Biomedical Applications of Biomolecules Isolated from Methanotrophic Bacteria in Wastewater Treatment Systems
Abstract
:1. Introduction
2. Methanotrophic Bacteria in Water Treatment Systems
2.1. Taxonomy and Phenotype
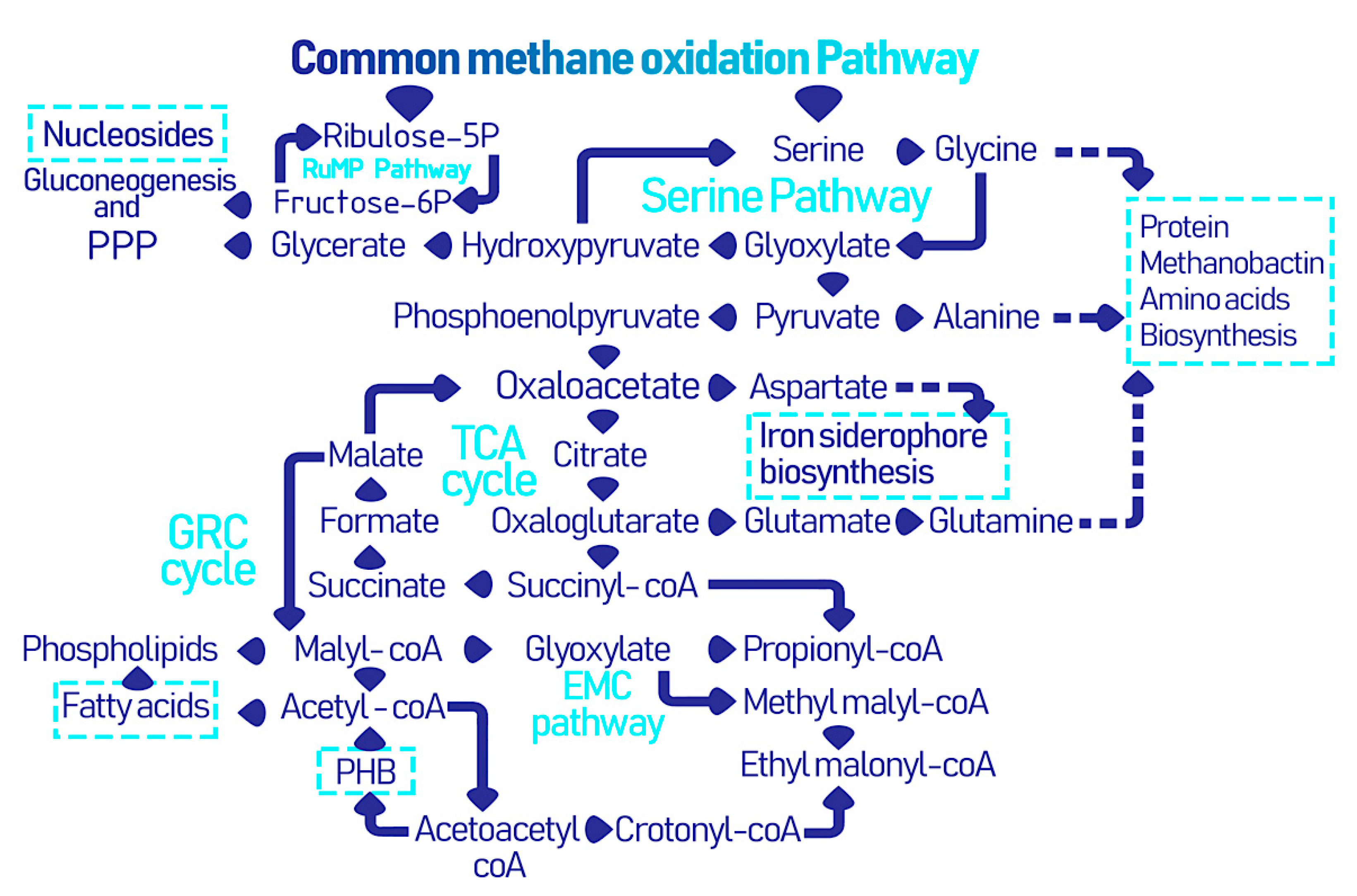
2.2. EcoPhysiology
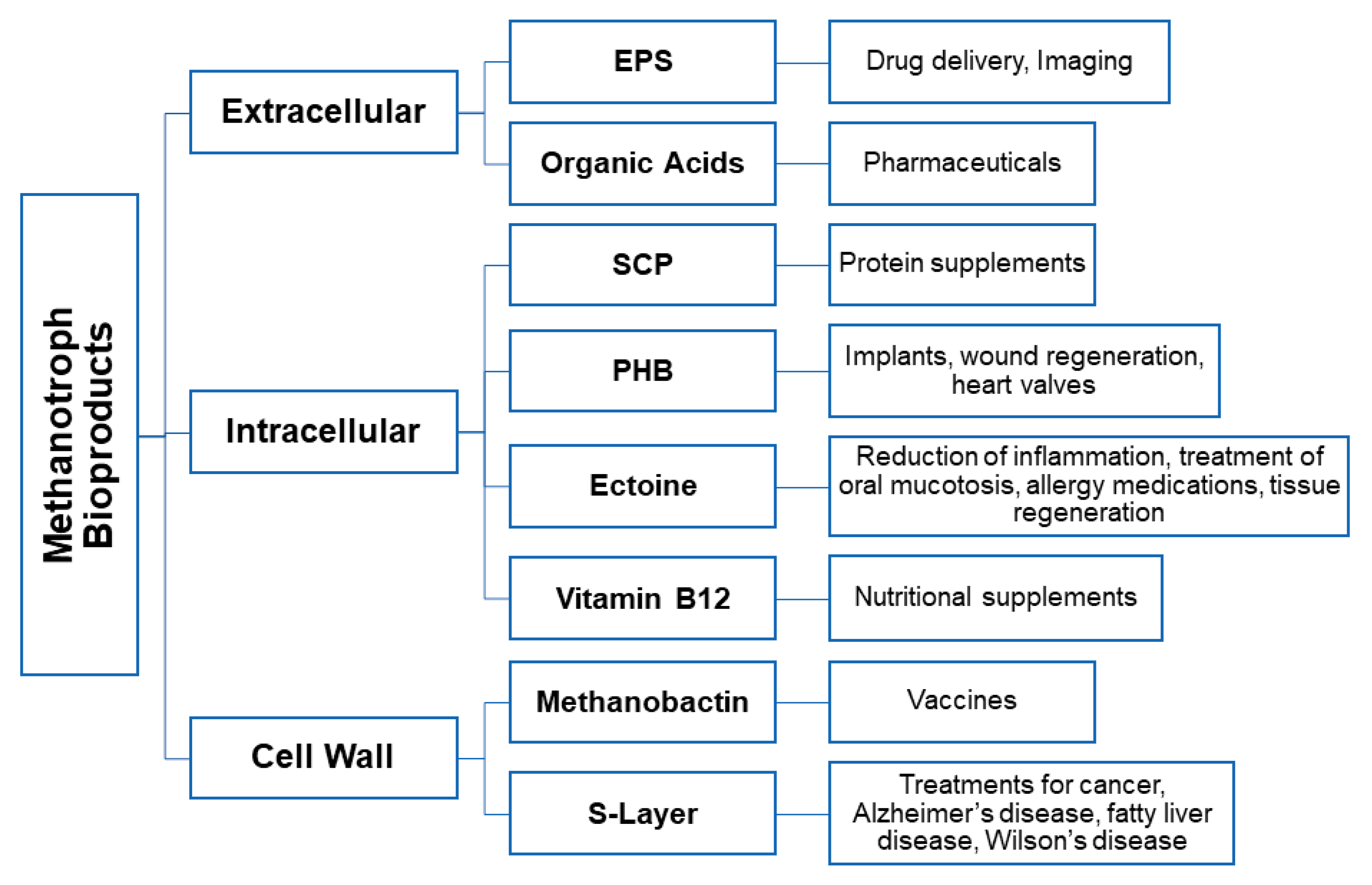
3. Microbially Recovered Resources from Methanotrophic Bacteria and Their Biomedical Applications
3.1. Exopolysaccharides
3.2. Polyhydroxyalkanoate
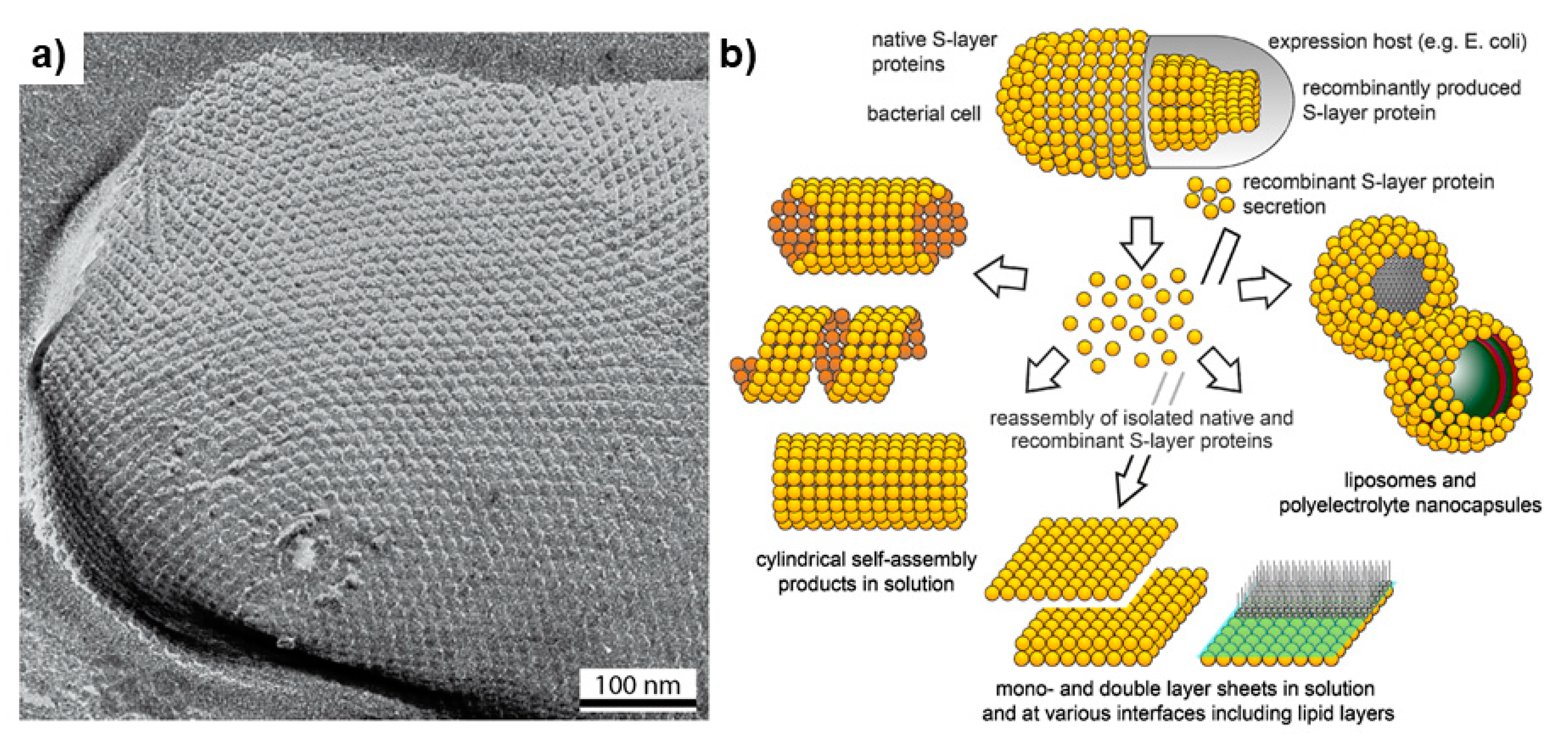
3.3. Surface Layers
3.4. Methanobactin
3.5. Antibacterial Proteins
3.6. Single Cell Protein (SCP)
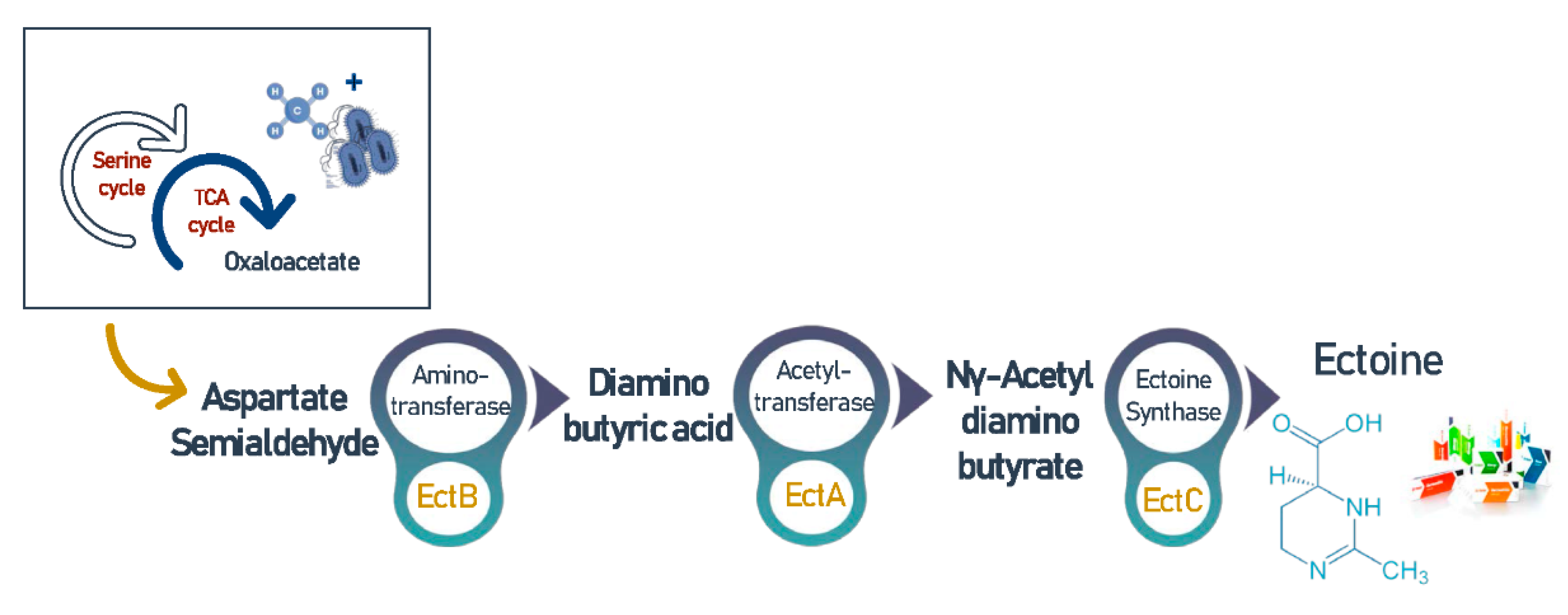
3.7. Ectoine
3.8. Carotenoids
4. Outlook and Practical Implications
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kohane, D.S.; Langer, R. Polymeric biomaterials in tissue engineering. Pediatr. Res. 2008, 63, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Doltra, A.; Dietrich, T.; Schneeweis, C.; Kelle, S.; Doltra, A.; Stawowy, P.; Fleck, E. Magnetic Resonance Imaging of Cardiovascular Fibrosis and Inflammation: From Clinical Practice to Animal Studies and Back. BioMed Res. Int. 2013, 2013, 676489. [Google Scholar] [CrossRef]
- Vaishnav, P.; Demain, A.L. Unexpected applications of secondary metabolites. Biotechnol. Adv. 2011, 29, 223–229. [Google Scholar] [CrossRef]
- Chin, Y.-W.; Balunas, M.J.; Chai, H.B.; Kinghorn, A.D. Drug discovery from natural sources. AAPS J. 2006, 8, E239–E253. [Google Scholar] [CrossRef] [Green Version]
- Subbarayappa, B.V. The roots of ancient medicine: An historical outline. J. Biosci. 2001, 26, 135–143. [Google Scholar] [CrossRef]
- Singh, B.P.; Rateb, M.E.; Rodriguez-Couto, S.; Polizeli, M. de L.T. de M.; Li, W.-J. Editorial: Microbial Secondary Metabolites: Recent Developments and Technological Challenges. Front. Microbiol. 2019, 10, 914. [Google Scholar] [CrossRef]
- Williams, S.F.; Martin, D.P. Applications of Polyhydroxyalkanoates (PHA) in Medicine and Pharmacy. Biopolym. Online 2005. [Google Scholar] [CrossRef]
- Narsing Rao, M.P.; Xiao, M.; Li, W.J. Fungal and bacterial pigments: Secondary metabolites with wide applications. Front. Microbiol. 2017, 8, 1113. [Google Scholar] [CrossRef]
- Baile, P.; Vidal, L.; Canals, A. A modified zeolite/iron oxide composite as a sorbent for magnetic dispersive solid-phase extraction for the preconcentration of nonsteroidal anti-inflammatory drugs in water and urine samples. J. Chromatogr. A 2019, 1603, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Sánchez, M.; Rocha, D.; Sánchez, B.; Rodríguez-Sanoja, R.; Sánchez, S.; et al. Production of microbial secondary metabolites: Regulation by the carbon source. Crit. Rev. Microbiol. 2010, 36, 146–167. [Google Scholar] [CrossRef]
- Anderson, A.J.; Dawes, E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990, 54, 450–472. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wright, G.D. An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 2011, 22, 552–558. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Prakash, V.; Ranjan, N. Marine fungi: A source of potential anticancer compounds. Front. Microbiol. 2018, 8, 2536. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Nomura, C.T.; Perrotta, J.A.; Stipanovic, A.J.; Nakas, J.P. Production and characterization of poly-3-hydroxybutyrate from biodiesel-glycerol by Burkholderia cepacia ATCC 17759. Biotechnol. Prog. 2010, 26, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y. Methanotrophs: Microbiology Fundamentals and Biotechnological Applications; Lee, E.Y., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; ISBN 9783030232603. [Google Scholar]
