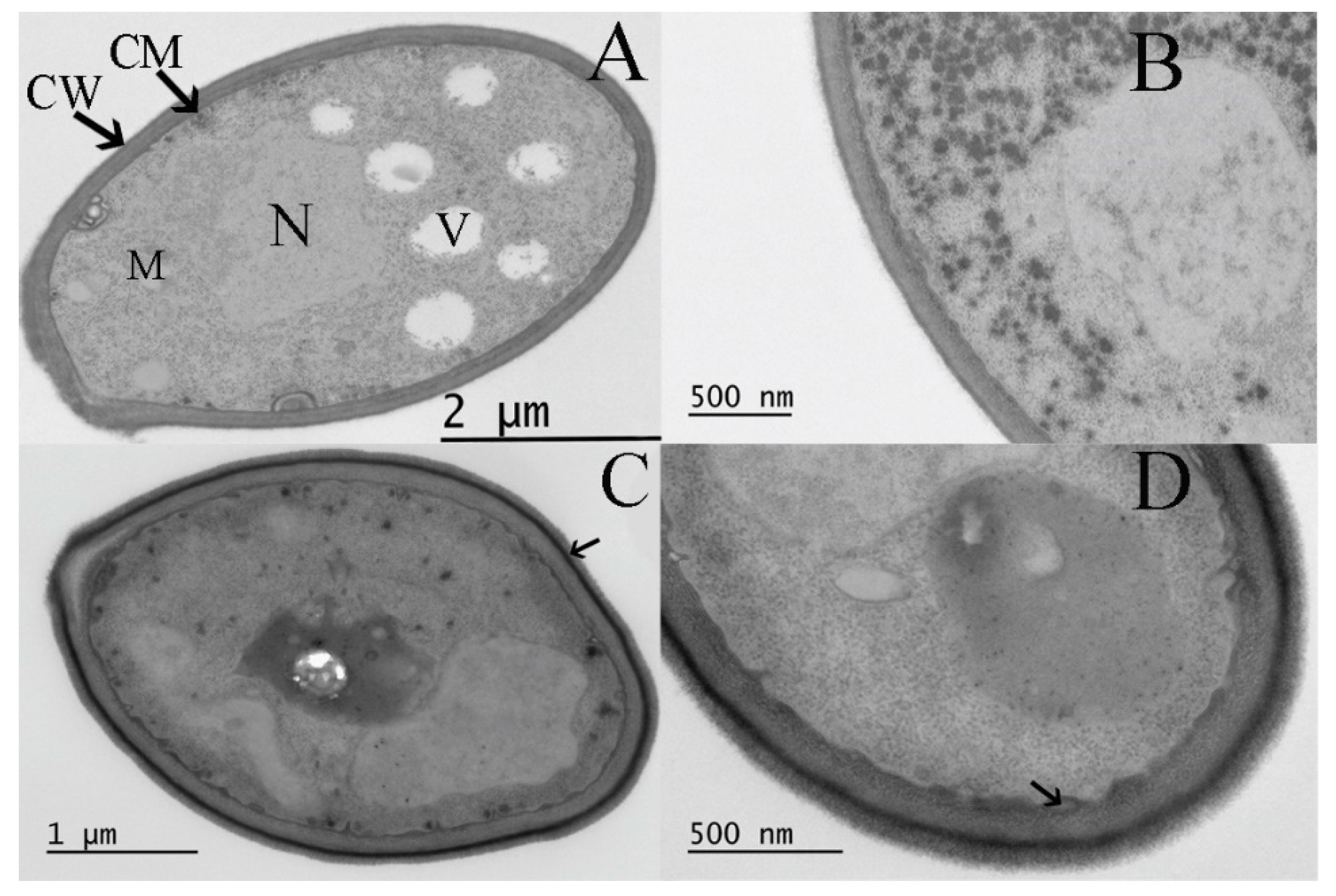
3.5. Effect of Curcumin on the Ultrastructure of S. boulardii
3.6. Evolution of pH and Bioactive Compounds
3.7. Sensorial Analysis of the Beers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bērziņa, L.; Mieriņa, I. Antiradical and Antioxidant Activity of Compounds Containing 1,3-Dicarbonyl Moiety: An Overview. Molecules 2023, 28, 6203. [Google Scholar] [CrossRef]
- Kunnumakkara, A.B.; Bordoloi, D.; Padmavathi, G.; Monisha, J.; Roy, N.K.; Prasad, S.; Aggarwal, B.B. Curcumin, the golden nutraceutical: Multitargeting for multiple chronic diseases. Br. J. Pharmacol. 2017, 174, 1325–1348. [Google Scholar] [CrossRef]
- Ghorbani, M.; Bigdeli, B.; Jalili-Baleh, L.; Baharifar, H.; Akrami, M.; Dehghani, S.; Goliaei, B.; Amani, A.; Lotfabadi, A.; Rashedi, H. Curcumin-lipoic acid conjugate as a promising anticancer agent on the surface of gold-iron oxide nanocomposites: A pH-sensitive targeted drug delivery system for brain cancer theranostics. Eur. J. Pharm. Sci. 2018, 114, 175–188. [Google Scholar] [CrossRef]
- Aquines, O.; Saavedra-Hernández, A.; Urbina-Arias, N.; Melchor-Martínez, E.M.; Sosa-Hernández, J.E.; Robledo-Padilla, F.; Iqbal, H.M.; Parra-Saldívar, R. In Silico Modeling Study of Curcumin Diffusion and Cellular Growth. Appl. Sci. 2022, 12, 9749. [Google Scholar] [CrossRef]
- Kang, C.; Wang, J.; Li, R.; Gong, J.; Wang, K.; Wang, Y.; Wang, Z.; He, R.; Li, F. Smart Targeted Delivery Systems for Enhancing Antitumor Therapy of Active Ingredients in Traditional Chinese Medicine. Molecules 2023, 28, 5955. [Google Scholar] [CrossRef]
- Somwar, R.; Perreault, M.; Kapur, S.; Taha, C.; Sweeney, G.; Ramlal, T.; Kim, D.Y.; Keen, J.; Côté, C.H.; Klip, A. Activation of p38 mitogen-activated protein kinase alpha and beta by insulin and contraction in rat skeletal muscle: Potential role in the stimulation of glucose transport. Diabetes 2000, 49, 1794–1800. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, J.M.; Kim, E.K.; Lee, J.O.; Lee, S.K.; Jung, J.H.; You, G.Y.; Park, S.H.; Suh, P.G.; Kim, H.S. Curcumin stimulates glucose uptake through AMPK-p38 MAPK pathways in L6 myotube cells. J. Cell. Physiol. 2010, 223, 771–778. [Google Scholar] [CrossRef]
- Fuentes, F.; Gomez, Y.; Paredes-Gonzalez, X.; Barve, A.; Nair, S.; Yu, S.; Saw, C.L.L.; Kong, A.N.T. Nrf2-mediated antioxidant and detoxifying enzyme induction by a combination of curcumin and sulforaphane. J. Chin. Pharm. Sci. 2016, 25, 559–569. [Google Scholar]
- Rijavec, T.; Zrimec, J.; van Spanning, R.; Lapanje, A. Natural microbial communities can be manipulated by artificially constructed biofilms. Adv. Sci. 2019, 6, 1901408. [Google Scholar] [CrossRef] [PubMed]
