Optimized Ribonucleoprotein Complexes Enhance Prime Editing Efficiency in Zebrafish
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Zebrafish Maintenance
2.3. pegRNA Generation
2.4. PE RNP Preparation and DNA Extraction
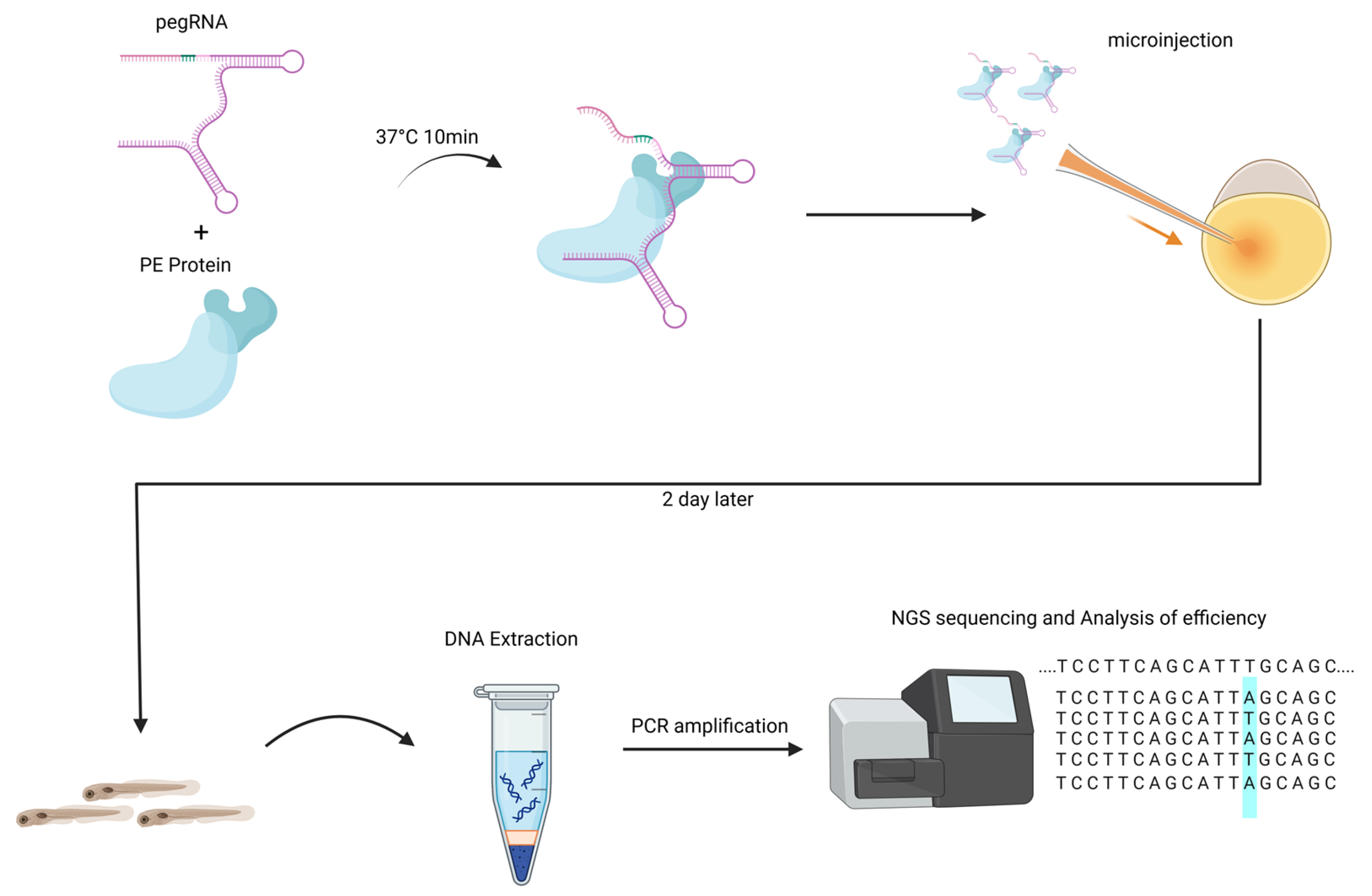
2.5. Microinjection and Image Acquisition in Zebrafish
2.6. Deep Amplicon Sequencing and Data Analysis
3. Results
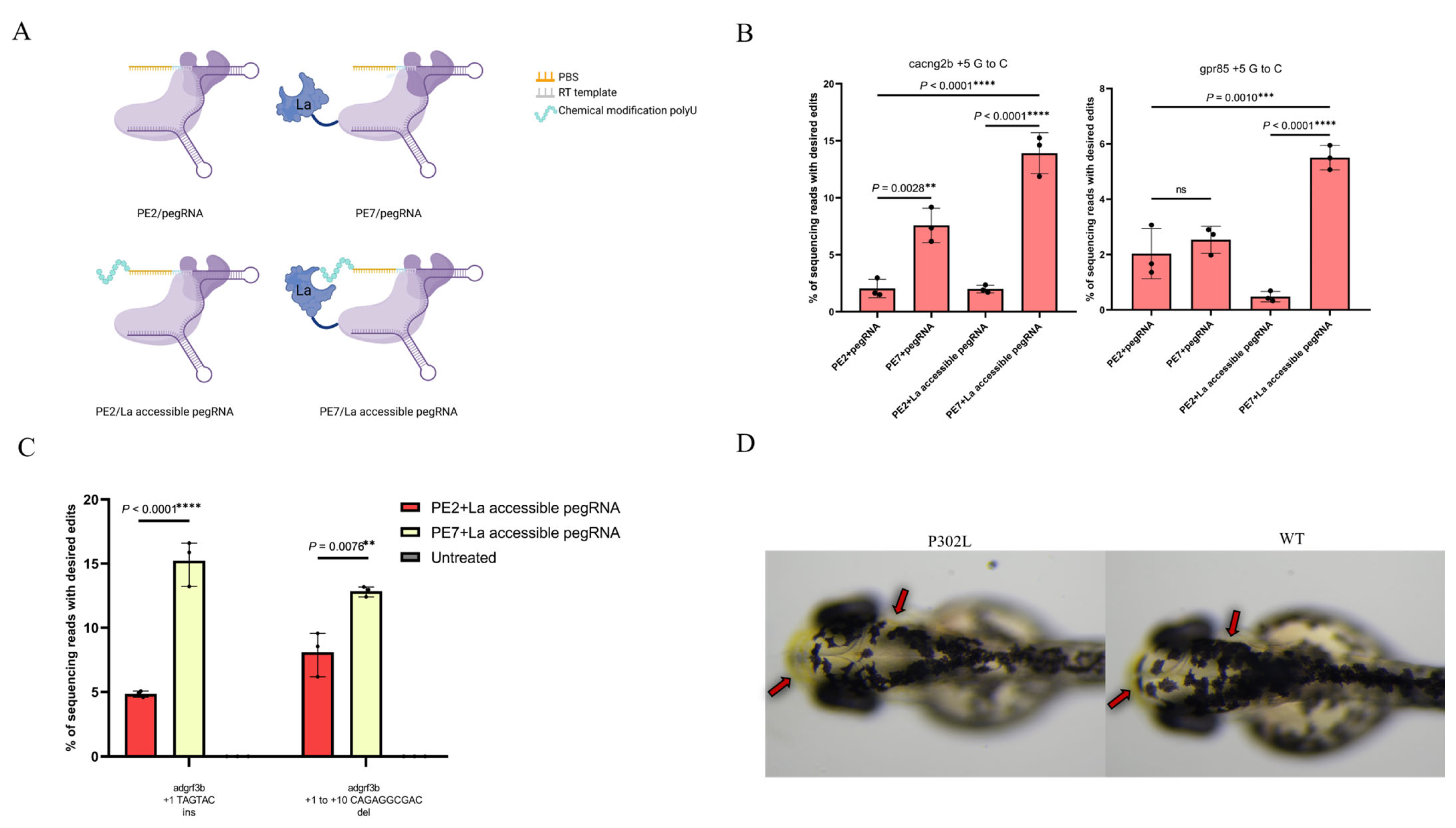
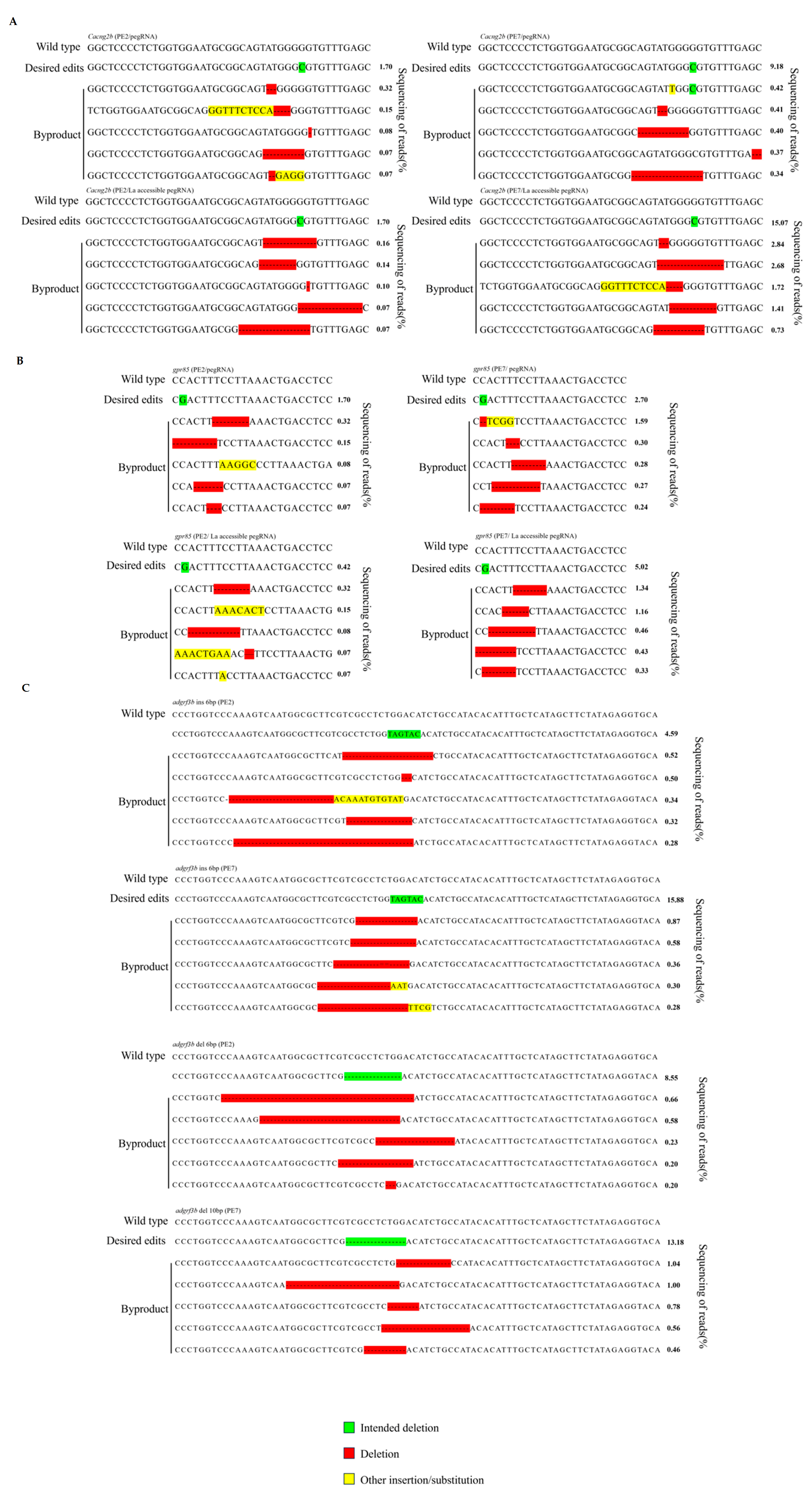
3.1. Optimized PE Significantly Improves the Efficiency of Prime Editing
3.2. Short Indels in Zebrafish
3.3. Editing Efficiency at Complex Disease Mutation tyr P302L
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, D. Gene editing nuclease and its application in tilapia. Sci. Bull. 2017, 62, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef]
- Zhou, L.; Li, M.; Wang, D. Role of sex steroids in fish sex determination and differentiation as revealed by gene editing. Gen. Comp. Endocrinol. 2021, 313, 113893. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.M.; Stevens, J.R.; Santos, E.M.; et al. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef]
- Li, X.Y.; Mei, J.; Ge, C.T.; Liu, X.L.; Gui, J.F. Sex determination mechanisms and sex control approaches in aquaculture animals. Sci. China Life Sci. 2022, 65, 1091–1122. [Google Scholar] [CrossRef]