- Valappil, S.P.; Misra, S.K.; Boccaccini, A.; Roy, I. Biomedical applications of polyhydroxyalkanoates, an overview of animal testing and in vivo responses. Expert Rev. Med. Devices 2006, 3, 853–868. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.; Rämä, T.; Oosterhuis, R.; Walker, A.; Pang, K.-L. The Neglected Marine Fungi, Sensu stricto, and Their Isolation for Natural Products’ Discovery. Mar. Drugs 2019, 17, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puyol, D.; Batstone, D.J.; Hülsen, T.; Astals, S.; Peces, M.; Krömer, J.O. Resource recovery from wastewater by biological technologies: Opportunities, challenges, and prospects. Front. Microbiol. 2017, 7, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Kehrein, P.; van Loosdrecht, M.; Osseweijer, P.; Garfí, M.; Dewulf, J.; Posada, J. A critical review of resource recovery from municipal wastewater treatment plants—market supply potentials, technologies and bottlenecks. Environ. Sci. Water Res. Technol. 2020, 6, 877–910. [Google Scholar] [CrossRef] [Green Version]
- Numberger, D.; Ganzert, L.; Zoccarato, L.; Mühldorfer, K.; Sauer, S.; Grossart, H.P.; Greenwood, A.D. Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Osunmakinde, C.O.; Selvarajan, R.; Mamba, B.B.; Msagati, T.A.M. Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis. Microorganisms 2019, 7, 506. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, F.; Tsunogai, U.; Suzuki, M.; Kosaka, A.; Machiyama, H.; Takai, K.; Nunoura, T.; Nealson, K.H.; Horikoshi, K. Characterization of C 1 -Metabolizing Prokaryotic Communities in Methane Seep Habitats at the Kuroshima Knoll, Southern 16S rRNA Genes. Appl. Environ. Microbiol. 2004, 70, 7445–7455. [Google Scholar] [CrossRef] [Green Version]
- Xin, J.; Zhang, Y.; Zhang, S.; Xia, C.; Li, S. Methanol production from CO2 by resting cells of the methanotrophic bacterium Methylosinus trichosporium IMV 3011. J. Basic Microbiol. 2007, 47, 426–435. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef]
- Vaksmaa, A.; Guerrero-Cruz, S.; van Alen, T.A.; Cremers, G.; Ettwig, K.F.; Lüke, C.; Jetten, M.S.M. Enrichment of anaerobic nitrate-dependent methanotrophic ‘Candidatus Methanoperedens nitroreducens’ archaea from an Italian paddy field soil. Appl. Microbiol. Biotechnol. 2017, 101, 7075–7084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Chen, Y.; Jiang, P.; Zhang, C.; Smith, T.J.; Murrell, J.C.; Xing, X.-H. Methanotrophs: Multifunctional bacteria with promising applications in environmental bioengineering. Biochem. Eng. J. 2010, 49, 277–288. [Google Scholar] [CrossRef]
- Yang, S.; Matsen, J.B.; Konopka, M.; Green-Saxena, A.; Clubb, J.; Sadilek, M.; Orphan, V.J.; Beck, D.; Kalyuzhnaya, M.G. Global Molecular Analyses of Methane Metabolism in Methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part II. Metabolomics and 13C-Labeling Study. Front. Microbiol. 2013, 4, 70. [Google Scholar] [CrossRef] [Green Version]
- Whittenbury, R.; Dalton, H. The Methylotrophic Bacteria. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin, Heidelberg, 1981; pp. 894–902. ISBN 978-3-662-13187-9. [Google Scholar]
- Kalyuzhnaya, M.G.; Khmelenina, V.N.; Kotelnikova, S.; Holmquist, L.; Pedersen, K.; Trotsenko, Y.A. Methylomonas scandinavica sp, nov., a new methanotrophic psychrotrophic bacterium isolated from deep igneous rock ground water of Sweden. Syst. Appl. Microbiol. 1999, 22, 565–572. [Google Scholar] [CrossRef]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strong, P.J.; Xie, S.; Clarke, W.P. Methane as a resource: Can the methanotrophs add value? Environ. Sci. Technol. 2015, 49, 4001–4018. [Google Scholar] [CrossRef] [PubMed]