- Magacz, M.; Alatorre-Santamaría, S.; Kędziora, K.; Klasa, K.; Mamica, P.; Pepasińska, W.; Lebiecka, M.; Kościelniak, D.; Pamuła, E.; Krzyściak, W. Modified Lactoperoxidase System as a Promising Anticaries Agent: In Vitro Studies on Streptococcus mutans Biofilms. Int. J. Mol. Sci. 2023, 24, 12136. [Google Scholar] [CrossRef] [PubMed]
- Nappi, F. Host-Bacterium Interaction Mechanisms in Staphylococcus aureus Endocarditis: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 11068. [Google Scholar] [CrossRef]
- Sousa, I.S.; Mello, T.P.; Pereira, E.P.; Granato, M.Q.; Alviano, C.S.; Santos, A.L.S.; Kneipp, L.F. Biofilm Formation by Chromoblastomycosis Fungi Fonsecaea pedrosoi and Phialophora verrucosa: Involvement with Antifungal Resistance. J. Fungi 2022, 8, 963. [Google Scholar] [CrossRef]
- Suiker, I.M.; Wösten, H.A. Spoilage yeasts in beer and beer products. Curr. Opin. Food Sci. 2022, 44, 100815. [Google Scholar] [CrossRef]
- Lolis, N.; Veldekis, D.; Moraitou, H.; Kanavaki, S.; Velegraki, A.; Triandafyllidis, C.; Tasioudis, C.; Pefanis, A.; Pneumatikos, I. Saccharomyces boulardii fungaemia in an intensive care unit patient treated with caspofungin. Crit. Care 2008, 12, 414. [Google Scholar]
- Lherm, T.; Monet, C.; Nougière, B.; Soulier, M.; Larbi, D.; Le Gall, C.; Caen, D.; Malbrunot, C. Seven cases of fungemia with Saccharomyces boulardii in critically ill patients. Intensive Care Med. 2002, 28, 797–801. [Google Scholar] [CrossRef]
- Reynolds, T.B.; Fink, G.R. Bakers’ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, W.; Gao, Y.; Zha, M.; Zhang, D.; Peng, X.; Zhang, H.; Wang, C.; Xu, C.; Zhou, T. Engineering Saccharomyces cerevisiae for improved biofilm formation and ethanol production in continuous fermentation. Biotechnol. Biofuels Bioprod. 2023, 16, 119. [Google Scholar] [CrossRef]
- Teunissen, A.; Steensma, H. the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 1995, 11, 1001–1013. [Google Scholar] [CrossRef]
- Van Mulders, S.E.; Christianen, E.; Saerens, S.M.; Daenen, L.; Verbelen, P.J.; Willaert, R.; Verstrepen, K.J.; Delvaux, F.R. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 178–190. [Google Scholar] [CrossRef]
- Hou, D.; Xu, X.; Wang, J.; Liu, C.; Niu, C.; Zheng, F.; Li, Q. Effect of environmental stresses during fermentation on brewing yeast and exploration on the novel flocculation-associated function of RIM15 gene. Bioresour. Technol. 2023, 379, 129004. [Google Scholar] [CrossRef]
