The Antioxidant Effect of Dietary Bioactives Arises from the Interplay between the Physiology of the Host and the Gut Microbiota: Involvement of Short-Chain Fatty Acids
Abstract
:1. Introduction
2. Importance of the Nrf2 Pathway and Its Link with Gut Microbiota
3. Composition of the Gut Microbiota
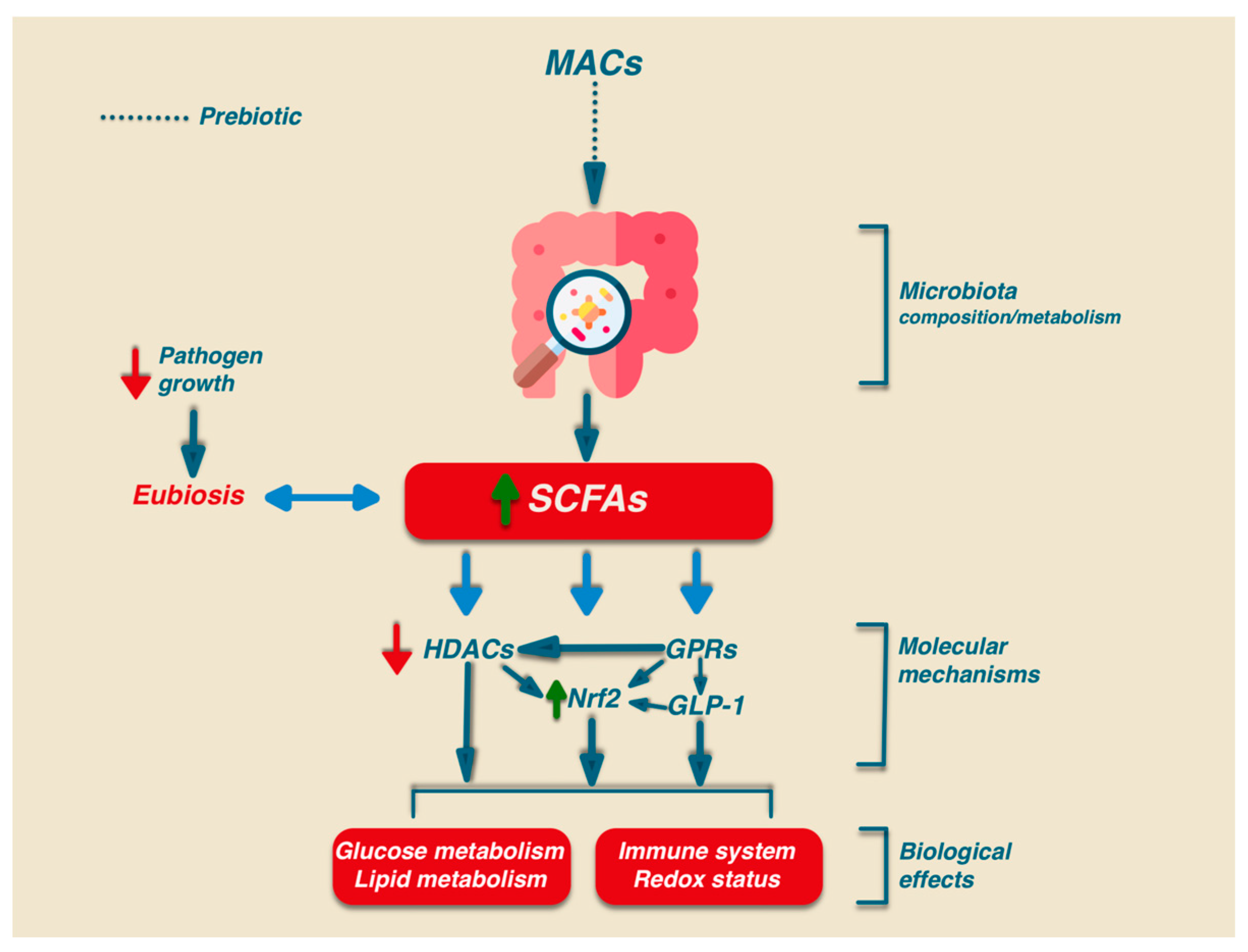
4. Effect of SCFAs on the Composition of the Gut Microbiota
4.1. Effect of SCFAs on Gut Homeostasis
4.2. Signaling Mechanisms Induced by SCFAs
5. Dietary Bioactive Molecules and the Gut Microbiota Composition
5.1. Effect of Microbiota-Accessible Carbohydrates (MACs) on Gut Homeostasis
| Treatment | Model | Microbiota Alteration/SCFA Production | Ref. |
|---|---|---|---|
| Metanalysis | Studies investigating the effect of dietary fiber on gut microbiota | ↑ Bifidobacterium ↑ SCFAs | [29] |
| Dietary Fiber | European children (Low-fiber diet) vs. Rural African village (High-fiber diet) | ↑ Bacteroidetes ↑ SCFAs | [92] |
| African Americans vs. Rural native Africans | ↑ Prevotella ↑ SCFAs | [94] | |
| Inulin | Mice with hyperuricemia vs. wild-type mice | ↑ microbial diversity ↑ SCFA-producing bacteria (Akkermansia and Ruminococcus). ↑ acetate, propionate, and butyrate | [95] |
| Nonalcoholic Fatty Liver Disease rat model | ↑ Bifidobacterium, Phascolarctobacterium, Blautia ↓ Acetate ↑ Propionate and Butyrate | [96] |
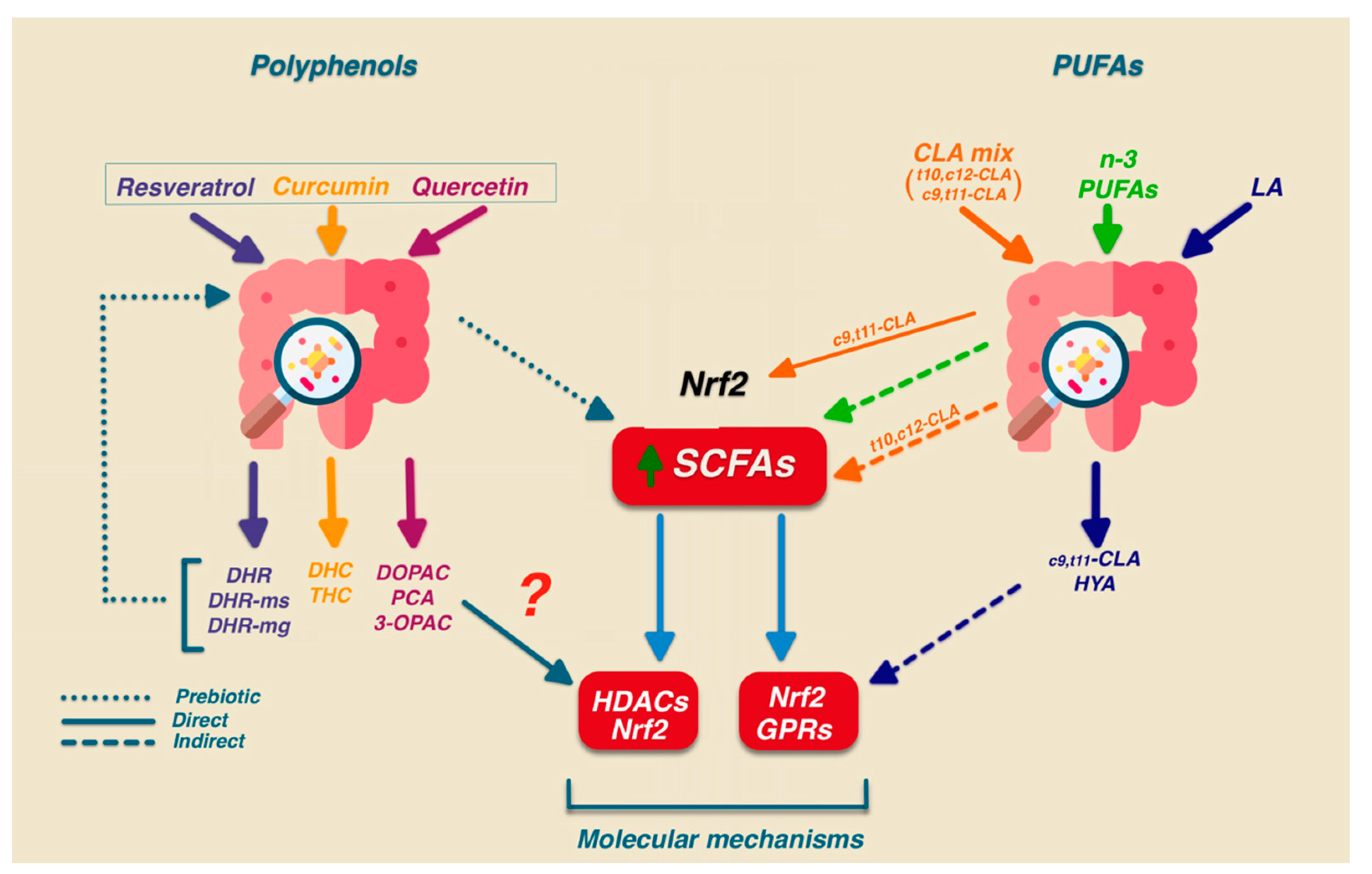
5.2. Effect of Polyphenols on Gut Homeostasis
5.3. Effect of Polyunsaturated Fatty Acids (PUFAs) and Conjugated Linoleic Acid (CLA) on Gut Homeostasis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sies, H. Oxidative eustress: On constant alert for redox homeostasis. Redox Biol. 2021, 41, 101867. [Google Scholar] [CrossRef]
- Kurtishi, A.; Rosen, B.; Patil, K.S.; Alves, G.W.; Møller, S.G. Cellular Proteostasis in Neurodegeneration. Mol. Neurobiol. 2019, 56, 3676–3689. [Google Scholar] [CrossRef]
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K. The intestinal microbiota and its role in human health and disease. J. Med. Investig. 2016, 63, 27–37. [Google Scholar] [CrossRef]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef]
- Sadovnikova, I.S.; Gureev, A.P.; Ignatyeva, D.A.; Gryaznova, M.V.; Chernyshova, E.V.; Krutskikh, E.P.; Novikova, A.G.; Popov, V.N. Nrf2/ARE Activators Improve Memory in Aged Mice via Maintaining of Mitochondrial Quality Control of Brain and the Modulation of Gut Microbiome. Pharmaceuticals 2021, 14, 607. [Google Scholar] [CrossRef]
- Suzuki, T.; Yamamoto, M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J. Biol. Chem. 2017, 292, 16817–16824. [Google Scholar] [CrossRef]
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Daiber, A.; Ghezzi, P.; León, R.; López, M.G.; Oliva, B.; et al. Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach. Pharmacol. Rev. 2018, 70, 348–383. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Hou, D.X. Multiple regulations of Keap1/Nrf2 system by dietary phytochemicals. Mol. Nutr. Food Res. 2016, 60, 1731–1755. [Google Scholar] [CrossRef]
- Abrescia, P.; Treppiccione, L.; Rossi, M.; Bergamo, P. Modulatory role of dietary polyunsaturated fatty acids in Nrf2-mediated redox homeostasis. Prog. Lipid Res. 2020, 80, 101066. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jiang, Z.; Lu, H.; Xu, Z.; Tong, R.; Shi, J.; Jia, G. Recent Advances of Natural Polyphenols Activators for Keap1-Nrf2 Signaling Pathway. Chem. Biodivers. 2019, 16, e1900400. [Google Scholar] [CrossRef]
- Bourlioux, P.; Koletzko, B.; Guarner, F.; Braesco, V. The intestine and its microflora are partners for the protection of the host: Report on the Danone Symposium “The Intelligent Intestine,” held in Paris, June 14, 2002. Am. J. Clin. Nutr. 2003, 78, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Rajoka, M.; Shi, J.; Mehwish, H.; Zhu, J.; Li, Q.; Shao, D.; Huang, Q.; Yang, H. Interaction between diet composition and gut microbiota and its impact on gastrointestinal tract health. Food Sci. Hum. Wellness 2017, 6, 121–130. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef]
- Xu, J.; Mahowald, M.A.; Ley, R.E.; Lozupone, C.A.; Hamady, M.; Martens, E.C.; Henrissat, B.; Coutinho, P.M.; Minx, P.; Latreille, P.; et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007, 5, e156. [Google Scholar] [CrossRef]
- Consortium, H.M.P. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Zhou, Y.; He, Y.; Liu, L.; Zhou, W.; Wang, P.; Hu, H.; Nie, Y.; Chen, Y. Alterations in Gut Microbial Communities Across Anatomical Locations in Inflammatory Bowel Diseases. Front. Nutr. 2021, 8, 615064. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.; Ursell, L.; Parfrey, L.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Tilg, H. Obesity, metabolic syndrome, and microbiota: Multiple interactions. J Clin. Gastroenterol. 2010, 44 (Suppl. 1), S16–S18. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Scheithauer, T.P.M.; Rampanelli, E.; Nieuwdorp, M.; Vallance, B.A.; Verchere, C.B.; van Raalte, D.H.; Herrema, H. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 2020, 11, 571731. [Google Scholar] [CrossRef]
- Angelucci, F.; Cechova, K.; Amlerova, J.; Hort, J. Antibiotics, gut microbiota, and Alzheimer’s disease. J. Neuroinflamm. 2019, 16, 108. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef]