- Luo, M.; Lu, G.; Yin, H.; Wang, L.; Atuganile, M.; Dong, Z. Fish pigmentation and coloration: Molecular mechanisms and aquaculture perspectives. Rev Aquac. 2021, 13, 2395–2440. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Gaudelli, N.; Komor, A.; Rees, H.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Carrington, B.; Weinstein, R.N.; Sood, R. BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish. Cells 2020, 9, 1690. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Zhang, Y.; Li, L.; Yang, Y.; Fei, J.-F.; Liu, Y.; Qin, W. SpG and SpRY variants expand the CRISPR toolbox for genome editing in zebrafish. Nat. Commun. 2022, 13, 3421. [Google Scholar] [CrossRef]
- Qin, W.; Lu, X.; Liu, Y.; Bai, H.; Li, S.; Lin, S. Precise A•T to G•C base editing in the zebrafish genome. BMC Biol. 2018, 16, 139. [Google Scholar] [CrossRef]
- Rosello, M.; Serafini, M.; Mignani, L.; Finazzi, D.; Giovannangeli, C.; Mione, M.C.; Concordet, J.-P.; Del Bene, F. Disease modeling by efficient genome editing using a near PAM-less base editor in vivo. Nat. Commun. 2022, 13, 3435. [Google Scholar] [CrossRef]
- Qin, W.; Liang, F.; Lin, S.J.; Petree, C.; Huang, K.; Zhang, Y.; Li, L.; Varshney, P.; Mourrain, P.; Liu, Y.; et al. ABE-ultramax for high-efficiency biallelic adenine base editing in zebrafish. Nat. Commun. 2024, 15, 5613. [Google Scholar] [CrossRef]