- AlSayed, A.; Fergala, A.; Eldyasti, A. Sustainable biogas mitigation and value-added resources recovery using methanotrophs intergrated into wastewater treatment plants. Rev. Environ. Sci. Biotechnol. 2018, 17, 351–393. [Google Scholar] [CrossRef]
- Mahmoud, A.M.A. Biological Conversion Process of Methane into Methanol Using Mixed Culture Methanotrophic Bacteria Enriched from Activated Sludge System; York University: Toronto, ON, Canada, 2017. [Google Scholar]
- Forster, D.; Dolan, J.R.; Dunthorn, M.; Bass, D.; Bittner, L.; Boutte, C.; Christen, R.; Claverie, J.; Decelle, J.; Edvardsen, B.; et al. Benthic protists: The under-charted majority. FEMS Microbiol. Ecol. 2016, 92, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kevbrina, M.V.; Okhapkina, A.A.; Akhlynin, D.S.; Kravchenko, I.K.; Nozhevnikova, A.N.; Gal’chenko, V.F.; Gal’chenko, V.F. Growth of Mesophilic Methanotrophs at Low Temperatures. Microbiology 2001, 70, 384–391. [Google Scholar] [CrossRef]
- Liebner, S.; Wagner, D. Abundance, distribution and potential activity of methane oxidizing bacteria in permafrost soils from the Lena Delta, Siberia. Environ. Microbiol. 2007, 9, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Wagner, D.; Lipski, A.; Embacher, A.; Gattinger, A. Methane fluxes in permafrost habitats of the Lena Delta: Effects of microbial community structure and organic matter quality. Environ. Microbiol. 2005, 7, 1582–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wartiainen, I.; Hestnes, A.G.; McDonald, I.R.; Svenning, M.M. Methylobacter tundripaludum sp. nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard islands, Norway (78° N). Int. J. Syst. Evol. Microbiol. 2006, 56, 109–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, M.O.; Rosenzweig, A.C. A tale of two methane monooxygenases. J. Biol. Inorg. Chem. 2017, 22, 307–319. [Google Scholar] [CrossRef]
- Korotkova, N.; Lidstrom, M.E.; Chistoserdova, L. Identification of genes involved in the glyoxylate regeneration cycle in Methylobacterium extorquens AM1, including two new genes, meaC and meaD. J. Bacteriol. 2005, 187, 1523–1526. [Google Scholar] [CrossRef] [Green Version]
- Borjesson, G.; Sundh, I.; Svensson, B. Microbial oxidation of CH4 at different temperatures in landfill cover soils. FEMS Microbiol. Ecol. 2004, 48, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Good, N.M.; Hu, B.; Yang, J.; Wang, Q.; Sadilek, M.; Yang, S. Metabolomics Revealed an Association of Metabolite Changes and Defective Growth in Methylobacterium extorquens AM1 Overexpressing ecm during Growth on Methanol. PLoS ONE 2016, 11, e0154043. [Google Scholar] [CrossRef]
- Korotkova, N.; Lidstrom, M.E. Connection between poly-β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 2001, 183, 1038–1046. [Google Scholar] [CrossRef] [Green Version]
- Babel, W. Pecularities of methylotrophs concerning overflow metabolism, especially the synthesis of polyhydroxyalkanoates. FEMS Microbiol. Lett. 1992, 103, 141–148. [Google Scholar] [CrossRef]
- Chiemchaisri, W.; Wu, J.S.; Visvanathan, C. Methanotrophic production of extracellular polysaccharide in landfill cover soils. Water Sci. Technol. 2001, 43, 151–158. [Google Scholar] [CrossRef]
- Malashenko, Y.R.; Pirog, T.P.; Romanovskaya, V.A.; Sokolov, I.G.; Grinberg, T.A. Search for methanotrophic producers of exopolysaccharides. Appl. Biochem. Microbiol. 2001, 37, 599–602. [Google Scholar] [CrossRef]
- Wilshusen, J.H.; Hettiaratchi, J.P.A.; De Visscher, A.; Saint-Fort, R. Methane oxidation and formation of EPS in compost: Effect of oxygen concentration. Environ. Pollut. 2004, 129, 305–314. [Google Scholar] [CrossRef]
