Pet Pyometra: Correlating Bacteria Pathogenicity to Endometrial Histological Changes
Abstract
:1. Introduction
2. Results
2.1. Bacterial Isolates from Intrauterine Content
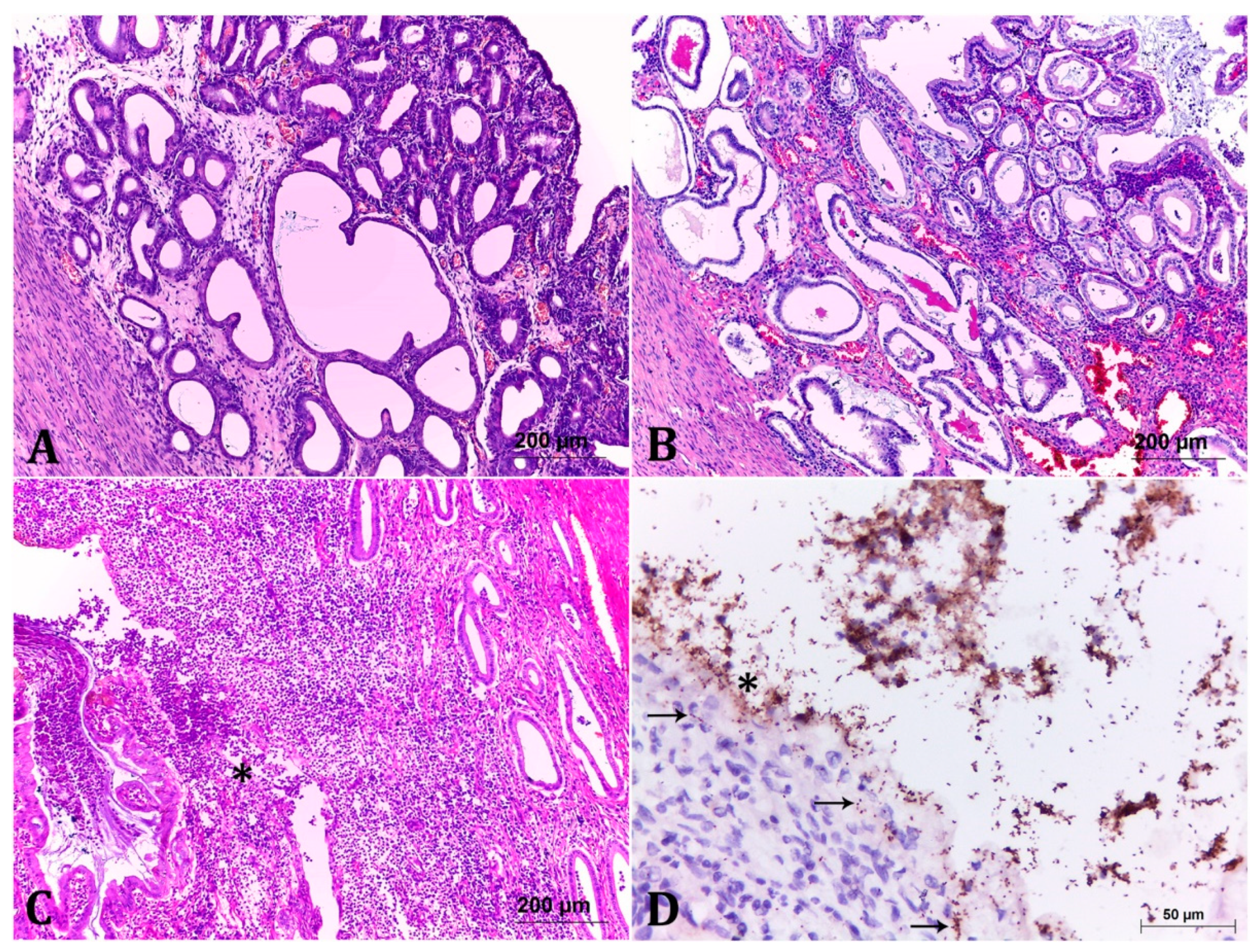
2.2. Classification of the Endometrial Histopathological Lesions from Pet Pyometra
2.3. Immunohistochemistry (IHC) Showed Important Aspects about E. coli Intrauterine Infection
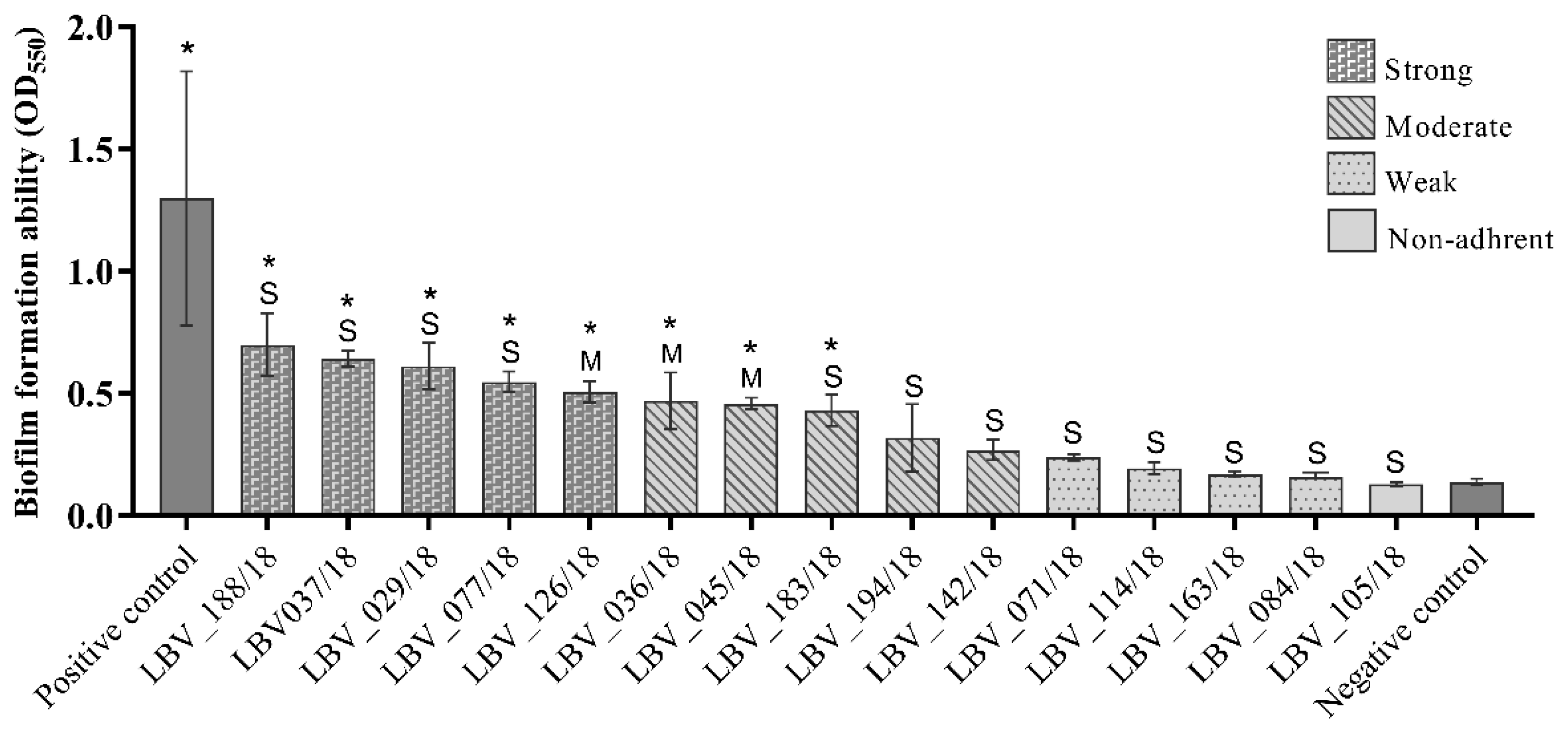
2.4. Extracellular Matrix Components Expression and In Vitro Biofilm Formation Were Widespread among the E. coli Strains
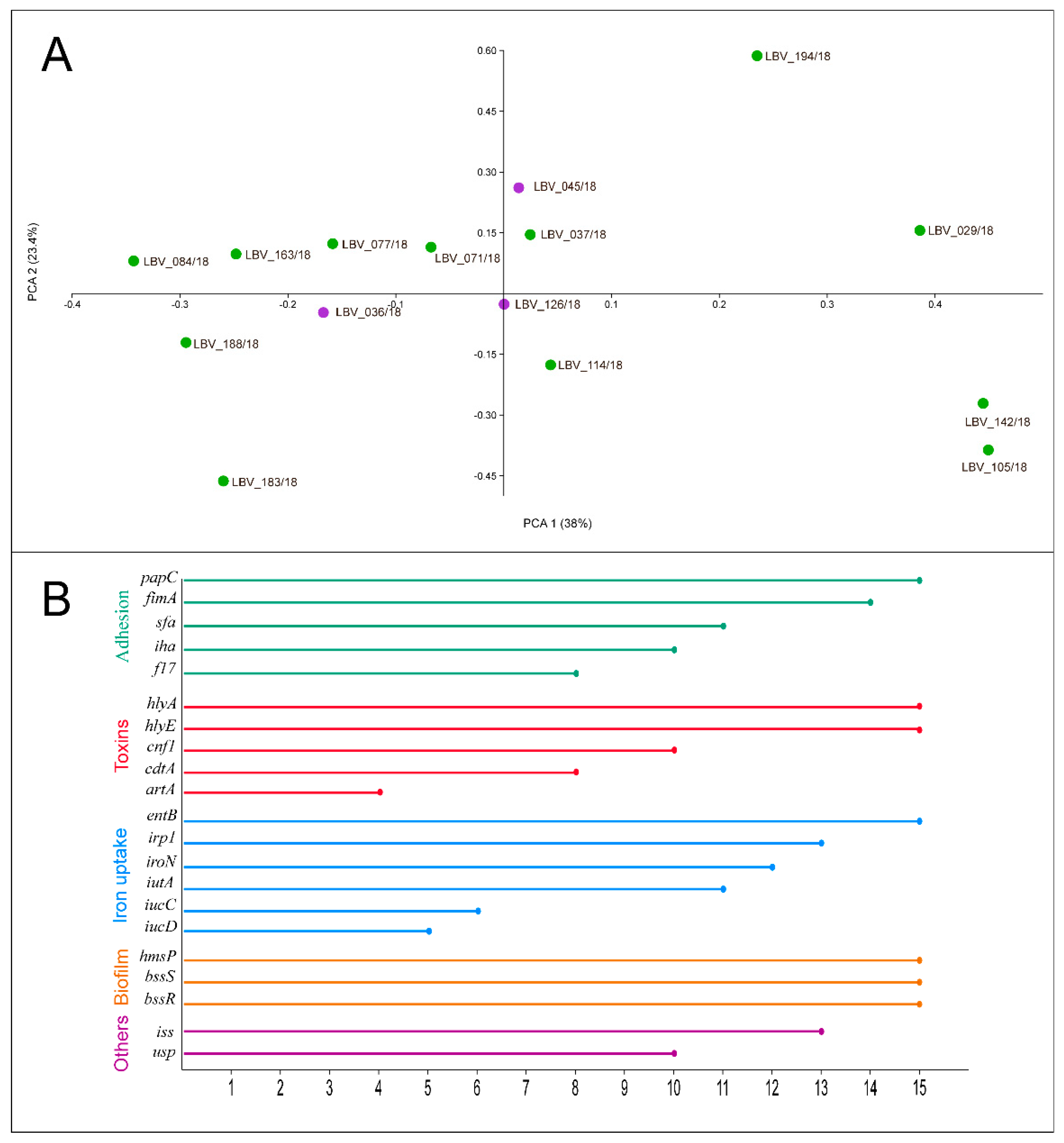
2.5. Virulence Genotyping of the E. coli Strains Highlights the Pathogenic Capacity of This Pathotype
3. Discussion
4. Materials and Methods
4.1. Collection of Samples from Pyometra Cases
4.2. Bacterial Isolation
4.3. Endometrial Histopathological Analyses
4.4. In Vitro Biofilm Formation Assay with the E. coli Strains
4.5. Expression of Extracellular Matrix Components in the E. coli Strains
4.6. Virulence Genotype of the E. coli Strains
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Qian, C.; Jiang, C.; Hou, J. The endometrium histopathology and cell ultrastructure in bitches with pyometra induced using progesterone and Escherichia coli. Tissue Cell 2020, 67. [Google Scholar] [CrossRef]
- Schlafer, D.H.; Foster, R.A. Female genital system. In Pathology of Domestic Animals, 6th ed.; Maxie, M.G., Ed.; Elsevier: St. Louise, MO, USA, 2016; Volume 3, pp. 390–392. [Google Scholar]
- De Bosschere, H.; Ducatelle, L.R.; Vermeirsch, H.; Van, W.; Broeck, D.; Coryn, M. Cystic endometrial hyperplasia-pyometra complex in the bitch: Should the two entities be disconnected? Theriogenology 2001, 55, 1509–1519. [Google Scholar] [CrossRef]