- Descamps, H.C.; Herrmann, B.; Wiredu, D.; Thaiss, C.A. The path toward using microbial metabolites as therapies. EBioMedicine 2019, 44, 747–754. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Mahowald, M.; Rey, F.; Seedorf, H.; Turnbaugh, P.; Fulton, R.; Wollam, A.; Shah, N.; Wang, C.; Magrini, V.; Wilson, R.; et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 2009, 106, 5859–5864. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef]
- Ojo, O.; Feng, Q.Q.; Ojo, O.O.; Wang, X.H. The Role of Dietary Fibre in Modulating Gut Microbiota Dysbiosis in Patients with Type 2 Diabetes: A Systematic Review and Meta-Analysis of Randomised Controlled Trials. Nutrients 2020, 12, 3239. [Google Scholar] [CrossRef]
- Imhann, F.; Vich Vila, A.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Takahashi, K.; Nishida, A.; Fujimoto, T.; Fujii, M.; Shioya, M.; Innaeda, H.; Lnatorni, O.; Bamba, S.; Andoh, A.; Sugimoto, M. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion 2016, 93, 59–65. [Google Scholar] [CrossRef]
- Rau, M.; Rehman, A.; Dittrich, M.; Groen, A.K.; Hermanns, H.M.; Seyfried, F.; Beyersdorf, N.; Dandekar, T.; Rosenstiel, P.; Geier, A. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United Eur. Gastroenterol. J. 2018, 6, 1496–1507. [Google Scholar] [CrossRef]
- Tsai, M.C.; Liu, Y.Y.; Lin, C.C.; Wang, C.C.; Wu, Y.J.; Yong, C.C.; Chen, K.D.; Chuah, S.K.; Yao, C.C.; Huang, P.Y.; et al. Gut Microbiota Dysbiosis in Patients with Biopsy-Proven Nonalcoholic Fatty Liver Disease: A Cross-Sectional Study in Taiwan. Nutrients 2020, 12, 820. [Google Scholar] [CrossRef]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Araujo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016, 63, 764–775. [Google Scholar] [CrossRef]
- Hirayama, M.; Ohno, K. Parkinson’s Disease and Gut Microbiota. Ann. Nutr. Metab. 2021, 77 (Suppl. 2), 28–35. [Google Scholar] [CrossRef]
- Chen, S.J.; Chen, C.C.; Liao, H.Y.; Lin, Y.T.; Wu, Y.W.; Liou, J.M.; Wu, M.S.; Kuo, C.H.; Lin, C.H. Association of Fecal and Plasma Levels of Short-Chain Fatty Acids with Gut Microbiota and Clinical Severity in Patients with Parkinson Disease. Neurology 2022, 98, e848–e858. [Google Scholar] [CrossRef]
- Yang, X.; Ai, P.; He, X.; Mo, C.; Zhang, Y.; Xu, S.; Lai, Y.; Qian, Y.; Xiao, Q. Parkinson’s Disease Is Associated with Impaired Gut-Blood Barrier for Short-Chain Fatty Acids. Mov. Disord. 2022, 37, 1634–1643. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef]
- Verhaar, B.J.H.; Hendriksen, H.M.A.; de Leeuw, F.A.; Doorduijn, A.S.; van Leeuwenstijn, M.; Teunissen, C.E.; Barkhof, F.; Scheltens, P.; Kraaij, R.; van Duijn, C.M.; et al. Gut Microbiota Composition Is Related to AD Pathology. Front. Immunol. 2021, 12, 794519. [Google Scholar] [CrossRef]
- Fernando, W.M.A.D.; Martins, I.J.; Morici, M.; Bharadwaj, P.; Rainey-Smith, S.R.; Lim, W.L.F.; Martins, R.N. Sodium Butyrate Reduces Brain Amyloid-β Levels and Improves Cognitive Memory Performance in an Alzheimer’s Disease Transgenic Mouse Model at an Early Disease Stage. J. Alzheimer’s Dis. 2020, 74, 91–99. [Google Scholar] [CrossRef]
- Barcenilla, A.; Pryde, S.; Martin, J.; Duncan, S.; Stewart, C.; Henderson, C.; Flint, H. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 2000, 66, 1654–1661. [Google Scholar] [CrossRef]
- Ohata, A.; Usami, M.; Miyoshi, M. Short-chain fatty acids alter tight junction permeability in intestinal monolayer cells via lipoxygenase activation. Nutrition 2005, 21, 838–847. [Google Scholar] [CrossRef]
- Rivera-Chávez, F.; Zhang, L.F.; Faber, F.; Lopez, C.A.; Byndloss, M.X.; Olsan, E.E.; Xu, G.; Velazquez, E.M.; Lebrilla, C.B.; Winter, S.E.; et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 2016, 19, 443–454. [Google Scholar] [CrossRef]
- Colgan, S.P.; Taylor, C.T. Hypoxia: An alarm signal during intestinal inflammation. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 281–287. [Google Scholar] [CrossRef]
- Bohnhoff, M.; Miller, C.P.; Martin, W.R. Resistance of the mouse’s intestinal tract to experimental salmonella infection. II. factors responsible for its loss following streptomycin treatment. J. Exp. Med. 1964, 120, 817–828. [Google Scholar] [CrossRef]
- Rolfe, R.D. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect. Immun. 1984, 45, 185–191. [Google Scholar] [CrossRef]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef]
- Gaudier, E.; Rival, M.; Buisine, M.P.; Robineau, I.; Hoebler, C. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol. Res. 2009, 58, 111–119. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Meesters, R.; van Eijk, H.; ten Have, G.; de Graaf, A.; Venema, K.; van Rossum, B.; Deutz, N. Application of liquid chromatography-mass spectrometry to measure the concentrations and study the synthesis of short chain fatty acids following stable isotope infusions. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007, 854, 57–62. [Google Scholar] [CrossRef]
- Moreau, N.; Goupry, S.; Antignac, J.; Monteau, F.; Le Bizec, B.; Champ, M.; Martin, L.; Dumon, H. Simultaneous measurement of plasma concentrations and C-13-enrichment of short-chain fatty acids, lactic acid and ketone bodies by gas chromatography coupled to mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 784, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef]