- Wang, J.; Salem, D.R.; Sani, R.K. Microbial polymers produced from methane: Overview of recent progress and new perspectives. In Microbial and Natural Macromolecules; Elsevier: Amsterdam, The Netherlands, 2021; pp. 117–142. [Google Scholar]
- Nwodo, U.U.; Green, E.; Okoh, A.I. Bacterial exopolysaccharides: Functionality and prospects. Int. J. Mol. Sci. 2012, 13, 14002–14015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.-C.; Wingender, J. Extracellular Polymeric Substances (EPS): Structural, Ecological and Technical Aspects. In Encyclopedia of Environmental Microbiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2003. [Google Scholar]
- Moscovici, M. Present and future medical applications of microbial exopolysaccharides. Front. Microbiol. 2015, 6, 1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fialho, A.M.; Moreira, L.M.; Granja, A.T.; Popescu, A.O.; Hoffmann, K.; Sá-Correia, I. Occurrence, production, and applications of gellan: Current state and perspectives. Appl. Microbiol. Biotechnol. 2008, 79, 889–900. [Google Scholar] [CrossRef]
- Lee, S.Y. Bacterial Polyhydroxyalkanoates. Biotechnol. Bioeng. 1996, 49, 1–14. [Google Scholar] [CrossRef]
- Madison, L.L.; Huisman, G.W. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol. Mol. Biol. Rev. 1999, 63, 21–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preusting, H.; Nijenhuis, A.; Witholt, B. Physical Characteristics of Poly(3-hydroxyalkanoates) and Poly(3-hydroxyalkenoates) Produced by Pseudomonas Oleovorans Grown on Aliphatic Hydrocarbons. Macromolecules 1990, 23, 4220–4224. [Google Scholar] [CrossRef]
- Yu, J. Production of PHA from starchy wastewater via organic acids. J. Biotechnol. 2001, 86, 105–112. [Google Scholar] [CrossRef]
- Misra, S.K.; Valappil, S.P.; Roy, I.; Boccaccini, A.R. Polyhydroxyalkanoate (PHA)/Inorganic Phase Composites for Tissue Engineering Applications. Biomacromolecules 2006, 7, 2249–2258. [Google Scholar] [CrossRef]
- Zhang, J.; Shishatskaya, E.I.; Volova, T.G.; da Silva, L.F.; Chen, G.Q. Polyhydroxyalkanoates (PHA) for therapeutic applications. Mater. Sci. Eng. C 2018, 86, 144–150. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Shao, W.; Jin, S.; Xu, T.; Jiang, X.; Yang, S.; Wang, Z.; Dai, J.; Wu, Q. Microgrooved poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) affects the phenotype of vascular smooth muscle cells through let-7a-involved regulation of actin dynamics. Biotechnol. Lett. 2014, 36, 2125–2133. [Google Scholar] [CrossRef]
- Wu, S.; Liu, Y.-L.; Cui, B.; Qu, X.-H.; Chen, G.-Q. Study on Decellularized Porcine Aortic Valve/Poly (3-hydroxybutyrate-co-3-hydroxyhexanoate) Hybrid Heart Valve in Sheep Model. Artif. Organs 2007, 31, 689–697. [Google Scholar] [CrossRef]
- Sodian, R.; Sperling, J.S.; Martin, D.P.; Egozy, A.; Stock, U.; Mayer, J.E.; Vacanti, J.P. Technical Report: Fabrication of a Trileaflet Heart Valve Scaffold from a Polyhydroxyalkanoate Biopolyester for Use in Tissue Engineering. Tissue Eng. 2000, 6, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Karahaliloglu, Z.; Ercan, B.; Taylor, E.N.; Chung, S.; Denkbąs, E.B.; Webster, T.J. Antibacterial nanostructured polyhydroxybutyrate membranes for guided bone regeneration. J. Biomed. Nanotechnol. 2015, 11, 2253–2263. [Google Scholar] [CrossRef]
- Gredes, T.; Gedrange, T.; Hinüber, C.; Gelinsky, M.; Kunert-Keil, C. Histological and molecular-biological analyses of poly(3-hydroxybutyrate) (PHB) patches for enhancement of bone regeneration. Ann. Anat. 2015, 199, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Strong, P.J.; Kalyuzhnaya, M.; Silverman, J.; Clarke, W.P. A methanotroph-based biorefinery: Potential scenarios for generating multiple products from a single fermentation. Bioresour. Technol. 2016, 215, 314–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.M.; Pum, D. S-layers: Principles and applications. FEMS Microbiol. Rev. 2014, 38, 823–864. [Google Scholar] [CrossRef]
