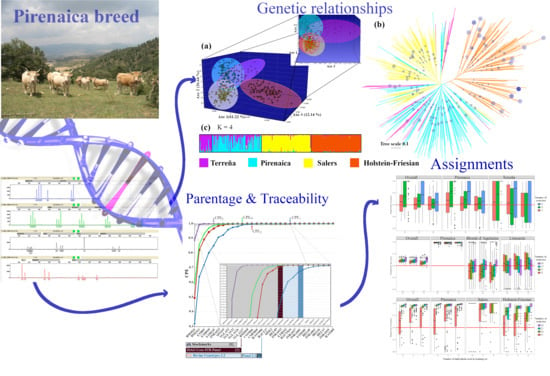
Genetic Characterization of the Local Pirenaica Cattle for Parentage and Traceability Purposes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Sample Genotyping
2.3. Statistical Analysis
3. Results and Discussion
3.1. Genetic Variations and Genetic Relationships
3.2. Individual Identification and Parentage Determination
3.3. Assignment Analysis of Breeds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Commission. Publication of an Application for Registration Pursuant to Article 6(2) of Regulation (EEC) No 2081/92 on the Protection of Geographical Indications and Designations of Origin; European Commission: Brussels, Belgium, 2003; Volume 46. [Google Scholar]
- BOE (Official Bulletin of the Spanish State). Real Decreto 2129/2008, de 26 de Diciembre, por el que se Establece el Programa Nacional de Conservación, Mejora y Fomento de las Razas Ganaderas; BOE: Madrid, Spain, 2008. [Google Scholar]
- Baldo, A.; Rogberg-Muñoz, A.; Prando, A.; Mello Cesar, A.S.; Lirón, J.P.; Sorarrain, N.; Ramelli, P.; Posik, D.M.; Pofcher, E.; Ripoli, M.V.; et al. Effect of consanguinity on Argentinean Angus beef DNA traceability. Meat Sci. 2010, 85, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Oceja, A.; Nuñez, C.; Baeta, M.; Gamarra, D.; de Pancorbo, M.M. Species identification in meat products: A new screening method based on high resolution melting analysis of cyt b gene. Food Chem. 2017, 2017, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Carolino, I.; Sousa, C.O.; Ferreira, S.; Carolino, N.; Silva, F.S.; da Gama, L.T. Implementation of a parentage control system in Portuguese beef-cattle with a panel of microsatellite markers. Genet. Mol. Biol. 2009, 32, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Maudet, C.; Luikart, G.; Taberlet, P. Genetic diversity and assignment tests among seven French cattle breeds based on microsatellite DNA analysis. J. Anim. Sci. 2002, 80, 942–950. [Google Scholar] [CrossRef]
- Shackell, G.H.; Tate, M.L.; Anderson, R.M. Installing a DNA-based traceability system in the meat industry. Proc. Assoc. Advmt. Anim. Breed. Genet. 2001, 14, 533–536. [Google Scholar]
- ISAG. Cattle Molecular Markers and Parentage Testing Workshop. Available online: https://www.isag.us/Docs/ISAG2006_CMMPT.pdf (accessed on 4 September 2020).
- ISAG. Cattle Molecular Markers and Parentage Testing Workshop. Available online: https://www.isag.us/Docs/ISAG2008_CattleParentage.pdf (accessed on 4 September 2020).
- FAO. Molecular Genetic Characterization of Animal Genetic Resources. Available online: http://www.fao.org/3/i2413e/i2413e00.htm (accessed on 4 September 2020).