- Mateus, L.; Henriques, S.; Merino, C.; Pomba, C.; Lopes da Costa, L.; Silva, E. Virulence genotypes of Escherichia coli canine isolates from pyometra, cystitis and fecal origin. Vet. Microbiol. 2013, 166, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Wernicki, A.; Krzyzanowski, J.; Puchalski, A. Characterization of Escherichia coli strains associated with canine pyometra. Pol. J. Vet. Sci. 2002, 5, 51–56. [Google Scholar]
- Hagman, R. Pyometra in small animals. Vet. Clin. North. Am. Small Anim Pract 2018, 48, 639–661. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Yoon, H.Y.; Lee, M.H.; Kim, J.H. Canine pyometra associated with Bacillus species: A case report. Vet. Med. 2018, 63, 143–149. [Google Scholar] [CrossRef] [Green Version]
- DeSilva, U.; Lyman, C.C.; Holyoak, G.R.; Meinkoth, K.; Wieneke, X.; Chillemi, K.A. Canine endometrial and vaginal microbiomes reveal distinct and complex ecosystems. PLoS ONE 2019, 14, e0210157. [Google Scholar] [CrossRef]
- Henriques, S.; Silva, E.; Lemsaddek, A.; Lopes-da-Costa, L.; Mateus, L. Genotypic and phenotypic comparison of Escherichia coli from uterine infections with different outcomes: Clinical metritis in the cow and pyometra in the bitch. Vet. Microbiol. 2014, 170, 109–116. [Google Scholar] [CrossRef]
- Lopes, C.E.; de Carli, S.; Weber, M.N.; Fonseca, A.C.V.; Tagliari, N.J.; Foresti, L.; Cibulski, S.P.; Mayer, F.Q.; Canal, C.W.; Siqueira, F.M. Insights on the genetic features of endometrial pathogenic Escherichia coli strains from pyometra in companion animals: Improving the knowledge about pathogenesis. Infect. Genet. Evol. 2020, 85, 104453. [Google Scholar] [CrossRef]
- Fiamengo, T.E.; Runcan, E.E.; Premanandan, C.; Blawut, B.; Coutinho da Silva, M.A. Evaluation of biofilm production by Escherichia coli isolated from clinical cases of canine pyometra. Top. Companion Anim. Med. 2020, 39, 100429. [Google Scholar] [CrossRef]