- Koller, M. Biodegradable and biocompatible polyhydroxy-alkanoates (PHA): Auspicious microbial macromolecules for pharmaceutical and therapeutic applications. Molecules 2018, 23, 362. [Google Scholar] [CrossRef] [Green Version]
- Sára, M.; Sleytr, U.B. S-layer proteins. J. Bacteriol. 2000, 182, 859–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A. Surface layers of methanotrophic bacteria. Microbiol. (Russian Fed.) 2013, 82, 529–541. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Bayley, H.; Sára, M.; Breitwieser, A.; Küpcü, S.; Mader, C.; Weigert, S.; Unger, F.M.; Messner, P.; Jahn-Schmid, B.; et al. VI. Applications of S-layers. FEMS Microbiol. Rev. 1997, 20, 151–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orata, F.D.; Meier-Kolthoff, J.P.; Sauvageau, D.; Stein, L.Y. Phylogenomic analysis of the gammaproteobacterial methanotrophs (order methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 2018, 9, 3162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darshan, M.; Nishith, D. Recovery and characterization of poly(3-Hydroxybutyric acid) synthesized in Staphylococcus epidermidis. Afr. J. Environ. Sci. Technol. 2014, 8, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Sleytr, U.B.; Messner, P.; Pum, D.; Sára, M. Crystalline bacterial cell surface layers. Mol. Microbiol. 1993, 10, 911–916. [Google Scholar] [CrossRef]
- Jahn-Schmid, B.; Graninger, M.; Glozik, M.; Küpcü, S.; Ebner, C.; Unger, F.M.; Sleytr, U.B.; Messner, P. Immunoreactivity of allergen (Bet v 1) conjugated to crystalline bacterial cell surface layers (S-layers). Immunotechnology 1996, 2, 103–113. [Google Scholar] [CrossRef]
- Volkel, D.; Zimmermann, K.; Breitwieser, A.; Pable, S.; Glatzel, M.; Scheiflinger, F.; Schwarz, H.P.; Sara, M.; Sleytr, U.B.; Dorner, F. Immunochemical detection of prion protein on dipsticks prepared with crystalline bacterial cell-surface layers. Transfusion 2003, 43, 1677–1682. [Google Scholar] [CrossRef]
- Breitwieser, A.; Mader, C.; Schocher, I.; Hoffmann-Sommergruber, K.; Aberer, W.; Scheiner, O.; Sleytr, U.B.; Sára, M. A novel dipstick developed for rapid Bet V 1-specific IgE detection: Recombinant allergen immobilized via a monoclonal antibody to crystalline bacterial cell-surface layers. Allergy 2008, 53, 786–793. [Google Scholar] [CrossRef]
- Badelt-Lichtblau, H.; Kainz, B.; Völlenkle, C.; Egelseer, E.-M.; Sleytr, U.B.; Pum, D.; Ilk, N. Genetic Engineering of the S-Layer Protein SbpA of Lysinibacillus sphaericus CCM 2177 for the Generation of Functionalized Nanoarrays. Bioconjug. Chem. 2009, 20, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Panwar, R.; Kumar, N.; Kashyap, V.; Singh, S.; Singh, H. Insights into Involvement of S-Layer Proteins of Probiotic Lactobacilli in Relation to Gut Health. Octa J. Environ. Res. 2017, 5, 228–245. [Google Scholar]
- Pum, D.; Toca-Herrera, J.L.; Sleytr, U.B. S-Layer protein self-assembly. Int. J. Mol. Sci. 2013, 14, 2484–2501. [Google Scholar] [CrossRef] [PubMed]
- Lonappan, L.; Brar, S.K.; Das, R.K.; Verma, M.; Surampalli, R.Y. Diclofenac and its transformation products: Environmental occurrence and toxicity—A review. Environ. Int. 2016, 96, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Semrau, J.D.; DiSpirito, A.A. Methanobactin: A Novel Copper-Binding Compound Produced by Methanotrophs. In Methanotrophs; Lee, E.Y., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; pp. 205–229. [Google Scholar]