- Vanhooydonck, M.; De Neef, E.; De Saffel, H.; Boel, A.; Willaert, A.; Callewaert, B.; Claes, K.B.M. Prime editing outperforms homology-directed repair as a tool for CRISPR-mediated variant knock-in in zebrafish. Lab Anim. 2025, 54, 165–172. [Google Scholar] [CrossRef]
- Chemello, F.; Chai, A.C.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Atmanli, A.; Mireault, A.A.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021, 7, eabg4910. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liang, S.Q.; Zheng, C.; Mintzer, E.; Zhao, Y.G.; Ponnienselvan, K.; Mir, A.; Sontheimer, E.J.; Gao, G.; Flotte, T.R.; et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 2021, 12, 2121. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 2021, 7, 1166–1187. [Google Scholar] [CrossRef]
- Zong, Y.; Liu, Y.; Xue, C.; Li, B.; Li, X.; Wang, Y.; Li, J.; Liu, G.; Huang, X.; Cao, X.; et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 2022, 40, 1394–1402. [Google Scholar] [CrossRef]
- Petri, K.; Zhang, W.; Ma, J.; Schmidts, A.; Lee, H.; Horng, J.E.; Kim, D.Y.; Kurt, I.C.; Clement, K.; Hsu, J.Y.; et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 2022, 40, 189–193. [Google Scholar] [CrossRef]
- Chen, P.J.; Liu, D.R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2023, 24, 161–177. [Google Scholar] [CrossRef]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef]
