cAMP Response Element Binding Protein 1 (CREB1) Promotes Monounsaturated Fatty Acid Synthesis and Triacylglycerol Accumulation in Goat Mammary Epithelial Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Goat Mammary Gland Tissue Collection
2.2. Cloning of the CDS Region of CREB1 and Bioinformatics Analysis
2.3. Generation of Adenovirus
2.4. Cell Culture and Treatment
2.5. RNA Extraction and Quantitative Real-Time PCR
2.6. Total Fatty Acid Extraction and Analysis
2.7. Measurement of Total Cellular TAG
2.8. Statistical Analysis
3. Results
3.1. Cloning and Bioinformatics Analysis of the CREB1 Gene CDS Region
3.2. Expression Patterns of CREB1 in Different Lactation Stages in Dairy Goat
3.3. CREB1 Overexpression Influences Expression of Genes Related to Lipogenesis in GMEC
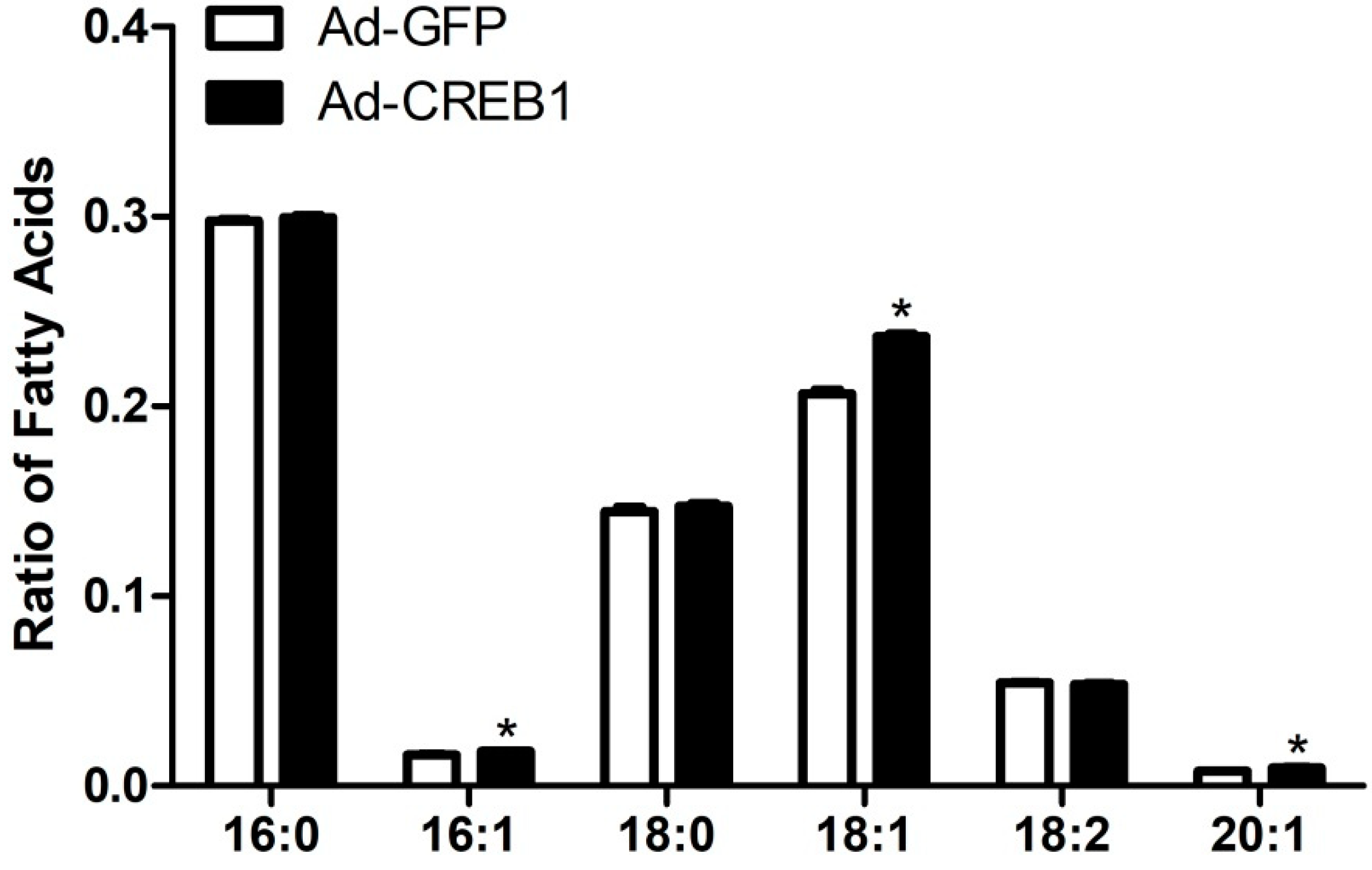
3.4. CREB1 Overexpression Alters Content of Fatty Acids
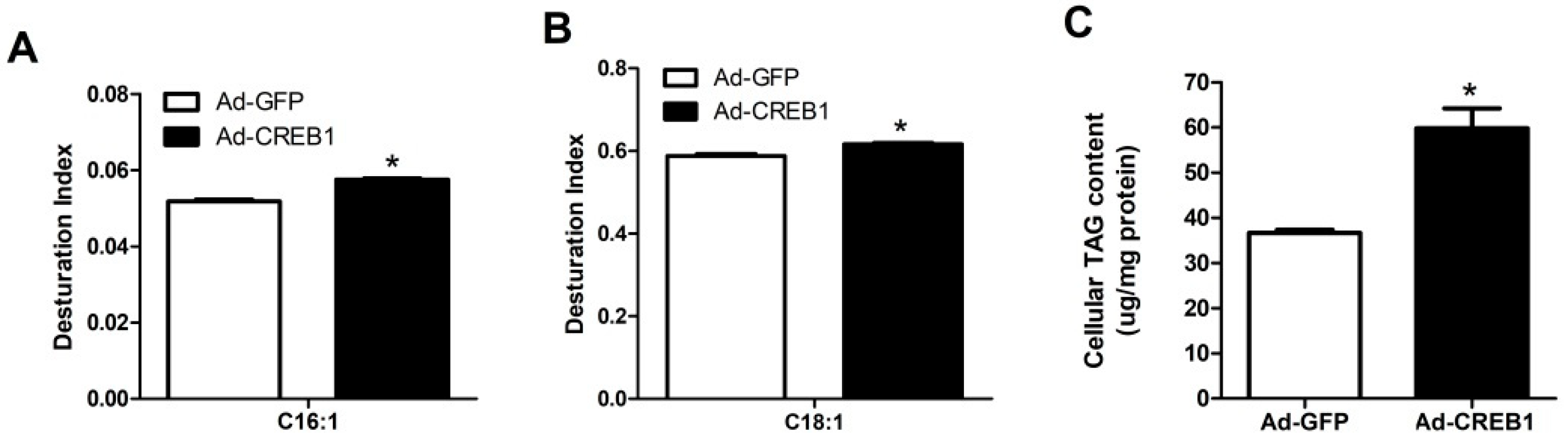
3.5. CREB1 Overexpression Upregulates the Desaturation Index of C16:1, C18:1 and TAG
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yao, D.; Luo, J.; He, Q.; Wu, M.; Shi, H.; Wang, H.; Wang, M.; Xu, H.; Loor, J.J. Thyroid hormone responsive (THRSP) promotes the synthesis of medium-chain fatty acids in goat mammary epithelial cells. J. Dairy Sci. 2016, 99, 3124–3133. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Yoshikawa, T.; Amemiya-Kudo, M.; Hasty, A.H.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Ohashi, K.; et al. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 2002, 43, 911–920. [Google Scholar]
- Reh, W.; Maga, E.; Collette, N.; Moyer, A.; Conrad-Brink, J.; Taylor, S.; Depeters, E.; Oppenheim, S.; Rowe, J.; Bondurant, R.; et al. Hot Topic: Using a Stearoyl-CoA Desaturase Transgene to Alter Milk Fatty Acid Composition. J. Dairy Sci. 2004, 87, 3510–3514. [Google Scholar] [CrossRef] [Green Version]
- Weiss-Hersh, K.; Garcia, A.L.; Marosvölgyi, T.; Szklenár, M.; Decsi, T.; Rühl, R. Saturated and monounsaturated fatty acids in membranes are determined by the gene expression of their metabolizing enzymes SCD1 and ELOVL6 regulated by the intake of dietary fat. Eur. J. Nutr. 2020, 59, 2759–2769. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Kato, T.; Atsumi, A.; Yamamoto, T.; Inoue, N.; Ishikawa, M.; Okada, S.; Ishigaki, N.; et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 2007, 13, 1193–1202. [Google Scholar] [CrossRef]
- Shi, H.; Wu, M.; Zhu, J.; Zhang, C.; Yao, D.; Luo, J.; Loor, J.J. Fatty acid elongase 6 plays a role in the synthesis of long-chain fatty acids in goat mammary epithelial cells. J. Dairy Sci. 2017, 100, 4987–4995. [Google Scholar] [CrossRef] [Green Version]