- Yajima, T. Contractile effect of short-chain fatty acids on the isolated colon of the rat. J. Physiol. 1985, 368, 667–678. [Google Scholar] [CrossRef]
- Plaisancié, P.; Dumoulin, V.; Chayvialle, J.A.; Cuber, J.C. Luminal peptide YY-releasing factors in the isolated vascularly perfused rat colon. J. Endocrinol. 1996, 151, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Fåk, F.; Jakobsdottir, G.; Kulcinskaja, E.; Marungruang, N.; Matziouridou, C.; Nilsson, U.; Stålbrand, H.; Nyman, M. The physico-chemical properties of dietary fibre determine metabolic responses, short-chain Fatty Acid profiles and gut microbiota composition in rats fed low- and high-fat diets. PLoS ONE 2015, 10, e0127252. [Google Scholar] [CrossRef]
- Kilner, J.; Corfe, B.; McAuley, M.; Wilkinson, S. A deterministic oscillatory model of microtubule growth and shrinkage for differential actions of short chain fatty acids. Mol. Biosyst. 2016, 12, 93–101. [Google Scholar] [CrossRef]
- Vincent, A.D.; Wang, X.Y.; Parsons, S.P.; Khan, W.I.; Huizinga, J.D. Abnormal absorptive colonic motor activity in germ-free mice is rectified by butyrate, an effect possibly mediated by mucosal serotonin. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G896–G907. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, T.; Suruga, K.; Oshima, Y.; Fukiharu, M.; Tsukamoto, Y.; Goda, T. Dietary acetic acid reduces serum cholesterol and triacylglycerols in rats fed a cholesterol-rich diet. Br. J. Nutr. 2006, 95, 916–924. [Google Scholar] [CrossRef] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-Generated Metabolites Promote Metabolic Benefits via Gut-Brain Neural Circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef]
- Ukai, M.; Tomura, A.; Ito, M. Cholesterol-synthesis in germfree and conventional rats. J. Nutr. 1976, 106, 1175–1183. [Google Scholar] [CrossRef]
- Wang, G.; Yu, Y.; Wang, Y.; Wang, J.; Guan, R.; Sun, Y.; Shi, F.; Gao, J.; Fu, X. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J. Cell. Physiol. 2019, 234, 17023–17049. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F.; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015, 29, 1395–1403. [Google Scholar] [CrossRef]
- Vijay, N.; Morris, M.E. Role of monocarboxylate transporters in drug delivery to the brain. Curr. Pharm. Des. 2014, 20, 1487–1498. [Google Scholar] [CrossRef]
- Daly, K.; Shirazi-Beechey, S. Microarray analysis of butyrate regulated genes in colonic epithelial cells. DNA Cell Biol. 2006, 25, 49–62. [Google Scholar] [CrossRef]
- Campos-Perez, W.; Martinez-Lopez, E. Effects of short chain fatty acids on metabolic and inflammatory processes in human health. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158900. [Google Scholar] [CrossRef] [PubMed]
- González-Bosch, C.; Boorman, E.; Zunszain, P.A.; Mann, G.E. Short-chain fatty acids as modulators of redox signaling in health and disease. Redox Biol. 2021, 47, 102165. [Google Scholar] [CrossRef] [PubMed]
- Visekruna, A.; Luu, M. The Role of Short-Chain Fatty Acids and Bile Acids in Intestinal and Liver Function, Inflammation, and Carcinogenesis. Front. Cell Dev. Biol. 2021, 9, 703218. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef]
- Karaki, S.; Mitsui, R.; Hayashi, H.; Kato, I.; Sugiya, H.; Iwanaga, T.; Furness, J.B.; Kuwahara, A. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006, 324, 353–360. [Google Scholar] [CrossRef]
- Ahmed, K.; Tunaru, S.; Offermanns, S. GPR109A, GPR109B and GPR81, a family of hydroxy-carboxylic acid receptors. Trends Pharmacol. Sci. 2009, 30, 557–562. [Google Scholar] [CrossRef]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef]
- Schotterl, S.; Brennenstuhl, H.; Naumann, U. Modulation of immune responses by histone deacetylase inhibitors. Crit. Rev. Oncog. 2015, 20, 139–154. [Google Scholar] [CrossRef]
- Weiss, U.; Möller, M.; Husseini, S.A.; Manderscheid, C.; Häusler, J.; Geisslinger, G.; Niederberger, E. Inhibition of HDAC Enzymes Contributes to Differential Expression of Pro-Inflammatory Proteins in the TLR-4 Signaling Cascade. Int. J. Mol. Sci. 2020, 21, 8943. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Chen, L.; Yang, W.; Huang, X.; Ma, C.; Chen, F.; Xiao, Y.; Zhao, Y.; Yao, S.; et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 2018, 9, 3555. [Google Scholar] [CrossRef]
- Salazar, N.; Arboleya, S.; Fernández-Navarro, T.; de Los Reyes-Gavilán, C.G.; Gonzalez, S.; Gueimonde, M. Age-Associated Changes in Gut Microbiota and Dietary Components Related with the Immune System in Adulthood and Old Age: A Cross-Sectional Study. Nutrients 2019, 11, 1765. [Google Scholar] [CrossRef]
- Matsumaru, D.; Motohashi, H. The KEAP1-NRF2 System in Healthy Aging and Longevity. Antioxidants 2021, 10, 1929. [Google Scholar] [CrossRef]
- Li, H.; Shang, Z.; Liu, X.; Qiao, Y.; Wang, K.; Qiao, J. Alleviates Enterotoxigenic. Front. Immunol. 2021, 12, 771826. [Google Scholar] [CrossRef]