- Gyles, C.L.; Fairbrother, J.M. Escherichia coli. In Pathogenesis of Bacterial Infection in Animals, 4th ed.; Gyles, C.L., Ed.; Blackwell Publishing: Ames, IA, USA, 2010; pp. 267–289. [Google Scholar]
- Brannon, J.R.; Dunigan, T.L.; Beebout, C.J.; Ross, T.; Wiebe, M.A.; Reynolds, W.S.; Hadjifrangiskou, M. Invasion of vaginal epithelial cells by Uropathogenic Escherichia coli. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Sheldon, I.M.; Rycroft, A.N.; Dogan, B.; Craven, M.; Bromfield, J.J.; Chandler, A.; Roberts, M.H.; Price, S.B.; Gilbert, R.O.; Simpson, K.W. Specific strains of Escherichia coli are pathogenic for the endometrium of cattle and cause Pelvic Inflammatory Disease in cattle and mice. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [Green Version]
- MacFaddin, J.F. Biochemical Tests for Identification of Medical Bacteria, 3rd ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2000; 928p. [Google Scholar]
- Bizzini, A.; Durussel, C.; Bille, J.; Greub, G.; Prod’hom, G. Performance of Matrix-assisted Laser Desorption Ionization-time of Flight Mass Spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 2010, 48, 1549–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neville, S.A.; LeCordier, A.; Ziochos, H.; Chater, M.J.; Gosbell, I.B.; Maley, M.W.; van Hal, S.J. Utility of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 2011, 49, 2980–2984. [Google Scholar] [CrossRef] [Green Version]