- Green, C.D.; Ozguden-Akkoc, C.G.; Wang, Y.; Jump, N.B.; Olson, L.K. Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species. J. Lipid Res. 2010, 51, 1871–1877. [Google Scholar] [CrossRef] [Green Version]
- Scott, R.; Bourtchuladze, R.; Gossweiler, S.; Dubnau, J.; Tully, T. CREB and the discovery of cognitive enhancers. J. Mol. Neurosci. 2002, 19, 171–177. [Google Scholar] [CrossRef]
- Della-Fazia, M.A.; Servillo, G.; Sassone-Corsi, P. Cyclic AMP signalling and cellular proliferation: Regulation of CREB and CREM. FEBS Lett. 1997, 410, 22–24. [Google Scholar] [CrossRef] [Green Version]
- Eortega-Martinez, S. A new perspective on the role of the CREB family of transcription factors in memory consolidation via adult hippocampal neurogenesis. Front. Mol. Neurosci. 2015, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Reusch, J.E.B.; Colton, L.A.; Klemm, D.J. CREB Activation Induces Adipogenesis in 3T3-L1 Cells. Mol. Cell. Biol. 2000, 20, 1008–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzig, S.; Hedrick, S.; Morantte, I.; Koo, S.-H.; Galimi, F.; Montminy, M. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-gamma. Nature 2003, 426, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.W.; Klemm, D.J.; Vinson, C.; Lane, M.D. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J. Biol. Chem. 2004, 279, 4471–4478. [Google Scholar] [CrossRef] [Green Version]
- Erion, D.M.; Ignatova, I.D.; Yonemitsu, S.; Nagai, Y.; Chatterjee, P.; Weismann, D.; Hsiao, J.J.; Zhang, D.; Iwasaki, T.; Stark, R.; et al. Prevention of Hepatic Steatosis and Hepatic Insulin Resistance by Knockdown of cAMP Response Element-Binding Protein. Cell Metab. 2009, 10, 499–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliner, J.D.; Andresen, J.M.; Hansen, S.K.; Zhou, S.; Tjian, R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 1996, 10, 2903–2911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Luo, J.; Tian, H.; Li, J.; Zhang, X.; Chen, Z.; Li, M.; Loor, J.J. Rapid communication: Lipid metabolic gene expression and triacylglycerol accumulation in goat mammary epithelial cells are decreased by inhibition of SREBP-1. J. Anim. Sci. 2018, 96, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Farr, V.; Stelwagen, K.; Cate, L.; Molenaar, A.J.; McFadden, T.; Davis, S. An Improved Method for the Routine Biopsy of Bovine Mammary Tissue. J. Dairy Sci. 1996, 79, 543–549. [Google Scholar] [CrossRef]