- Yan, J.; Oyler-Castrillo, P.; Ravisankar, P.; Ward, C.C.; Levesque, S.; Jing, Y.; Simpson, D.; Zhao, A.; Li, H.; Yan, W.; et al. Improving prime editing with an endogenous small RNA-binding protein. Nature 2024, 628, 639–647. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, S.; Zong, Y.; Yu, H.; Zhu, Z.; Liu, G.; Kou, L.; Wang, Y.; Qiu, J.-L.; Li, J.; et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021, 39, 923–927. [Google Scholar] [CrossRef]
- Rothgangl, T.; Tálas, A.; Ioannidi, E.I.; Weber, Y.; Böck, D.; Matsushita, M.; Villiger, E.A.; Schmidheini, L.; Moon, W.J.; Lin, P.J.C.; et al. Treatment of a metabolic liver disease in mice with a transient prime editing approach. Nat. Biomed. Eng. 2025. [Google Scholar] [CrossRef]
- Lei, X.; Huang, A.; Chen, D.; Wang, X.; Ji, R.; Wang, J.; Zhang, Y.; Zhang, Y.; Lu, S.; Zhang, K.; et al. Rapid generation of long, chemically modified pegRNAs for prime editing. Nat. Biotechnol. 2024, 43, 1156–1167. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, Y.; Huang, S.; Qu, S.; Cheng, D.; Yao, Y.; Ji, Q.; Wang, X.; Huang, X.; Liu, J. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nat. Commun. 2022, 13, 1856. [Google Scholar] [CrossRef]