- Saguez, C.; Viterbo, D.; Descorps-Declère, S.; Cormack, B.P.; Dujon, B.; Richard, G.-F. Functional variability in adhesion and flocculation of yeast megasatellite genes. Genetics 2022, 221, iyac042. [Google Scholar] [CrossRef]
- Bouyx, C.; Schiavone, M.; Teste, M.-A.; Dague, E.; Sieczkowski, N.; Julien, A.; François, J.M. The dual role of amyloid-β-sheet sequences in the cell surface properties of FLO11-encoded flocculins in Saccharomyces cerevisiae. Elife 2021, 10, e68592. [Google Scholar] [CrossRef]
- Bouyx, C.; Schiavone, M.; François, J.M. FLO11, a developmental gene conferring impressive adaptive plasticity to the yeast Saccharomyces cerevisiae. Pathogens 2021, 10, 1509. [Google Scholar] [CrossRef]
- Collins, J.H.; Kunyeit, L.; Weintraub, S.; Sharma, N.; White, C.; Haq, N.; Anu-Appaiah, K.; Rao, R.P.; Young, E.M. Genetic basis for probiotic yeast phenotypes revealed by nanopore sequencing. G3 Genes Genomes Genet. 2023, 13, jkad093. [Google Scholar] [CrossRef]
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A Saccharomyces gene family involved in invasive growth, cell–cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, L.; Schulze, F.; Braus, G.H. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell–cell and cell–substrate adherence of S. cerevisiae S288c. Mol. Microbiol. 2007, 66, 1276–1289. [Google Scholar] [CrossRef]
- Perpetuini, G.; Battistelli, N.; Tittarelli, F.; Suzzi, G.; Tofalo, R. Influence of FLO1 and FLO5 genes on aroma profile of sparkling wines. LWT 2021, 146, 111407. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Khoury, Z.H.; Vila, T.; Puthran, T.R.; Sultan, A.S.; Montelongo-Jauregui, D.; Melo, M.A.S.; Jabra-Rizk, M.A. The role of Candida albicans secreted polysaccharides in augmenting Streptococcus mutans adherence and mixed biofilm formation: In vitro and in vivo studies. Front. Microbiol. 2020, 11, 307. [Google Scholar] [CrossRef]
- Alalwan, H.; Rajendran, R.; Lappin, D.F.; Combet, E.; Shahzad, M.; Robertson, D.; Nile, C.J.; Williams, C.; Ramage, G. The anti-adhesive effect of curcumin on Candida albicans biofilms on denture materials. Front. Microbiol. 2017, 8, 659. [Google Scholar] [CrossRef]
- Mohammadi, K.; Saris, P.E.J. Biofilm Formation of Probiotic Saccharomyces cerevisiae var. boulardii on Glass Surface during Beer Bottle Ageing. Beverages 2022, 8, 77. [Google Scholar]
- Kovač, B.; Piletić, K.; Kovačević Ganić, N.; Gobin, I. The Effectiveness of Benzalkonium Chloride as an Active Compound on Selected Foodborne Pathogens Biofilm. Hygiene 2022, 2, 226–235. [Google Scholar] [CrossRef]