- Kenney, G.E.; Rosenzweig, A.C. Chalkophores. Annu. Rev. Biochem. 2018, 87, 645–676. [Google Scholar] [CrossRef]
- Kenney, G.E.; Dassama, L.M.K.; Pandelia, M.E.; Gizzi, A.S.; Martinie, R.J.; Gao, P.; DeHart, C.J.; Schachner, L.F.; Skinner, O.S.; Ro, S.Y.; et al. The biosynthesis of methanobactin. Science (80-.) 2018, 359, 1411–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiSpirito, A.A.; Semrau, J.D.; Murrell, J.C.; Gallagher, W.H.; Dennison, C.; Vuilleumier, S. Methanobactin and the Link between Copper and Bacterial Methane Oxidation. Microbiol. Mol. Biol. Rev. 2016, 80, 387–409. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, R.; Rosenzweig, A.C. Copper methanobactin: A molecule whose time has come. Curr. Opin. Chem. Biol. 2008, 12, 245–249. [Google Scholar] [CrossRef] [Green Version]
- Dassama, L.M.K.; Kenney, G.E.; Rosenzweig, A.C. Methanobactins: From genome to function. Metallomics 2017, 9, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Semrau, J.D.; DiSpirito, A.A.; Obulisamy, P.K.; Kang-Yun, C.S. Methanobactin from methanotrophs: Genetics, structure, function and potential applications. FEMS Microbiol. Lett. 2020, 367, fnaa045. [Google Scholar] [CrossRef]
- Pashkova, N.I.; Starostina, N.G.; Tsiomenko, A.B. A secretory protein involved in the antagonistic interactions between methanotrophic bacteria. Biochemistry 1997, 62, 386–390. [Google Scholar] [PubMed]
- Starostina, N.G.; Pashkova, N.I.; Tsiomenko, A.B. Detection and partial characterization of bacteriocin in the methanotrophic bacterium Methylobacter bovis. Biochemistry 1998, 63, 1122–1125. [Google Scholar] [PubMed]
- Kalyuzhnaya, M.G.; Puri, A.W.; Lidstrom, M.E. Metabolic engineering in methanotrophic bacteria. Metab. Eng. 2015, 29, 142–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najafpour, G.D. Single-Cell Protein. In Biochemical Engineering and Biotechnology; Najafpour, G.D., Ed.; Elsevier: Amsterdam, 2007; pp. 332–341. ISBN 978-0-444-52845-2. [Google Scholar]
- García-Garigay, M.; Gómez-Ruiz, L.; Cruz-Guerrero, A.E.; Bárzana, E. Single-Cell Protein|Algae. In Encylopedia Food Science and Nutrition; Academic Press: Cambridge, MA, USA, 2003; pp. 5269–5276. [Google Scholar]
- Litchfield, J.H. Single-cell proteins. Science (80-.) 1983, 219, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.M.; Da Silva, A.; Rocha, R.; Lúcia, C.; Ribeiro, G.; Da, R.; Rodrigues, S.; Almeida, L.; Soares, S. Nutritional evaluation of single-cell protein produced by Spirulina platensis. Afr. J. Food Sci. 2011, 5, 799–805. [Google Scholar] [CrossRef]
- Reshetnikov, A.S.; Khmelenina, V.N.; Mustakhimov, I.I.; Trotsenko, Y.A. Genes and enzymes of Ectoine biosynthesis in halotolerant methanotrophs. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2011; Volume 495, pp. 15–30. [Google Scholar]
- Czech, L.; Hermann, L.; Stöveken, N.; Richter, A.A.; Höppner, A.; Smits, S.H.J.; Heider, J.; Bremer, E. Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: Genetics, phylogenomics, biochemistry, and structural analysis. Genes (Basel) 2018, 9, 177. [Google Scholar] [CrossRef] [Green Version]
- Pastor, J.M.; Salvador, M.; Argandoña, M.; Bernal, V.; Reina-Bueno, M.; Csonka, L.N.; Iborra, J.L.; Vargas, C.; Nieto, J.J.; Cánovas, M. Ectoines in cell stress protection: Uses and biotechnological production. Biotechnol. Adv. 2010, 28, 782–801. [Google Scholar] [CrossRef]
- Trotsenko, Y.A.; Khmelenina, V.N. Biology of extremophilic and extremotolerant methanotrophs. Arch. Microbiol. 2002, 177, 123–131. [Google Scholar] [CrossRef]
- Kanapathipillai, M.; Lentzen, G.; Sierks, M.; Park, C.B. Ectoine and hydroxyectoine inhibit aggregation and neurotoxicity of Alzheimer’s β-amyloid. FEBS Lett. 2005, 579, 4775–4780. [Google Scholar] [CrossRef] [Green Version]