- De Lorenzo, C.; de Andrade, C.P.; Machado, V.S.L.; Bianchi, M.V.; Rolim, V.M.; Cruz, R.A.S.; Driemeier, D. Piglet colibacillosis diagnosis based on Multiplex Polymerase Chain Reaction and Immunohistochemistry of paraffin-embedded tissues. J. Vet. Sci. 2018, 19, 27–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified Microtiter-plate test for quantification of Staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Solomon, E.B.; Niemira, B.A.; Sapers, G.M.; Annous, B.A. Biofilm formation, cellulose production, and Curli biosynthesis by Salmonella originating from produce, animal, and clinical sources. J. Food Prot. 2005, 68, 906–912. [Google Scholar] [CrossRef]
- Bokranz, W.; Wang, X.; Tschäpe, H.; Römling, U. Expression of cellulose and Curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 2005, 54, 1171–1182. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past: Paleontological Statistics Software package for education and data analysis. Palaeontol. Electronica 2001, 4, 1–9. Available online: http://palaeo-electronica.org/2001_1/past/issue1_01.htm (accessed on 23 May 2021).
- Legendre, P.; Legendre, L. Ordination in reduced space. In Numerical Ecology, 2nd ed.; Legendre, L., Ed.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 424–443. [Google Scholar]
- Abramson, J.H. WINPEPI updated: Computer programs for epidemiologists, and their teaching potential. Epidemiol. Perspect. Innov. 2011, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Sample ID | Endometrial Severity Degree | Host | Bacterial Isolation * |
|---|---|---|---|
| LBV_024/18 | Mild | Canine | Bacillus sp. + Staphylococcus sp. |
| LBV_038/18 | Mild | Feline | No growth |
| LBV_041/18 | Mild | Canine | No growth |
| LBV_043/18 | Mild | Canine | Enterococcus sp. |
| LBV_036/18 | Moderate | Feline | Escherichia coli |
| LBV_045/18 | Moderate | Canine | Escherichia coli |
| LBV_054/18 | Moderate | Canine | Streptococcus sp. |
| LBV_070/18 | Moderate | Feline | Enterococcus sp. |