- Mello, T.P.; Aor, A.C.; Gonçalves, D.S.; Seabra, S.H.; Branquinha, M.H.; Santos, A.L. Assessment of biofilm formation by Scedosporium apiospermum, S. aurantiacum, S. minutisporum and Lomentospora prolificans. Biofouling 2016, 32, 737–749. [Google Scholar] [CrossRef]
- Kesavelu Sr, D.; Rohit, A.; Karunasagar, I.; Karunasagar, I. Composition and laboratory correlation of commercial probiotics in India. Cureus 2020, 12, e11334. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, W.; Lu, J.; Zhao, M. Phenolic profiles and antioxidant activities of commercial beers. Food Chem. 2010, 119, 1150–1158. [Google Scholar] [CrossRef]
- Opalek, M.; Wloch-Salamon, D. Aspects of multicellularity in Saccharomyces cerevisiae yeast: A review of evolutionary and physiological mechanisms. Genes 2020, 11, 690. [Google Scholar] [CrossRef]
- Willaert, R.G.; Kayacan, Y.; Devreese, B. The Flo adhesin family. Pathogens 2021, 10, 1397. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; García, R.; Pavón-Vergés, M.; Rodríguez-Peña, J.M.; Arroyo, J. Control of gene expression via the yeast CWI pathway. Int. J. Mol. Sci. 2022, 23, 1791. [Google Scholar] [CrossRef]
- Speranza, B.; Corbo, M.R.; Campaniello, D.; Altieri, C.; Sinigaglia, M.; Bevilacqua, A. Biofilm formation by potentially probiotic Saccharomyces cerevisiae strains. Food Microbiol. 2020, 87, 103393. [Google Scholar] [CrossRef]
- Nishimura, A.; Nakagami, K.; Kan, K.; Morita, F.; Takagi, H. Arginine inhibits Saccharomyces cerevisiae biofilm formation by inducing endocytosis of the arginine transporter Can1. Biosci. Biotechnol. Biochem. 2022, 86, 1300–1307. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, F.; Yu, Y.; Ding, S.; Chen, T.; Sun, W.; Liang, C.; Yu, B.; Ying, H.; Liu, D. Effect of quorum-sensing molecule 2-phenylethanol and ARO genes on Saccharomyces cerevisiae biofilm. Appl. Microbiol. Biotechnol. 2021, 105, 3635–3648. [Google Scholar] [CrossRef]
- Azad, G.K.; Singh, V.; Golla, U.; Tomar, R.S. Depletion of cellular iron by curcumin leads to alteration in histone acetylation and degradation of Sml1p in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e59003. [Google Scholar] [CrossRef]
- Zia, A.; Farkhondeh, T.; Pourbagher-Shahri, A.M.; Samarghandian, S. The role of curcumin in aging and senescence: Molecular mechanisms. Biomed. Pharmacother. 2021, 134, 111119. [Google Scholar] [CrossRef]
- Minear, S.; O’Donnell, A.F.; Ballew, A.; Giaever, G.; Nislow, C.; Stearns, T.; Cyert, M.S. Curcumin inhibits growth of Saccharomyces cerevisiae through iron chelation. Eukaryot. Cell 2011, 10, 1574–1581. [Google Scholar] [CrossRef]