- Zhang, Y.; Qin, W.; Lu, X.; Xu, J.; Huang, H.; Bai, H.; Li, S.; Lin, S. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 2017, 8, 118. [Google Scholar] [CrossRef]
- Ponnienselvan, K.; Liu, P.; Nyalile, T.; Oikemus, S.; Maitland, S.A.; Lawson, N.D.; Luban, J.; Wolfe, S.A. Reducing the inherent auto-inhibitory interaction within the pegRNA enhances prime editing efficiency. Nucleic Acids Res. 2023, 51, 6966–6980. [Google Scholar] [CrossRef]
- Zhang, W.; Petri, K.; Ma, J.; Lee, H.; TsaiJ, C.-L.; Joung, K.; Yeh, J.-R.J. Enhancing CRISPR prime editing by reducing misfolded pegRNA interactions. Elife 2024, 12, RP90948. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; Lin, Q. Optimized Ribonucleoprotein Complexes Enhance Prime Editing Efficiency in Zebrafish. Animals 2025, 15, 2295. https://doi.org/10.3390/ani15152295
Qin L, Lin Q. Optimized Ribonucleoprotein Complexes Enhance Prime Editing Efficiency in Zebrafish. Animals. 2025; 15(15):2295. https://doi.org/10.3390/ani15152295
Chicago/Turabian StyleQin, Lang, and Qiupeng Lin. 2025. "Optimized Ribonucleoprotein Complexes Enhance Prime Editing Efficiency in Zebrafish" Animals 15, no. 15: 2295. https://doi.org/10.3390/ani15152295
APA StyleQin, L., & Lin, Q. (2025). Optimized Ribonucleoprotein Complexes Enhance Prime Editing Efficiency in Zebrafish. Animals, 15(15), 2295. https://doi.org/10.3390/ani15152295