| LBV_072/18 | Moderate | Canine | Escherichia coli + Staphylococcus pseudintermedius |
| LBV_115/18 | Moderate | Canine | No growth |
| LBV_121/18 | Moderate | Canine | Streptococcus sp. |
| LBV_126/18 | Moderate | Feline | Escherichia coli |
| LBV_179/18 | Moderate | Canine | Enterococcus sp. |
| LBV_029/18 | Severe | Canine | Escherichia coli |
| LBV_034/18 | Severe | Feline | Staphylococcus pseudintermedius + Streptococcus sp. |
| LBV_037/18 | Severe | Canine | Escherichia coli |
| LBV_053/18 | Severe | Canine | No growth |
| LBV_071/18 | Severe | Feline | Escherichia coli |
| LBV_077/18 | Severe | Canine | Escherichia coli |
| LBV_084/18 | Severe | Canine | Escherichia coli |
| LBV_105/18 | Severe | Canine | Escherichia coli |
| LBV_112/18 | Severe | Canine | Streptococcus sp. |
| LBV_114/18 | Severe | Feline | Escherichia coli |
| LBV_133/18 | Severe | Canine | Enterococcus sp. |
| LBV_142/18 | Severe | Canine | Escherichia coli |
| LBV_163/18 | Severe | Canine | Escherichia coli |
| LBV_183/18 | Severe | Canine | Escherichia coli |
| LBV_188/18 | Severe | Canine | Escherichia coli |
| LBV_189/18 | Severe | Canine | Pseudomonas aeruginosa |
| LBV_194/18 | Severe | Canine | Escherichia coli |
| LBV213/18 | NA | Canine | Negative control |
| LBV235/18 | NA | Canine | Negative control |
| LBV256/18 | NA | Feline | Negative control |
| LBV262/18 | NA | Canine | Negative control |
| LBV249/18 | NA | Canine | Negative control |
| LBV214/18 | NA | Feline | Negative control |
| LBV240/18 | NA | Feline | Negative control |
| LBV256/18 | NA | Feline | Negative control |
| E. coli Strain | Host | Histopathology Severity Degree | Phylo-Group | Curli Fimbriae | Cellulose Production | Biofilm Formation |
|---|---|---|---|---|---|---|
| LBV_036/18 | F | Moderate | B2 | rdar | Positive | Moderately |