- Shi, H.; Zhao, W.; Luo, J.; Yao, D.; Sun, Y.; Li, J.; Loor, J.; Shi, H. Peroxisome proliferator-activated receptor γ1 and γ2 isoforms alter lipogenic gene networks in goat mammary epithelial cells to different extents. J. Dairy Sci. 2014, 97, 5437–5447. [Google Scholar] [CrossRef]
- Yao, D.; Luo, J.; He, Q.; Xu, H.; Li, J.; Shi, H.; Wang, H.; Chen, Z.; Loor, J.J. Liver X receptor α promotes the synthesis of monounsaturated fatty acids in goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase 1 in an SREBP-1-dependent manner. J. Dairy Sci. 2016, 99, 6391–6402. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.B.; Luo, J.; Yao, D.W.; Zhu, J.J.; Xu, H.F.; Shi, H.P. and Loor, J.J. Peroxisome proliferator-activated receptor-Y stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase. J. Dairy Sci. 2013, 96, 7844–7853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Luo, J.; Zhao, W.; Yang, Y.; Tian, H.; Shi, H.; Bionaz, M. Overexpression of SREBP1 (sterol regulatory element binding protein 1) promotes de novo fatty acid synthesis and triacylglycerol accumulation in goat mammary epithelial cells. J. Dairy Sci. 2016, 99, 783–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Luo, J.; Zhang, L.; Zhu, J. MicroRNAs Synergistically Regulate Milk Fat Synthesis in Mammary Gland Epithelial Cells of Dairy Goats. Gene Expr. 2013, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genom. 2007, 29, 312–319. [Google Scholar] [CrossRef]
- Xu, H.; Luo, J.; Wang, H.; Zhang, T.; Tian, H.; Yao, D.; Loor, J.J.; Wang, H. Sterol regulatory element binding protein-1 (SREBP-1)c promoter: Characterization and transcriptional regulation by mature SREBP-1 and liver X receptor α in goat mammary epithelial cells. J. Dairy Sci. 2016, 99, 1595–1604. [Google Scholar] [CrossRef] [Green Version]
- Loor, J.J.; Herbein, J.H. Reduced Fatty Acid Synthesis and Desaturation Due to Exogenous trans10, cis12-CLA in Cows Fed Oleic or Linoleic Oil. J. Dairy Sci. 2003, 86, 1354–1369. [Google Scholar] [CrossRef] [Green Version]
- Hegyi, K.; Falus, A.; Toth, S. Elevated CREB activity in embryonic fibroblasts of gene-targeted histamine deficient mice. Inflamm. Res. 2007, 56, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Vankoningsloo, S.; De Pauw, A.; Houbion, A.; Tejerina, S.; Demazy, C.; De Longueville, F.; Bertholet, V.; Renard, P.; Remacle, J.; Holvoet, P.; et al. CREB activation induced by mitochondrial dysfunction triggers triglyceride accumulation in 3T3-L1 preadipocytes. J. Cell Sci. 2006, 119, 1266–1282. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Luo, J.; Xu, H.; Wang, M.; Zhu, J.; Shi, H.; Haile, A.B.; Wang, H.; Sun, Y. Fatty acid synthase promoter: Characterization, and transcriptional regulation by sterol regulatory element binding protein-1 in goat mammary epithelial cells. Gene 2015, 561, 157–164. [Google Scholar] [CrossRef]