- Dong, W.; Jia, Y.; Liu, X.; Zhang, H.; Li, T.; Huang, W.; Chen, X.; Wang, F.; Sun, W.; Wu, H. Sodium butyrate activates NRF2 to ameliorate diabetic nephropathy possibly via inhibition of HDAC. J. Endocrinol. 2017, 232, 71–83. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Z.; Zhang, H.; Liang, W.; Huang, W.; Li, Y.; Wang, Z.; Wang, J.; Jia, Y.; Liu, B.; et al. Sodium butyrate attenuates diabetes-induced aortic endothelial dysfunction via P300-mediated transcriptional activation of Nrf2. Free Radic. Biol. Med. 2018, 124, 454–465. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Sun, S.; Araki, Y.; Hanzawa, F.; Umeki, M.; Kojima, T.; Nishimura, N.; Ikeda, S.; Mochizuki, S.; Oda, H. High sucrose diet-induced dysbiosis of gut microbiota promotes fatty liver and hyperlipidemia in rats. J. Nutr. Biochem. 2021, 93, 108621. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Topping, D.L.; Clifton, P.M. Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001, 81, 1031–1064. [Google Scholar] [CrossRef] [PubMed]
- Martinez, K.B.; Leone, V.; Chang, E.B. Western diets, gut dysbiosis, and metabolic diseases: Are they linked? Gut Microbes 2017, 8, 130–142. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Schnorr, S.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yu, Y.; Li, H.; Ding, X.; Li, X.; Jing, X.; Chen, J.; Liu, G.; Lin, Y.; Jiang, C.; et al. Inulin supplementation ameliorates hyperuricemia and modulates gut microbiota in Uox-knockout mice. Eur. J. Nutr. 2021, 60, 2217–2230. [Google Scholar] [CrossRef]
- Yang, Z.; Su, H.; Lv, Y.; Tao, H.; Jiang, Y.; Ni, Z.; Peng, L.; Chen, X. Inulin intervention attenuates hepatic steatosis in rats via modulating gut microbiota and maintaining intestinal barrier function. Food Res. Int. 2023, 163, 112309. [Google Scholar] [CrossRef]
- Williamson, G.; Clifford, M. Role of the small intestine, colon and microbiota in determining the metabolic fate of polyphenols. Biochemical. Pharmacol. 2017, 139, 24–39. [Google Scholar] [CrossRef]
- Manach, C.; Williamson, G.; Morand, C.; Scalbert, A.; Remesy, C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005, 81, 230S–242S. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mateos, A.; Vauzour, D.; Krueger, C.; Shanmuganayagam, D.; Reed, J.; Calani, L.; Mena, P.; Del Rio, D.; Crozier, A. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: An update. Arch. Toxicol. 2014, 88, 1803–1853. [Google Scholar] [CrossRef]
- Cassidy, A.; Minihane, A. The role of metabolism (and the microbiome) in defining the clinical efficacy of dietary flavonoids. Am. J. Clin. Nutr. 2017, 105, 10–22. [Google Scholar] [CrossRef]
- Aura, A. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem. Rev. 2008, 7, 407–429. [Google Scholar] [CrossRef]
- Heleno, S.; Martins, A.; Queiroz, M.; Ferreira, I. Bioactivity of phenolic acids: Metabolites versus parent compounds: A review. Food Chem. 2015, 173, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Koudoufio, M.; Desjardins, Y.; Feldman, F.; Spahis, S.; Delvin, E.; Levy, E. Insight into Polyphenol and Gut Microbiota Crosstalk: Are Their Metabolites the Key to Understand Protective Effects against Metabolic Disorders? Antioxidants 2020, 9, 982. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, T.; Sela, D.A.; Xiao, J.; Boyacioglu, D.; Chen, F.; Capanoglu, E. The Reciprocal Interactions between Polyphenols and Gut Microbiota and Effects on Bioaccessibility. Nutrients 2016, 8, 78. [Google Scholar] [CrossRef]
- Catalkaya, G.; Venema, K.; Lucini, L.; Rocchetti, G.; Delmas, D.; Daglia, M.; De Filippis, A.; Xiao, H.H.; Quiles, J.L.; Xiao, J.; et al. Interaction of dietary polyphenols and gut microbiota:Microbial metabolism of polyphenols, influence on the gut microbiota, and implications on host health. Food Front. 2020, 1, 109–133. [Google Scholar] [CrossRef]
- Carbonell-Capella, J.M.; Buniowska, M.; Barba, F.J.; Esteve, M.J.; Frígola, A. Analytical Methods for Determining Bioavailability and Bioaccessibility of Bioactive Compounds from Fruits and Vegetables: A Review. Compr. Rev. Food Sci. Food Saf. 2014, 13, 155–171. [Google Scholar] [CrossRef]
- Wightman, E.L.; Haskell-Ramsay, C.F.; Reay, J.L.; Williamson, G.; Dew, T.; Zhang, W.; Kennedy, D.O. The effects of chronic trans-resveratrol supplementation on aspects of cognitive function, mood, sleep, health and cerebral blood flow in healthy, young humans. Br. J. Nutr. 2015, 114, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Mahale, J.; Singh, R.; Howells, L.; Britton, R.; Khan, S.; Brown, K. Detection of Plasma Curcuminoids from Dietary Intake of Turmeric-Containing Food in Human Volunteers. Mol. Nutr. Food Res. 2018, 62, e1800267. [Google Scholar] [CrossRef]
- Marin, L.; Miguelez, E.; Villar, C.; Lombo, F. Bioavailability of Dietary Polyphenols and Gut Microbiota Metabolism: Antimicrobial Properties. BioMed Res. Int. 2015, 2015, 905215. [Google Scholar] [CrossRef]
- Burapan, S.; Kim, M.; Han, J. Curcuminoid Demethylation as an Alternative Metabolism by Human Intestinal Microbiota. J. Agric. Food Chem. 2017, 65, 3305–3310. [Google Scholar] [CrossRef]
- Chiou, Y.; Wu, J.; Huang, Q.; Shahidi, F.; Wang, Y.; Ho, C.; Pan, M. Metabolic and colonic microbiota transformation may enhance the bioactivities of dietary polyphenols. J. Funct. Foods 2014, 7, 3–25. [Google Scholar] [CrossRef]
- Griffiths, L.A.; Barrow, A. Metabolism of flavonoid compounds in germ-free rats. Biochem. J. 1972, 130, 1161–1162. [Google Scholar] [CrossRef] [PubMed]