- Arora, A.; Ha, C.; Park, C.B. Inhibition of insulin amyloid formation by small stress molecules. FEBS Lett. 2004, 564, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Furusho, K.; Yoshizawa, T.; Shoji, S. Ectoine alters subcellular localization of inclusions and reduces apoptotic cell death induced by the truncated Machado-Joseph disease gene product with an expanded polyglutamine stretch. Neurobiol. Dis. 2005, 20, 170–178. [Google Scholar] [CrossRef]
- Rieckmann, T.; Gatzemeier, F.; Christiansen, S.; Rothkamm, K.; Münscher, A. The inflammation-reducing compatible solute ectoine does not impair the cytotoxic effect of ionizing radiation on head and neck cancer cells. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Casale, M.; Moffa, A.; Carbone, S.; Fraccaroli, F.; Costantino, A.; Sabatino, L.; Lopez, M.A.; Baptista, P.; Cassano, M.; Rinaldi, V. Topical Ectoine: A Promising Molecule in the Upper Airways Inflammation—A Systematic Review. BioMed Res. Int. 2019, 2019, 1–10. [Google Scholar] [CrossRef]
- Dwivedi, M.; Brinkkötter, M.; Harishchandra, R.K.; Galla, H.J. Biophysical investigations of the structure and function of the tear fluid lipid layers and the effect of ectoine. Part B: Artificial lipid films. Biochim. Biophys. Acta-Biomembr. 2014, 1838, 2716–2727. [Google Scholar] [CrossRef] [Green Version]
- Bunger, J.; Driller, H.-J.; Martin, R. Use of Ecotine or Ecotine Derivatives in Comsntic Formulations. U.S. Patent 6,602,514,B1, 5 August 2003. [Google Scholar]
- Bilstein, A.; Scherner, O.; Lentzen, G. Composition Containing Ecotine or Hydroxyecotine as an Active Substance for Promoting the Regeneration of Injured Body Tissue. U.S. Patent 20170189435A1, 24 March 2017. [Google Scholar]
- Niyogi, K.K.; Bjo¨rkmanbjo¨rkman, O.; Grossman, A.R. The roles of specific xanthophylls in photoprotection. Plant Biol. 1997, 94, 14162–14167. [Google Scholar] [CrossRef] [Green Version]
- Clinton, S.K. Lycopene: Chemistry, Biology, and Implications for Human Health and Disease. Nutr. Rev. 2009, 56, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Nabi, F.; Arain, M.A.; Rajput, N.; Alagawany, M.; Soomro, J.; Umer, M.; Soomro, F.; Wang, Z.; Ye, R.; Liu, J. Health benefits of carotenoids and potential application in poultry industry: A review. J. Anim. Physiol. Anim. Nutr. (Berl.) 2020, 104, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Böhm, V. Carotenoids. Antioxidants 2019, 8, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapiero, H.; Townsend, D.M.; Tew, K.D. The role of carotenoids in the prevention of human pathologies. Biomed. Pharmacother. 2004, 58, 100. [Google Scholar] [CrossRef]
- Guo, W.; Li, D.; He, R.; Wu, M.; Chen, W.; Gao, F.; Zhang, Z.; Yao, Y.; Yu, L.; Chen, S. Synthesizing value-added products from methane by a new Methylomonas. J. Appl. Microbiol. 2017, 123, 1214–1227. [Google Scholar] [CrossRef]
- Rissanen, T.; Voutilainen, S.; Nyyssonen, K.; Salonen, J.T. Lycopene, Atherosclerosis, and Coronary Heart Disease. Exp. Biol. Med. 2002, 227, 900–907. [Google Scholar] [CrossRef]
- Cardoso, L.A.C.; Karp, S.G.; Vendruscolo, F.; Kanno, K.Y.F.; Zoz, L.I.C.; Carvalho, J.C. Biotechnological Production of Carotenoids and Their Applications in Food and Pharmaceutical Products. In Carotenoids; Cvetkovic, D.J., Nikolic, G.S., Eds.; InTechOpen: London, UK, 2017; pp. 125–141. [Google Scholar]