- Van de Goor, L.H.P.; Koskinen, M.T.; Van Haeringen, W.A. Population studies of 16 bovine STR loci for forensic purposes. Int. J. Leg. Med. 2009, 125, 111–119. [Google Scholar] [CrossRef]
- Arana, A.; Soret, B.; Lasa, I.; Alfonso, L. Meat traceability using DNA markers: Application to the beef industry. Meat Sci. 2002, 61, 367–373. [Google Scholar] [CrossRef]
- Cañón, J.; Alexandrino, P.; Bessa, I.; Carleos, C.; Carretero, Y.; Dunner, S.; Ferran, N.; Garcia, D.; Jordana, J.; Laloë, D.; et al. Genetic diversity measures of local European beef cattle breeds for conservation purposes. Genet. Sel. Evol. 2001, 33, 311–332. [Google Scholar] [CrossRef]
- Rendo, F.; Iriondo, M.; Jugo, B.M.; Aguirre, A.; Mazón, L.I.; Vicario, A.; Gómez, M.; Estonba, A. Analysis of the genetic structure of endangered bovine breeds from the Western Pyrenees using DNA microsatellite markers. Biochem. Genet. 2004, 42, 99–108. [Google Scholar] [CrossRef]
- Gamarra, D.; López-Oceja, A.; Jiménez-moreno, S.; de Pancorbo, M.M. Forensic efficacy of twelve STRs in Spanish cattle. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, 253–255. [Google Scholar] [CrossRef]
- Gamarra, D.; Lopez-Oceja, A.; de Pancorbo, M.M. Genetic characterization and founder effect analysis of recently introduced Salers cattle breed population. Animal 2016, 11, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Amigues, Y.; Boitard, S.; Bertrand, C.; Sancristobal, M.; Rocha, D. Genetic characterization of the Blonde d’Aquitaine cattle breed using microsatellite markers and relationship with three other French cattle populations. J. Anim. Breed. Genet. 2011, 128, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Gamarra, D.; Aldai, N.; Arakawa, A.; Barron, L.J.R.; López-Oceja, A.; de Pancorbo, M.M.; Taniguchi, M. Distinct correlations between lipogenic gene expression and fatty acid composition of subcutaneous fat among cattle breeds. BMC Vet. Res. 2018, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E.A. Performing the exact test of hardy-weinberg proportion for multiple alleles. Biometrics 1992. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Available online: https://kimura.univ-montp2.fr/genetix/ (accessed on 4 September 2020).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Weir, B. Genetic Data Analysis II: Methods for Discrete Population Genetic Data; Sinauer Associates, Ed.; Sinauer Associates is an Imprint of Oxford University Press: Oxford, UK, 1996. [Google Scholar]
- Jamieson, A.; Taylor, S.C.S. Comparisons of three probability formulae for parentage exclusion. Anim. Genet. 1997, 28, 397–400. [Google Scholar] [CrossRef]
- Rannala, B.; Mountain, J.L. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 1997, 94, 197–201. [Google Scholar] [CrossRef]
- Paetkau, D.; Calvert, W.; Stirling, I.; Strobeck, C. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 1995, 4, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Alapetite, A.; Cornuet, J.M.; Paetkau, D.; Baudouin, L.; Estoup, A. GENECLASS2: A software for genetic assignment and first-generation migrant detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.Y.; Marschall, E.A.; Sovic, M.G.; Fries, A.C.; Gibbs, H.L.; Ludsin, S.A. assignPOP: An r package for population assignment using genetic, non-genetic, or integrated data in a machine-learning framework. Methods Ecol. Evol. 2018, 9, 439–446. [Google Scholar] [CrossRef]
- Anderson, E.C. Assessing the power of informative subsets of loci for population assignment: Standard methods are upwardly biased. Mol. Ecol. Resour. 2010, 10, 701–710. [Google Scholar] [CrossRef]