- Yao, D.; Luo, J.; He, Q.; Shi, H.; Li, J.; Wang, H.; Xu, H.; Chen, Z.; Yi, Y.; Loor, J.J. SCD1 Alters Long-Chain Fatty Acid (LCFA) Composition and Its Expression Is Directly Regulated by SREBP-1 and PPARγ 1 in Dairy Goat Mammary Cells. J. Cell. Physiol. 2017, 232, 635–649. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 Are the Most Abundant Isoforms in Bovine Mammary Tissue and Their Expression Is Affected by Stage of Lactation. J. Nutr. 2008, 138, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wu, L.; Wang, X.; Xin, Y.; Zan, L. Tissue expression analysis, cloning and characterization of the 5′-regulatory region of the bovine FABP3 gene. Mol. Biol. Rep. 2016, 43, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Lengi, A.J.; Corl, B. Bovine Brain Region-Specific Stearoyl-CoA Desaturase Expression and Fatty Acid Composition. Lipids 2015, 50, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Shang, P.; Zheng, F.; Han, F.; Song, Y.; Pan, Z.; Yu, S.; Zhuang, X.; Chen, S. Lipin1 mediates cognitive impairment in fld mice via PKD-ERK pathway. Biochem. Biophys. Res. Commun. 2020, 525, 286–291. [Google Scholar] [CrossRef]
- Zhang, W.; Zhong, W.; Sun, Q.; Sun, X.; Zhou, Z. Adipose-specific lipin1 overexpression in mice protects against alcohol-induced liver injury. Sci. Rep. 2018, 8, 408. [Google Scholar] [CrossRef] [Green Version]
- Chandran, K.; Goswami, S.; Sharma-Walia, N. Implications of a peroxisome proliferator-activated receptor alpha (PPARα) ligand clofibrate in breast cancer. Oncotarget 2015, 7, 15577–15599. [Google Scholar] [CrossRef]
- Csaki, L.S.; Dwyer, J.R.; Fong, L.G.; Tontonoz, P.; Young, S.G.; Reue, K. Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog. Lipid Res. 2013, 52, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Luz, A.L.; Kassotis, C.D.; Stapleton, H.M.; Meyer, J.N. The high-production volume fungicide pyraclostrobin induces triglyceride accumulation associated with mitochondrial dysfunction, and promotes adipocyte differentiation independent of PPARγ activation, in 3T3-L1 cells. Toxicology 2018, 393, 150–159. [Google Scholar] [CrossRef]
- Reusch, J.E.B.; Klemm, D.J. Inhibition of cAMP-response Element-binding Protein Activity Decreases Protein Kinase B/Akt Expression in 3T3-L1 Adipocytes and Induces Apoptosis. J. Biol. Chem. 2002, 277, 1426–1432. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |



| Gene1 | Accession No. | Primer Sequences (5′-3′) | Source |
|---|---|---|---|