- Elvira-Torales, L.I.; García-Alonso, J.; Periago-Castón, M.J. Nutritional importance of carotenoids and their effect on liver health: A review. Antioxidants 2019, 8, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milani, A.; Basirnejad, M.; Shahbazi, S.; Bolhassani, A. Carotenoids: Biochemistry, pharmacology and treatment. Br. J. Pharmacol. 2017, 174, 1290–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatt, T.; Patel, K. Carotenoids: Potent to Prevent Diseases Review. Nat. Products Bioprospect. 2020, 10, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.W.; Yao, H.; Stead, K.; Wang, T.; Tao, L.; Cheng, Q.; Sharpe, P.L.; Suh, W.; Nagel, E.; Arcilla, D.; et al. Construction of the astaxanthin biosynthetic pathway in a methanotrophic bacterium Methylomonas sp. strain 16a. J. Ind. Microbiol. Biotechnol. 2007, 34, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Bonartsev, A.P.; Bonartseva, G.A.; Shaitan, K.V.; Kirpichnikov, M.P. Poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate)-based biopolymer systems. Biochem. Suppl. Ser. B Biomed. Chem. 2011, 5, 10–21. [Google Scholar] [CrossRef]
- Sahdev, S.; Sunil, A.E.; Khattar, K.; Kulvinder, A.E.; Saini, S. Production of active eukaryotic proteins through bacterial expression systems: A review of the existing biotechnology strategies. Mol. Cell. Biochem. 2008, 307, 249–264. [Google Scholar] [CrossRef]
- Swartz, J.R. Advances in Escherichia coli production of therapeutic proteins. Curr. Opin. Biotechnol. 2001, 12, 195–201. [Google Scholar] [CrossRef]
- García-Gómez, E.; González-Pedrajo, B.; Camacho-Arroyo, I.; Regina Mena Barreto Silva, F. Role of Sex Steroid Hormones in Bacterial-Host Interactions. BioMed Res. Int. 2013, 2013, 928290. [Google Scholar] [CrossRef]
- Ariede, M.B.; Candido, T.M.; Jacome, A.L.M.; Velasco, M.V.R.; de Carvalho, J.C.M.; Baby, A.R. Cosmetic attributes of algae—A review. Algal Res. 2017, 25, 483–487. [Google Scholar] [CrossRef]
- Chen, D.; Hou, Q.; Zhong, L.; Zhao, Y.; Li, M.; Fu, X. Bioactive Molecules for Skin Repair and Regeneration: Progress and Perspectives. Stem Cells Int. 2019, 2019, 6789823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dario, P.; Carrozza, M.C.; Benvenuto, A. Micro-systems in biomedical applications. J. Micromechanics Microengineering 2000, 10, 235–244. [Google Scholar] [CrossRef] [Green Version]

| Carotenoid | Disease(s) | Mode of Action | Reference(s) |
|---|---|---|---|
| α-carotenes, β-carotene | Diabetes, dyslipidemia, and hyperhomocysteinemia |
| [108,109,116] |
| Astaxanthin, lutein, β-cryptoxanthin, lycopene | Heart disease |
| [117] |
| Lutein and zeaxanthin | Macular degeneration and cataract |
| [106] |
| Astaxanthin, β-carotene and lycopene | Alzheimer’s, Huntington’s, Parkinson’s, and amyotrophic lateral sclerosis (ALS) |
| [107] |
| β-cryptoxanthin | Osteoporosis |
| [114] |
| β-cryptoxanthin, lycopene and β-carotene | Ovarian, breast, prostate, cervical, and liver cancer |
| [110] |
| Astaxanthin, β-carotene, lycopene, lutein, and β-cryptoxanthin | Liver damage |
| [114] |
| β-carotene, canthaxanthin and lycopene | Melanoma |
| [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salem, R.; ElDyasti, A.; Audette, G.F. Biomedical Applications of Biomolecules Isolated from Methanotrophic Bacteria in Wastewater Treatment Systems. Biomolecules 2021, 11, 1217. https://doi.org/10.3390/biom11081217
Salem R, ElDyasti A, Audette GF. Biomedical Applications of Biomolecules Isolated from Methanotrophic Bacteria in Wastewater Treatment Systems. Biomolecules. 2021; 11(8):1217. https://doi.org/10.3390/biom11081217
Chicago/Turabian StyleSalem, Rana, Ahmed ElDyasti, and Gerald F. Audette. 2021. "Biomedical Applications of Biomolecules Isolated from Methanotrophic Bacteria in Wastewater Treatment Systems" Biomolecules 11, no. 8: 1217. https://doi.org/10.3390/biom11081217