| LBV_045/18 | C | Moderate | B2 | rdar | Positive | Moderately |
| LBV_126/18 | F | Moderate | B2 | saw | Negative | Strongly |
| LBV_029/18 | C | Severe | B2 | rdar | Positive | Strongly |
| LBV_037/18 | C | Severe | B2 | rdar | Positive | Strongly |
| LBV_071/18 | F | Severe | B2 | rdar | Positive | Weakly |
| LBV_077/18 | C | Severe | B2 | rdar | Positive | Strongly |
| LBV_084/18 | C | Severe | B2 | saw | Negative | Weakly |
| LBV_114/18 | C | Severe | B2 | rdar | Positive | Weakly |
| LBV_183/18 | F | Severe | B2 | rdar | Positive | Moderately |
| LBV_188/18 | C | Severe | B2 | rdar | Positive | Strongly |
| LBV_163/18 | C | Severe | Clade I | rdar | Positive | Weakly |
| LBV_105/18 | C | Severe | Clade I | rdar | Positive | Non-adherent |
| LBV_194/18 | C | Severe | Unknown | saw | Negative | Moderately |
| LBV_142/18 | C | Severe | F | saw | Positive | Moderately |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, C.E.; De Carli, S.; Riboldi, C.I.; De Lorenzo, C.; Panziera, W.; Driemeier, D.; Siqueira, F.M. Pet Pyometra: Correlating Bacteria Pathogenicity to Endometrial Histological Changes. Pathogens 2021, 10, 833. https://doi.org/10.3390/pathogens10070833
Lopes CE, De Carli S, Riboldi CI, De Lorenzo C, Panziera W, Driemeier D, Siqueira FM. Pet Pyometra: Correlating Bacteria Pathogenicity to Endometrial Histological Changes. Pathogens. 2021; 10(7):833. https://doi.org/10.3390/pathogens10070833
Chicago/Turabian StyleLopes, Cassiane Elisabete, Silvia De Carli, Camila Imperico Riboldi, Cíntia De Lorenzo, Welden Panziera, David Driemeier, and Franciele Maboni Siqueira. 2021. "Pet Pyometra: Correlating Bacteria Pathogenicity to Endometrial Histological Changes" Pathogens 10, no. 7: 833. https://doi.org/10.3390/pathogens10070833