| ACACA | XM_005693156.1 | F: CTCCAACCTCAACCACTACGG | [21] |
| R: GGGGAATCACAGAAGCAGCC | |||
| AGPAT6 | NM_001083669.1 | F: AAGCAAGTTGCCCATCCTCA | [21] |
| R: AAACTGTGGCTCCAATTTCGA | |||
| CREB1 | MK158073.1 | F: CACTCAGCCAGGCACTACCA | This manuscript |
| R: GGAAGACGCCATAACAACCC | |||
| DGAT1 | XM_005688895.1 | F: CCACTGGGACCTGAGGTGTC | [21] |
| R: GCATCACCACACACCAATTCA | |||
| ELOVL6 | NM_001102155.1 | F: GGAAGCCTTTAGTGCTCTGGTC | [22] |
| R: ATTGTATCTCCTAGTTCGGGTGC | |||
| FABP3 | NM_001285701.1 | F: GATGAGACCACGGCAGATG | [21] |
| R: GTCAACTATTTCCCGCACAAG | |||
| FASN | DQ915966.3 | F: GGGCTCCACCACCGTGTTCCA | [21] |
| R: GCTCTGCTGGGCCTGCAGCTG | |||
| GPAM | AY515690 | F: ATTGACCCTTGGCACGATAG | [21] |
| R: GATGAGACCACGGCAGATG | |||
| LPL | DQ997818 | F: AGGACACTTGCCACCTCATTC | [21] |
| R: TTGGAGTCTGGTTCCCTCTTGTA | |||
| LPIN1 | NM_002707716 | F: TCCCTGCTCGGACGTAATTG | [23] |
| R: TGGCCACCAGAATAAAGCATG | |||
| MRPL39 | XM_005674737.1 | F: AGGTTCTCTTTTGTTGGCATCC | [24] |
| R: TTGGTCAGAGCCCCAGAAGT | |||
| RPS9 | DT860044 | F: CCTCGACCAAGAGCTGAAG | [24] |
| R: CCTCCAGACCTCACGTTTGTTC | |||
| SCD1 | GU947654 | F: CCATCGCCTGTGGAGTCAC | [21] |
| R: GTCGGATAAATCTAGCGTAGCA | |||
| SREBF1 | HM443643.1 | F: ACGCCATCGAGAAACGCTAC | [21] |
| R: GTGCGCAGACTCAGGTTCTC | |||
| UXT | XM_005700842.1 | F: TGTGGCCCTTGGATATGGTT | [24] |
| R: GGTTGTCGCTGAGCTCTGTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, D.; Yang, C.; Ma, J.; Chen, L.; Luo, J.; Ma, Y.; Loor, J.J. cAMP Response Element Binding Protein 1 (CREB1) Promotes Monounsaturated Fatty Acid Synthesis and Triacylglycerol Accumulation in Goat Mammary Epithelial Cells. Animals 2020, 10, 1871. https://doi.org/10.3390/ani10101871
Yao D, Yang C, Ma J, Chen L, Luo J, Ma Y, Loor JJ. cAMP Response Element Binding Protein 1 (CREB1) Promotes Monounsaturated Fatty Acid Synthesis and Triacylglycerol Accumulation in Goat Mammary Epithelial Cells. Animals. 2020; 10(10):1871. https://doi.org/10.3390/ani10101871
Chicago/Turabian StyleYao, Dawei, Chunlei Yang, Jing Ma, Lili Chen, Jun Luo, Yi Ma, and Juan. J. Loor. 2020. "cAMP Response Element Binding Protein 1 (CREB1) Promotes Monounsaturated Fatty Acid Synthesis and Triacylglycerol Accumulation in Goat Mammary Epithelial Cells" Animals 10, no. 10: 1871. https://doi.org/10.3390/ani10101871
APA StyleYao, D., Yang, C., Ma, J., Chen, L., Luo, J., Ma, Y., & Loor, J. J. (2020). cAMP Response Element Binding Protein 1 (CREB1) Promotes Monounsaturated Fatty Acid Synthesis and Triacylglycerol Accumulation in Goat Mammary Epithelial Cells. Animals, 10(10), 1871. https://doi.org/10.3390/ani10101871