- Deepa, A.; Naveena, K.; Anindya, R. DNA repair activity of Fe (II)/2OG-dependent dioxygenases affected by low iron level in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, foy014. [Google Scholar] [CrossRef]
- Stępień, K.; Wojdyła, D.; Nowak, K.; Mołoń, M. Impact of curcumin on replicative and chronological aging in the Saccharomyces cerevisiae yeast. Biogerontology 2020, 21, 109–123. [Google Scholar] [CrossRef]
- López-Balladares, O.; Espinoza-Montero, P.J.; Fernández, L. Electrochemical Evaluation of Cd, Cu, and Fe in Different Brands of Craft Beers from Quito, Ecuador. Foods 2023, 12, 2264. [Google Scholar] [CrossRef]
- Silva, S.; Oliveira, A.I.; Cruz, A.; Oliveira, R.F.; Almeida, R.; Pinho, C. Physicochemical Properties and Antioxidant Activity of Portuguese Craft Beers and Raw Materials. Molecules 2022, 27, 8007. [Google Scholar] [CrossRef] [PubMed]
- Echave, J.; Barral, M.; Fraga-Corral, M.; Prieto, M.A.; Simal-Gandara, J. Bottle aging and storage of wines: A review. Molecules 2021, 26, 713. [Google Scholar] [CrossRef]
- Paramera, E.I.; Konteles, S.J.; Karathanos, V.T. Microencapsulation of curcumin in cells of Saccharomyces cerevisiae. Food Chem. 2011, 125, 892–902. [Google Scholar] [CrossRef]
- Paramera, E.I.; Konteles, S.J.; Karathanos, V.T. Stability and release properties of curcumin encapsulated in Saccharomyces cerevisiae, β-cyclodextrin and modified starch. Food Chem. 2011, 125, 913–922. [Google Scholar] [CrossRef]
- Beauvais, A.; Loussert, C.; Prevost, M.C.; Verstrepen, K.; Latgé, J.P. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 2009, 9, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Canetta, E.; Adya, A.K.; Walker, G.M. Atomic force microscopic study of the effects of ethanol on yeast cell surface morphology. FEMS Microbiol. Lett. 2006, 255, 308–315. [Google Scholar] [CrossRef]
- Ma, M.; Han, P.; Zhang, R.; Li, H. Ultrastructural changes of Saccharomyces cerevisiae in response to ethanol stress. Can. J. Microbiol. 2013, 59, 589–597. [Google Scholar] [CrossRef]
- Dvořáková, M.; Douanier, M.; Jurková, M.; Kellner, V.; Dostálek, P. Comparison of antioxidant activity of barley (Hordeum vulgare L.) and malt extracts with the content of free phenolic compounds measured by high performance liquid chromatography coupled with CoulArray detector. J. Inst. Brew. 2008, 114, 150–159. [Google Scholar] [CrossRef]
- Krofta, K.; Mikyška, A.; Hašková, D. Antioxidant characteristics of hops and hop products. J. Inst. Brew. 2008, 114, 160–166. [Google Scholar] [CrossRef]
- Piazzon, A.; Forte, M.; Nardini, M. Characterization of phenolics content and antioxidant activity of different beer types. J. Agric. Food Chem. 2010, 58, 10677–10683. [Google Scholar] [CrossRef]