- Hehemann, J.H.; Correc, G.; Barbeyron, T.; Helbert, W.; Czjzek, M.; Michel, G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef] [PubMed]
- van Duynhoven, J.; Vaughan, E.E.; Jacobs, D.M.; Kemperman, R.A.; van Velzen, E.J.; Gross, G.; Roger, L.C.; Possemiers, S.; Smilde, A.K.; Doré, J.; et al. Metabolic fate of polyphenols in the human superorganism. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4531–4538. [Google Scholar] [CrossRef]
- Wu, L.; Lyu, Y.; Srinivasagan, R.; Wu, J.; Ojo, B.; Tang, M.; El-Rassi, G.D.; Metzinger, K.; Smith, B.J.; Lucas, E.A.; et al. Astaxanthin-Shifted Gut Microbiota Is Associated with Inflammation and Metabolic Homeostasis in Mice. J. Nutr. 2020, 150, 2687–2698. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, M.; Uehara, O.; Ogasa, S.; Sano, T.; Kubota, A.; Kojima, H.; Tanaka, T.; Maeda, H.; Miyashita, K.; Mutoh, M. Alteration of fecal microbiota by fucoxanthin results in prevention of colorectal cancer in AOM/DSS mice. Carcinogenesis 2021, 42, 210–219. [Google Scholar] [CrossRef]
- Xia, H.; Liu, C.; Li, C.C.; Fu, M.; Takahashi, S.; Hu, K.Q.; Aizawa, K.; Hiroyuki, S.; Wu, G.; Zhao, L.; et al. Dietary Tomato Powder Inhibits High-Fat Diet-Promoted Hepatocellular Carcinoma with Alteration of Gut Microbiota in Mice Lacking Carotenoid Cleavage Enzymes. Cancer Prev. Res. 2018, 11, 797–810. [Google Scholar] [CrossRef]
- Wang, X.; Liu, F.; Cui, Y.; Yin, Y.; Li, S.; Li, X. Apple Polyphenols Extracts Ameliorate High Carbohydrate Diet-Induced Body Weight Gain by Regulating the Gut Microbiota and Appetite. J. Agric. Food Chem. 2022, 70, 196–210. [Google Scholar] [CrossRef]
- Cladis, D.P.; Simpson, A.M.R.; Cooper, K.J.; Nakatsu, C.H.; Ferruzzi, M.G.; Weaver, C.M. Blueberry polyphenols alter gut microbiota & phenolic metabolism in rats. Food Funct. 2021, 12, 2442–2456. [Google Scholar] [CrossRef]
- Guo, X.; Xu, Y.; Geng, R.; Qiu, J.; He, X. Curcumin Alleviates Dextran Sulfate Sodium-Induced Colitis in Mice Through Regulating Gut Microbiota. Mol. Nutr. Food Res. 2022, 66, e2100943. [Google Scholar] [CrossRef]
- Zhang, F.; Zhou, Y.; Chen, H.; Jiang, H.; Zhou, F.; Lv, B.; Xu, M. Curcumin Alleviates DSS-Induced Anxiety-Like Behaviors via the Microbial-Brain-Gut Axis. Oxid. Med. Cell. Longev. 2022, 2022, 6244757. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Shin, Y.; Kim, S.H.; Jin, M.; Choi, J.J. Dietary Epigallocatechin-3-Gallate Alters the Gut Microbiota of Obese Diabetic. J. Med. Food 2020, 23, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Li, Y.L.; Zhang, X.; Wang, K.B.; Huang, J.A.; Liu, Z.H.; Zhu, M.Z. Polyphenols from Fu Brick Tea Reduce Obesity via Modulation of Gut Microbiota and Gut Microbiota-Related Intestinal Oxidative Stress and Barrier Function. J. Agric. Food Chem. 2021, 69, 14530–14543. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.Y.; Chi, Y.Y.; Han, J.X.; Kong, P.; Liang, Z.; Wang, D.; Xiang, H.; Xie, Q. Litchi chinensis seed prevents obesity and modulates the gut microbiota and mycobiota compositions in high-fat diet-induced obese zebrafish. Food Funct. 2022, 13, 2832–2845. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Wang, L.; Liu, M.; Wu, X.; Lu, Q.; Liu, R. The underlying mechanism of A-type procyanidins from peanut skin on DSS-induced ulcerative colitis mice by regulating gut microbiota and metabolism. J. Food Biochem. 2022, 46, e14103. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Shen, X.; Chu, Q.; Zheng, X. Pomegranate fruit pulp polyphenols reduce diet-induced obesity with modulation of gut microbiota in mice. J. Sci. Food Agric. 2022, 102, 1968–1977. [Google Scholar] [CrossRef]
- Liu, D.; Ji, Y.; Wang, K.; Guo, Y.; Wang, H.; Zhang, H.; Li, L.; Li, H.; Cui, S.W. Purple sweet potato anthocyanin extract regulates redox state related to gut microbiota homeostasis in obese mice. J. Food Sci. 2022, 87, 2133–2146. [Google Scholar] [CrossRef]
- Chen, C.; Wang, H.; Hong, T.; Huang, X.; Xia, S.; Zhang, Y.; Chen, X.; Zhong, Y.; Nie, S. Effects of tea polysaccharides in combination with polyphenols on dextran sodium sulfate-induced colitis in mice. Food Chem. X 2022, 13, 100190. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, J.; Liu, M.; Qiu, S.; Yi, S.; Yu, W.; Liu, T.; Huang, X.; Ning, F. Monofloral Triadica Cochinchinensis Honey Polyphenols Improve Alcohol-Induced Liver Disease by Regulating the Gut Microbiota of Mice. Front. Immunol. 2021, 12, 673903. [Google Scholar] [CrossRef]
- Logan, I.E.; Shulzhenko, N.; Sharpton, T.J.; Bobe, G.; Liu, K.; Nuss, S.; Jones, M.L.; Miranda, C.L.; Vasquez-Perez, S.; Pennington, J.M.; et al. Xanthohumol Requires the Intestinal Microbiota to Improve Glucose Metabolism in Diet-Induced Obese Mice. Mol. Nutr. Food Res. 2021, 65, e2100389. [Google Scholar] [CrossRef]
- Zlatanos, S.N.; Laskaridis, K.; Sagredos, A. Conjugated linoleic acid content of human plasma. Lipids Health Dis. 2008, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.L.; Leitch, J.; Blake, R.J.; Garg, R. Long-chain n-3 polyunsaturated fatty acid incorporation into human atrium following fish oil supplementation. Lipids 2006, 41, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Murru, E.; Carta, G.; Manca, C.; Saebo, A.; Santoni, M.; Mostallino, R.; Pistis, M.; Banni, S. Dietary Phospholipid-Bound Conjugated Linoleic Acid and Docosahexaenoic Acid Incorporation Into Fetal Liver and Brain Modulates Fatty Acid and. Front. Nutr. 2022, 9, 834066. [Google Scholar] [CrossRef] [PubMed]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, Y.; Gao, H.; Li, D.; Jiang, R.; Ge, L.; Tong, C.; Xu, K. Associations among Dietary Omega-3 Polyunsaturated Fatty Acids, the Gut Microbiota, and Intestinal Immunity. Mediat. Inflamm. 2021, 2021, 8879227. [Google Scholar] [CrossRef]