- Wang, J. The computer program structure for assigning individuals to populations: Easy to use but easier to misuse. Mol. Ecol. Resour. 2016, 17, 981–990. [Google Scholar] [CrossRef]
- MacHugh, D.E. Molecular Biogeography and Genetic Structure of Domesticated Cattle. Ph.D. Thesis, Trinity College, University of Dublin, Dublin, Ireland, 1996. [Google Scholar]
- Beja-Pereira, A.; Alexandrino, P.; Bessa, I.; Carretero, Y.; Dunner, S.; Ferrand, N.; Jordana, J.; Laloe, D.; Moazami-Goudarzi, K.; Sanchez, A.; et al. Genetic characterization of Southwestern European bovine breeds: A historical and biogeographical reassessment with a set of 16 microsatellites. J. Hered. 2003, 94, 243–250. [Google Scholar] [CrossRef]
- Ginja, C.; Telo Da Gama, L.; Penedo, M.C.T. Analysis of STR markers reveals high genetic structure in portuguese native cattle. J. Hered. 2010, 101, 201–210. [Google Scholar] [CrossRef]
- Lopez-Oceja, A.; Muro-Verde, A.; Gamarra, D.; Cardoso, S.; De Pancorbo, M.M. New Q lineage found in bovine (Bos taurus) of Iberian Peninsula. Mitochondrial DNA 2016, 27, 3597–3601. [Google Scholar] [CrossRef]
- Zilhao, J. The spread of agro-pastoral economies across Mediterranean Europe: A view from the far west. J. Mediterr. Archaeol. 1993, 6, 5–63. [Google Scholar] [CrossRef]
- Cymbron, T.; Loftus, R.T.; Malheiro, M.I.; Bradley, D.G. Mitochondrial sequence variation suggests an African influence in Portuguese cattle. Proc. R. Soc. B Biol. Sci. 1999, 266, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Martín-Burriel, I.; García-Muro, E.; Zaragoza, P. Genetic diversity analysis of six Spanish native cattle breeds using microsatellites. Anim. Genet. 1999, 30, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Mendizábal, J.A.; Purroy, A.; Aranguren, F.J.; Eguinoa, P.; Arana, A. Evolución de la morfología en la raza vacuna Pirenaica. Arch. Zootec. 1998, 47, 387–395. [Google Scholar]
- Dalvit, C.; De Marchi, M.; Dal Zotto, R.; Zanetti, E.; Meuwissen, T.; Cassandro, M. Genetic characterization of the Burlina cattle breed using microsatellites markers. J. Anim. Breed. Genet. 2008, 125, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhu, C.; Xu, Z.; Jiang, X.; Yang, S.; Chen, A. Microsatellite markers for animal identification and meat traceability of six beef cattle breeds in the Chinese market. Food Control 2017, 78, 469–475. [Google Scholar] [CrossRef]
- Suh, S.; Kim, Y.S.; Cho, C.Y.; Byun, M.J.; Choi, S.B.; Ko, Y.G.; Lee, C.W.; Jung, K.S.; Bae, K.H.; Kim, J.H. Assessment of genetic diversity, relationships and structure among Korean native cattle breeds using microsatellite markers. Asian Australas. J. Anim. Sci. 2014. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statiscal Learning with Applications in R; Springer: New York, NY, USA, 2013; ISBN 9780387781884. [Google Scholar]
- Fernández, M.E.; Goszczynski, D.E.; Lirón, J.P.; Villegas-Castagnasso, E.E.; Carino, M.H.; Ripoli, M.V.; Rogberg-Muñoz, A.; Posik, D.M.; Peral-García, P.; Giovambattista, G. Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability and assessment of parentage in an inbred Angus herd. Genet. Mol. Biol. 2013, 36, 185–191. [Google Scholar] [CrossRef]
- ISAG. ISAG Cattle Core and Additional SNP Panel. Available online: https://www.isag.us/committees.asp?autotry=true&ULnotkn=true (accessed on 5 March 2020).
- Tian, F.; Sun, D.; Zhang, Y. Establishment of paternity testing system using microsatellite markers in Chinese Holstein. J. Genet. Genom. 2008. [Google Scholar] [CrossRef]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNPs and other types of molecular markers. Genet. Sel. Evol. GSE 2008, 40, 241–264. [Google Scholar] [CrossRef]



| Marker | k | Ho | He | PIC | MP | PE-1P | PE-2P | PE-SI | HW |