- Ulloa, P.A.; Vidal, J.; Ávila, M.I.; Labbe, M.; Cohen, S.; Salazar, F.N. Effect of the addition of propolis extract on bioactive compounds and antioxidant activity of craft beer. J. Chem. 2017, 2017, 6716053. [Google Scholar] [CrossRef]
- Serea, D.; Horincar, G.; Constantin, O.E.; Aprodu, I.; Stănciuc, N.; Bahrim, G.E.; Stanciu, S.; Rapeanu, G. Value-Added White Beer: Influence of Red Grape Skin Extract on the Chemical Composition, Sensory and Antioxidant Properties. Sustainability 2022, 14, 9040. [Google Scholar] [CrossRef]


| Gene | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| FLO1 | ACAGAGACAACAAAGCAAACC | ACACACACCAGATTCGCAG |
| FLO5 | ACCCCAACAAACGTAACCCT | GGTGCTAGCTGTTGTTGGAG |
| FLO8 | CGTATCCAGATTCAATTCCTCC | GCTGTTCACTATTCGTTGCC |
| FLO9 | ACAACAGAGCAAACCACAG | ACCGTAACAACATCATTCACAG |
| FLO10 | CTACACAACACCCACCAAC | AAGGCAACATTACCTCCAAC |
| FLO11 | AACCAAGTCCATCCCAACC | GCGAGTAGCAACCACATAAAG |
| 18S | TTAATGACCCACTCGGCAC | CACCACCACCCACAAAATC |
| Fermentation | Bottle Aging | |||
|---|---|---|---|---|
| Parameter | Control | B/CUR | Control | B/CUR |
| pH | 4.57 a | 4.74 b | 4.52 a | 4.32 c |
| TPC (mg GAE/L) | 229 ± 18 a | 283 ± 22 b | 209 ± 15 a | 261 ± 24 b |
| TFC (mg QE/L) | 15.2 ± 2.6 a | 23.6 ± 2 b | 12.4 ± 3.3 a | 19.1 ± 2.5 b |
| Attributes | B/CUR | Control |
|---|---|---|
| Appearance | 8.3 ± 0.48 a | 7.4 ± 0.48 b |
| Aroma | 7.0 ± 0.90 a | 7.1 ± 0.67 a |
| Taste | 7.7 ± 0.76 a | 7.1 ± 0.83 b |
| Mouthfeel | 6.8 ± 0.85 a | 6.5 ± 0.61 a |
| Aftertaste and finish | 7.3 ± 0.89 a | 6.61 ± 0.77 b |
| Overall impression | 8.1 ± 0.67 a | 7.6 ± 0.69 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi, K.; Saris, P.E.J. Antibiofilm Effect of Curcumin on Saccharomyces boulardii during Beer Fermentation and Bottle Aging. Biomolecules 2023, 13, 1367. https://doi.org/10.3390/biom13091367
Mohammadi K, Saris PEJ. Antibiofilm Effect of Curcumin on Saccharomyces boulardii during Beer Fermentation and Bottle Aging. Biomolecules. 2023; 13(9):1367. https://doi.org/10.3390/biom13091367
Chicago/Turabian StyleMohammadi, Khosrow, and Per Erik Joakim Saris. 2023. "Antibiofilm Effect of Curcumin on Saccharomyces boulardii during Beer Fermentation and Bottle Aging" Biomolecules 13, no. 9: 1367. https://doi.org/10.3390/biom13091367
APA StyleMohammadi, K., & Saris, P. E. J. (2023). Antibiofilm Effect of Curcumin on Saccharomyces boulardii during Beer Fermentation and Bottle Aging. Biomolecules, 13(9), 1367. https://doi.org/10.3390/biom13091367