- Robertson, R.C.; Seira Oriach, C.; Murphy, K.; Moloney, G.M.; Cryan, J.F.; Dinan, T.G.; Ross, R.P.; Stanton, C. Deficiency of essential dietary n-3 PUFA disrupts the caecal microbiome and metabolome in mice. Br. J. Nutr. 2017, 118, 959–970. [Google Scholar] [CrossRef]
- Menni, C.; Zierer, J.; Pallister, T.; Jackson, M.A.; Long, T.; Mohney, R.P.; Steves, C.J.; Spector, T.D.; Valdes, A.M. Omega-3 fatty acids correlate with gut microbiome diversity and production of N-carbamylglutamate in middle aged and elderly women. Sci. Rep. 2017, 7, 11079. [Google Scholar] [CrossRef]
- Noriega, B.S.; Sanchez-Gonzalez, M.A.; Salyakina, D.; Coffman, J. Understanding the Impact of Omega-3 Rich Diet on the Gut Microbiota. Case Rep. Med. 2016, 2016, 3089303. [Google Scholar] [CrossRef]
- Kaliannan, K.; Wang, B.; Li, X.Y.; Kim, K.J.; Kang, J.X. A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci. Rep. 2015, 5, 11276. [Google Scholar] [CrossRef]
- Younge, N.; Yang, Q.; Seed, P.C. Enteral High Fat-Polyunsaturated Fatty Acid Blend Alters the Pathogen Composition of the Intestinal Microbiome in Premature Infants with an Enterostomy. J. Pediatr. 2017, 181, 93–101.e6. [Google Scholar] [CrossRef]
- Mokkala, K.; Röytiö, H.; Munukka, E.; Pietilä, S.; Ekblad, U.; Rönnemaa, T.; Eerola, E.; Laiho, A.; Laitinen, K. Gut Microbiota Richness and Composition and Dietary Intake of Overweight Pregnant Women Are Related to Serum Zonulin Concentration, a Marker for Intestinal Permeability. J. Nutr. 2016, 146, 1694–1700. [Google Scholar] [CrossRef] [PubMed]
- Watson, H.; Mitra, S.; Croden, F.C.; Taylor, M.; Wood, H.M.; Perry, S.L.; Spencer, J.A.; Quirke, P.; Toogood, G.J.; Lawton, C.L.; et al. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut 2018, 67, 1974–1983. [Google Scholar] [CrossRef] [PubMed]
- Balfegó, M.; Canivell, S.; Hanzu, F.A.; Sala-Vila, A.; Martínez-Medina, M.; Murillo, S.; Mur, T.; Ruano, E.G.; Linares, F.; Porras, N.; et al. Effects of sardine-enriched diet on metabolic control, inflammation and gut microbiota in drug-naïve patients with type 2 diabetes: A pilot randomized trial. Lipids Health Dis. 2016, 15, 78. [Google Scholar] [CrossRef] [PubMed]
- Mujico, J.R.; Baccan, G.C.; Gheorghe, A.; Díaz, L.E.; Marcos, A. Changes in gut microbiota due to supplemented fatty acids in diet-induced obese mice. Br. J. Nutr. 2013, 110, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Hougen, H.; Vollmer, A.C.; Hiebert, S.M. Gut bacteria profiles of Mus musculus at the phylum and family levels are influenced by saturation of dietary fatty acids. Anaerobe 2012, 18, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; O’ Doherty, R.M.; Murphy, E.F.; Wall, R.; O’ Sullivan, O.; Nilaweera, K.; Fitzgerald, G.F.; Cotter, P.D.; Ross, R.P.; Stanton, C. Impact of dietary fatty acids on metabolic activity and host intestinal microbiota composition in C57BL/6J mice. Br. J. Nutr. 2014, 111, 1905–1917. [Google Scholar] [CrossRef]
- Caesar, R.; Tremaroli, V.; Kovatcheva-Datchary, P.; Cani, P.D.; Bäckhed, F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015, 22, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef]
- Pariza, M.W. Perspective on the safety and effectiveness of conjugated linoleic acid. Am. J. Clin. Nutr. 2004, 79, 1132S–1136S. [Google Scholar] [CrossRef]
- Marques, T.M.; Wall, R.; O’Sullivan, O.; Fitzgerald, G.F.; Shanahan, F.; Quigley, E.M.; Cotter, P.D.; Cryan, J.F.; Dinan, T.G.; Ross, R.P.; et al. Dietary trans-10, cis-12-conjugated linoleic acid alters fatty acid metabolism and microbiota composition in mice. Br. J. Nutr. 2015, 113, 728–738. [Google Scholar] [CrossRef]
- den Hartigh, L.J.; Gao, Z.; Goodspeed, L.; Wang, S.; Das, A.K.; Burant, C.F.; Chait, A.; Blaser, M.J. Obese Mice Losing Weight Due to trans-10,cis-12 Conjugated Linoleic Acid Supplementation or Food Restriction Harbor Distinct Gut Microbiota. J. Nutr. 2018, 148, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Chidambaram, S.B.; Essa, M.M.; Rathipriya, A.G.; Bishir, M.; Ray, B.; Mahalakshmi, A.M.; Tousif, A.H.; Sakharkar, M.K.; Kashyap, R.S.; Friedland, R.P.; et al. Gut dysbiosis, defective autophagy and altered immune responses in neurodegenerative diseases: Tales of a vicious cycle. Pharmacol. Ther. 2021, 231, 107988. [Google Scholar] [CrossRef] [PubMed]
- Custers; Emma, E.M.; Kiliaan; Amanda, J. Dietary lipids from body to brain. Prog. Lipid Res. 2022, 85, 101144. [Google Scholar] [CrossRef]
- Gorissen, L.; Raes, K.; Weckx, S.; Dannenberger, D.; Leroy, F.; De Vuyst, L.; De Smet, S. Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl. Microbiol. Biotechnol. 2010, 87, 2257–2266. [Google Scholar] [CrossRef]
- Bergamo, P.; Luongo, D.; Maurano, F.; Mazzarella, G.; Stefanile, R.; Rossi, M. Conjugated linoleic acid enhances glutathione synthesis and attenuates pathological signs in MRL/MpJ-Fas(lpr) mice. J. Lipid Res. 2006, 47, 2382–2391. [Google Scholar] [CrossRef]