|---|---|---|---|---|---|---|---|---|---|
| BM1824 | 4 | 0.711 | 0.745 | 0.694 | 0.114 | 0.319 | 0.668 | 0.592 | NS |
| BM2113 | 8 | 0.816 | 0.824 | 0.797 | 0.062 | 0.469 | 0.821 | 0.646 | NS |
| ETH03 | 8 | 0.588 | 0.621 | 0.582 | 0.181 | 0.224 | 0.597 | 0.513 | NS |
| ETH010 | 6 | 0.746 | 0.710 | 0.658 | 0.160 | 0.295 | 0.649 | 0.570 | NS |
| ETH225 | 6 | 0.684 | 0.639 | 0.604 | 0.161 | 0.241 | 0.623 | 0.527 | NS |
| INRA023 | 7 | 0.737 | 0.769 | 0.729 | 0.088 | 0.373 | 0.736 | 0.610 | NS |
| SPS115 | 6 | 0.596 | 0.581 | 0.522 | 0.235 | 0.182 | 0.503 | 0.481 | NS |
| TGLA53 | 11 | 0.754 | 0.820 | 0.791 | 0.063 | 0.462 | 0.815 | 0.643 | NS |
| TGLA122 | 11 | 0.746 | 0.768 | 0.734 | 0.084 | 0.387 | 0.758 | 0.611 | NS |
| TGLA126 | 5 | 0.658 | 0.608 | 0.556 | 0.201 | 0.203 | 0.545 | 0.502 | NS |
| TGLA227 | 11 | 0.860 | 0.865 | 0.845 | 0.039 | 0.559 | 0.882 | 0.672 | NS |
| BM1818 | 7 | 0.728 | 0.783 | 0.750 | 0.080 | 0.401 | 0.770 | 0.620 | NS |
| CSSM66 | 12 | 0.732 | 0.777 | 0.755 | 0.078 | 0.422 | 0.808 | 0.619 | NS |
| CSRM60 | 7 | 0.652 | 0.632 | 0.602 | 0.163 | 0.240 | 0.633 | 0.523 | NS |
| ILSTS006 | 7 | 0.719 | 0.806 | 0.774 | 0.070 | 0.432 | 0.789 | 0.634 | NS |
| HAUT27 | 7 | 0.693 | 0.745 | 0.702 | 0.111 | 0.342 | 0.706 | 0.594 | NS |
| MM12 | 11 | 0.842 | 0.804 | 0.777 | 0.086 | 0.445 | 0.810 | 0.634 | NS |
| HEL09 | 7 | 0.754 | 0.798 | 0.764 | 0.075 | 0.417 | 0.776 | 0.629 | NS |
| INRA032 | 8 | 0.693 | 0.747 | 0.710 | 0.099 | 0.351 | 0.725 | 0.597 | NS |
| ETH152 | 6 | 0.788 | 0.746 | 0.701 | 0.124 | 0.336 | 0.696 | 0.594 | NS |
| HAUT024 | 8 | 0.772 | 0.737 | 0.691 | 0.123 | 0.327 | 0.687 | 0.589 | NS |
| INRA037 | 9 | 0.268 | 0.663 | 0.601 | 0.231 | 0.250 | 0.588 | 0.536 | *** |
| INRA005 | 4 | 0.604 | 0.642 | 0.564 | 0.198 | 0.206 | 0.500 | 0.518 | NS |
| ETH185 | 8 | 0.536 | 0.596 | 0.554 | 0.222 | 0.202 | 0.563 | 0.496 | NS |
| HEL05 | 8 | 0.561 | 0.567 | 0.535 | 0.217 | 0.185 | 0.555 | 0.477 | NS |
| INRA063 | 4 | 0.563 | 0.576 | 0.506 | 0.243 | 0.172 | 0.469 | 0.474 | NS |
| HEL01 | 5 | 0.554 | 0.558 | 0.505 | 0.254 | 0.165 | 0.486 | 0.466 | NS |
| HEL13 | 5 | 0.482 | 0.469 | 0.421 | 0.352 | 0.113 | 0.400 | 0.401 | NS |
| INRA035 | 3 | 0.298 | 0.427 | 0.384 | 0.397 | 0.091 | 0.357 | 0.370 | NS |
| ILST005 | 2 | 0.351 | 0.372 | 0.302 | 0.463 | 0.069 | 0.237 | 0.319 | NS |
| Terreña | Pirenaica | Salers | Holstein-Friesian | |
|---|---|---|---|---|
| Terreña | - | 0.04277 | 0.14579 | 0.14030 |
| Pirenaica | 0.04102 | - | 0.11512 | 0.14115 |
| Salers | 0.12724 | 0.10324 | - | 0.24822 |
| Holstein-Friesian | 0.12304 | 0.12369 | 0.19886 | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gamarra, D.; Taniguchi, M.; Aldai, N.; Arakawa, A.; Lopez-Oceja, A.; de Pancorbo, M.M. Genetic Characterization of the Local Pirenaica Cattle for Parentage and Traceability Purposes. Animals 2020, 10, 1584. https://doi.org/10.3390/ani10091584
Gamarra D, Taniguchi M, Aldai N, Arakawa A, Lopez-Oceja A, de Pancorbo MM. Genetic Characterization of the Local Pirenaica Cattle for Parentage and Traceability Purposes. Animals. 2020; 10(9):1584. https://doi.org/10.3390/ani10091584
Chicago/Turabian StyleGamarra, David, Masaaki Taniguchi, Noelia Aldai, Aisaku Arakawa, Andres Lopez-Oceja, and Marian M. de Pancorbo. 2020. "Genetic Characterization of the Local Pirenaica Cattle for Parentage and Traceability Purposes" Animals 10, no. 9: 1584. https://doi.org/10.3390/ani10091584
APA StyleGamarra, D., Taniguchi, M., Aldai, N., Arakawa, A., Lopez-Oceja, A., & de Pancorbo, M. M. (2020). Genetic Characterization of the Local Pirenaica Cattle for Parentage and Traceability Purposes. Animals, 10(9), 1584. https://doi.org/10.3390/ani10091584