- Bergamo, P.; Cocca, E.; Monaco, A.; Cozzolino, V.; Boscaino, F.; Ferrandino, I.; Maurano, F.; Rossi, M. Protective effect of Rumenic acid rich cow’s milk against colitis is associated with the activation of Nrf2 pathway in a murine model. Prostaglandins Leukot. Essent. Fat. Acids 2017, 125, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Bergamo, P.; Luongo, D.; Miyamoto, J.; Cocca, E.; Kishino, S.; Ogawa, J.; Tanabe, S.; Rossi, M. Immunomodulatory activity of a gut microbial metabolite of dietary linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, associated with improved antioxidant/detoxifying defences. J. Funct. Foods 2014, 11, 192–202. [Google Scholar] [CrossRef]
- Furumoto, H.; Nanthirudjanar, T.; Kume, T.; Izumi, Y.; Park, S.B.; Kitamura, N.; Kishino, S.; Ogawa, J.; Hirata, T.; Sugawara, T. 10-Oxo-trans-11-octadecenoic acid generated from linoleic acid by a gut lactic acid bacterium Lactobacillus plantarum is cytoprotective against oxidative stress. Toxicol. Appl. Pharmacol. 2016, 296, 1–9. [Google Scholar] [CrossRef]
- Miyamoto, J.; Mizukure, T.; Park, S.B.; Kishino, S.; Kimura, I.; Hirano, K.; Bergamo, P.; Rossi, M.; Suzuki, T.; Arita, M.; et al. A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40-MEK-ERK pathway. J. Biol. Chem. 2015, 290, 2902–2918. [Google Scholar] [CrossRef]
- Yokoji-Takeuchi, M.; Takahashi, N.; Yamada-Hara, M.; Sulijaya, B.; Tsuzuno, T.; Aoki-Nonaka, Y.; Tabeta, K.; Kishino, S.; Ogawa, J.; Yamazaki, K. A bacterial metabolite induces Nrf2-mediated anti-oxidative responses in gingival epithelial cells by activating the MAPK signaling pathway. Arch. Oral Biol. 2020, 110, 104602. [Google Scholar] [CrossRef]
- Druart, C.; Neyrinck, A.M.; Dewulf, E.M.; De Backer, F.C.; Possemiers, S.; Van de Wiele, T.; Moens, F.; De Vuyst, L.; Cani, P.D.; Larondelle, Y.; et al. Implication of fermentable carbohydrates targeting the gut microbiota on conjugated linoleic acid production in high-fat-fed mice. Br. J. Nutr. 2013, 110, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Fleck, A.K.; Hucke, S.; Teipel, F.; Eschborn, M.; Janoschka, C.; Liebmann, M.; Wami, H.; Korn, L.; Pickert, G.; Hartwig, M.; et al. Dietary conjugated linoleic acid links reduced intestinal inflammation to amelioration of CNS autoimmunity. Brain 2021, 144, 1152–1166. [Google Scholar] [CrossRef] [PubMed]
| Disease | Model | Microbiota Alteration Production of SCFAs | Ref. | |
|---|---|---|---|---|
| Diabetes | Randomized clinical trial High-fiber diet | Type 2 diabetes ↓ SCFAs High fiber intake ↑ SCFAs ↑ SCFA-producing bacteria | [28] | |
| Meta analysis Dietary fiber | ↑ Butyrate, propionate ↑ Bifidobacterium | [29] | ||
| Inflammatory Bowel Disease (IBD) | 313 patients | ↓ Acetate-to-butyrate converter Firmicutes (Roseburia) ↓ Propionate ↑ Pathogens (Enterobacteriaceae, Proteobacteria) | [30] | |
| 127 patients 87 healthy controls | ↓ Butyrate-producing bacteria (Firmicutes) ↓ SCFAs (acetate, propionate, butyrate) | [31] | ||
| 10 inactive Crohn patients 10 healthy controls | ↓ SCFA-producing bacteria ↓ Roseburia inulinivorans, ↓ Ruminococcus torques, ↓ Clostridium lavalense, ↓ Bacteroides uniformis ↓ Faecalibacterium prausnitzii | [32] | ||
| Nonalcoholic Fatty Liver Disease | 14 nonalcoholic fatty liver, 18 nonalcoholic steatohepatitis 27 healthy controls | ↑ SCFA levels ↑ SCFA-producing bacteria (Fusobacteriaceae, Prevotellaceae) | [33] | |
| 25 nonalcoholic fatty liver 25 nonalcoholic steatohepatitis 25 healthy donors | ↓ Ruminococcaceae ↓ Clostridiales ↑ Bacteroidetes ↓ Firmicutes | [34] | ||
| 30 patients F0/1 fibrosis stage 27 patients F ≥ 2 fibrosis | ↑ Bacteroidetes (F ≥ 2) ↑ Ruminococcus (F ≥ 2) ↓ Prevotella | [35] | ||
| Neurodegeneration | Parkinson’s Disease | Nonparametric meta-analysis | ↑ Akkermansia ↓ Fecal SCFAs (acetate, propionate, butyrate) | [36] |
| 96 patients 85 controls | ↓ Fecal SCFAs ↑ Plasma SCFAs ↑ Pro-inflammatory bacteria | [37] | ||
| 95 patients 33 controls | ↓ Fecal SCFAs (propionic acetic, butyric) ↑ Plasma SCFA (propionic acetic) | [38] | ||
| Alzheimer’s Disease | 25 patients | ↓ Firmicutes, Bifidobacterium ↑ Bacteroidetes | [39] | |
| 33 dementia 22 mild cognitive impairment 120 subjective cognitive decline | ↓ SCFA-producing bacteria (Ruminococcus, Eubacterium) ↑ AD biomarkers (Amyloid-β1-42 and p-tau concentrations) | [40] | ||
| Mouse model Sodium butyrate supplementation | ↓ Amyloid-β1-42 protein (40%) | [41] | ||
| Phylum | Family | Genus | FFAR3 (GPR41) | FFAR2 (GPR43) | GPR109A | |
|---|---|---|---|---|---|---|
| Firmicutes | Lachnospiraceae | Coprococcus | ACETATE | + + | + + | + + |
| Barnesiella | ||||||
| Ruminococcaceae | ||||||
| Akkermansia | ||||||
| Prevotella | ||||||
| Bifidobacterium | ||||||
| Bacteroidetes | Bacteroidaceae | Bacteroides | PROPIONATE | + | + + | + |
| Prevotellaceae | Prevotella | |||||
| Rikenellaceae | Alistipes | |||||
| Firmicutes | Eubacterium | |||||
| Blautia | ||||||
| Coprococcus | ||||||
| Veillonellaceae | Dialister | |||||
| Acidaminococcaceae | Phascolarctobacterium | |||||
| Verrucomicrobia | Verrucomicrobiaceae | Akkermansia | ||||
| Firmicutes | Lachnospiraceae | Eubacterium | BUTYRATE | + + | + + | + + |
| Roseburia | ||||||
| Clostridium | ||||||
| Eubacterium | ||||||
| Anaerostipes | ||||||
| Coprococcus | ||||||
| Ruminococcaceae | Faecalibacterium | |||||
| Subdoligranulum | ||||||
| Erysipelotrichaceae | Holdemanella |
| Component | Animal Model | Effect on Gut Microbes | Ref. |
|---|---|---|---|
| Astaxanthin | β-carotene oxygenase 2 knockout mice | ↑ Mucispirillum schaedleri, Akkermansia, Muciniphila | [115] |
| Fucoxanthin | azoxymethane/dextran sulfate sodium treated mice | ↑ Lachnospiraceae, ↓ Bacteroidlales, Rikenellaceae | [116] |
| Tomato powder | BCO1/BCO2 double knockout mice | ↑ Lactobacillus, Bifidobacterium, ↓ Bacteroides, Mucispirillum | [117] |
| Apple polyphenol extract | Wild-type mice | ↑ Verrucomicrobia, Akkermansia | [118] |
| Blueberry extract | Sprague–Dawley rats | ↑ Diversity of gut microbes | [119] |
| Curcumin | Wild-type mice | ↑ Akkermansia, Roseburia, Coprococcus | [120] |
| Wild-type mice | ↑ Muribaculaceae, ↓ Bacteroides, Ruminococcaceae | [121] | |
| Epicatechin gallate | Obese diabetic mice | ↑ Firmicutes: Bacteroidetes ratio, ↑ Lactobacillius | [122] |
| Fu brick tea | Donor rats | ↑ Akkermansia maciniphilla, Bacteroides, Alloprevotella | [123] |
| Litchi chinensis seed extract | Zebrafish | ↑ Trichococcus, Muribaculaceae, Lactobacillus, ↓ Micrococcaceae, Staphyllococcus | [124] |
| Peanut skin procyanidin | DSS-induced ulcerative colitis in mice | ↑ Lachnospiraceae, Roseburia, ↓ Bacteroides, Helicobacter, Parabacteroides | [125] |
| Pomegranate fruit pulp | Wild-type mice | ↑ Akkermansia maciniphilla, Parabacteroides distsonis, Bacteroides acidifaciens | [126] |
| Purple sweet potato anthocyanin extract | Obese mice | ↑ Lactobacillus, Bifidobacterium, Akkermansia | [127] |
| Tea polyphenols and polysaccharides | DSS-induced colitis in mice | ↑ Lactobacillus, ↓ Proteobacteria, Enterobacteriaceae | [128] |
| Triadica cochinchinensis honey polyphenol | Cefixime-treated mice | ↓ Firmicutes/Bacteroidetes | [129] |
| Xanthohumol | Male Tac: SW mice | ↓ Porphyromonadaceae, Lachnospiraceae, Lactobacillaceae, ↑ A. muciniphila, P. goldsteinii, A. finegoldii | [130] |
| Fatty Acids | Effect on Gut Microbes | Ref. | ||
|---|---|---|---|---|
| Clinical studies | Omega-3 rich diet | A 45-year-old male consuming 600 mg of omega-3 (daily for 14 days) | ↓ Species diversity ↑ Butyrate-producing bacteria (Eubacterium, Roseburia, Anaerostipes, Coprococcus, Subdoligranulum, Pseudobutyrivibrio) | [138] |
| Enteral supplementation of a fish and safflower blended oil | 32 premature infants with enterostomy (10 weeks) | ↓ pathogenic bacteria (Streptococcus, Clostridium, Escherichia, Pantoea, Serratia, and Citrobacter genera) | [140] | |
| Omega-3 rich diet | Pregnant women | ↑ F. prausnitzii species of the Firmicutes phylum ↓ Bacteroides genus of the Bacteroidetes phylum | [141] | |
| DHA/EPA | 20 Healthy volunteers (8 wks, 4 g/day) | ↑ SCFA-producing bacteria (Bifidobacterium, Roseburia lactobacillus) | [142] | |
| Estimated food intake of omega-3 fatty acids | 876 female twins | ↑ n3-PUFA ↑ SCFA-producing bacteria (Lachnospiraceae family) | [137] | |
| Omega-3 (sardines) (~3 g of EPA + DHA) | 32 patients with type 2 diabetes 100 g of sardines (5 days per week for 6 months) | ↓ Firmicutes/Bacteroidetes ratio, ↑ Prevotella genus in the omega-3 group | [143] | |
| Animal models | n-3 PUFA | male C57BL/6 mice n-3 supplemented (n3+) n-3 deficient (n3−) vs control (CONT) | (n3−) ↓ SCFAs vs. CONT (n3+) ↓ Butyrate vs. CONT | [136] |
| EPA-DHA | HFD-induced obese mice + EPA-DHA | ↑ Firmicutes | [144] | |
| PUFAs omega-6 (n6) omega-3 (n3) | Wild-type mice fed + n3 or n6/(14 wks) | ↓ proportion of Bacteroidetes phylum | [145] | |
| palm oil (PO), olive oil (OO) flaxseed/fish oil (FOO) compared with mice fed a low-fat diet (LF) | C57BL/6J mice fed with High-fat diet (HF+ PO, OO or FOO) compared with mice fed LF | HF+PO ↓ Bacteroidetes comp. to HF+OO HF+FFO ↑ Bifidobacterium comp. to LF | [146] | |
| High-fat diet (45%) with fish oil (FO) or lard (L) | C57Bl/6 Wild-type germ-free mice | FO ↑ Lactobacillus genus and Akkermansia muciniphila sp. L ↑ Bilophila genus of the Proteobacteri phylum | [147] | |
| Omega-3 PUFAs | male Sprague–Dawley rats | ↑ Bifidobacteria | [148] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuciniello, R.; Di Meo, F.; Filosa, S.; Crispi, S.; Bergamo, P. The Antioxidant Effect of Dietary Bioactives Arises from the Interplay between the Physiology of the Host and the Gut Microbiota: Involvement of Short-Chain Fatty Acids. Antioxidants 2023, 12, 1073. https://doi.org/10.3390/antiox12051073
Cuciniello R, Di Meo F, Filosa S, Crispi S, Bergamo P. The Antioxidant Effect of Dietary Bioactives Arises from the Interplay between the Physiology of the Host and the Gut Microbiota: Involvement of Short-Chain Fatty Acids. Antioxidants. 2023; 12(5):1073. https://doi.org/10.3390/antiox12051073
Chicago/Turabian StyleCuciniello, Rossana, Francesco Di Meo, Stefania Filosa, Stefania Crispi, and Paolo Bergamo. 2023. "The Antioxidant Effect of Dietary Bioactives Arises from the Interplay between the Physiology of the Host and the Gut Microbiota: Involvement of Short-Chain Fatty Acids" Antioxidants 12, no. 5: 1073. https://doi.org/10.3390/antiox12051073








