Transcriptome Analysis Unveils the Effects of Proline on Gene Expression in the Yeast Komagataella phaffii
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast and Bacterial Strains
2.2. Media and Cultivation Conditions
2.3. Oligonucleotides
2.4. Molecular Methods
2.5. Enzymatic Assays
2.6. Gene Nomenclature
2.7. The Construction of K. phaffii P1AP-GS115 Strain
2.8. The Introduction of Gene Deletions in K. phaffii
2.9. RNA-Sequencing
2.10. Bioinformatic Analysis
3. Results
3.1. The Analysis of KpPUT1 Gene Expression in K. phaffii
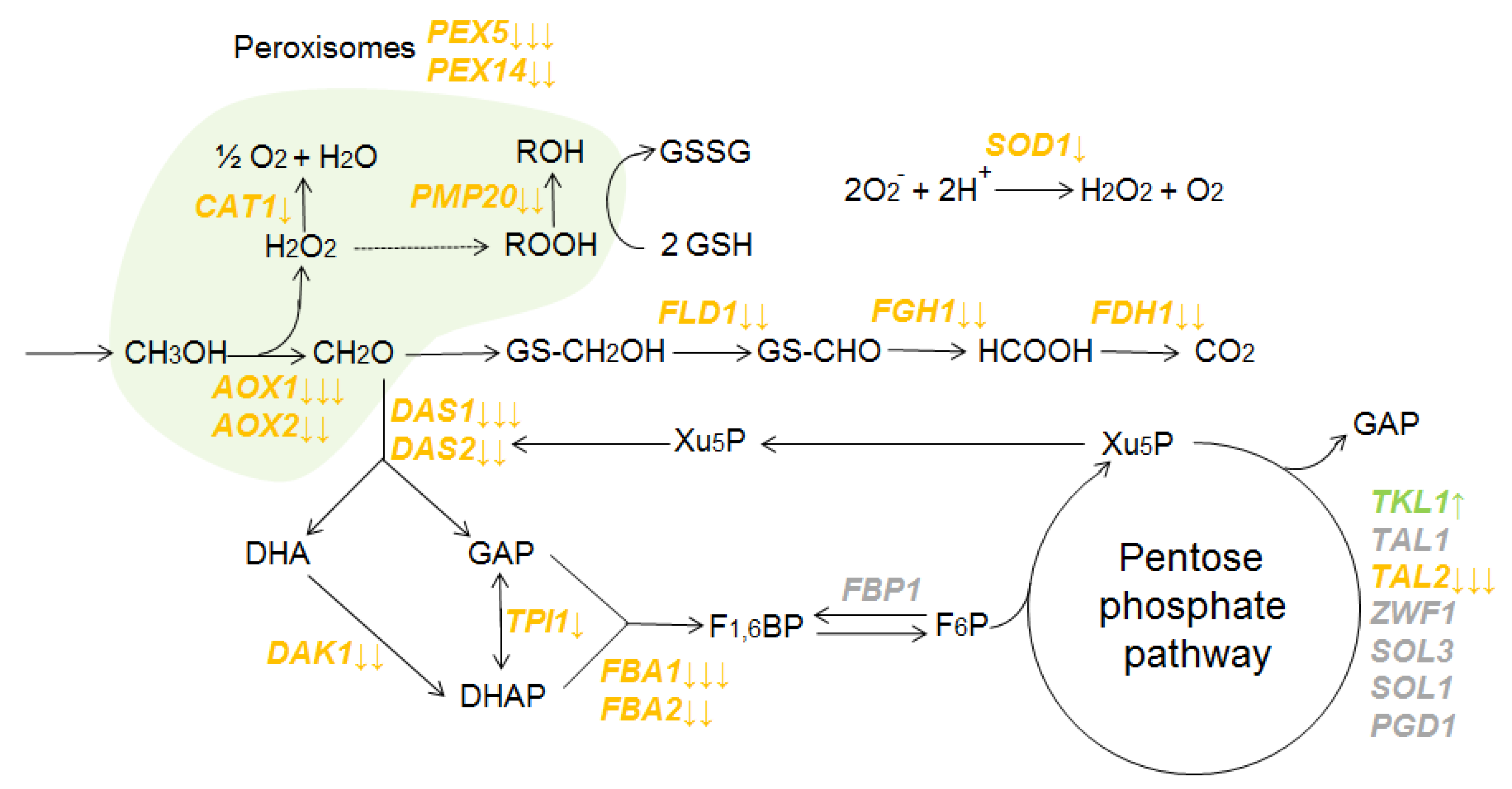
3.2. The Analysis of Gene Expression of K. phaffii Yeast on the Transcriptome Level
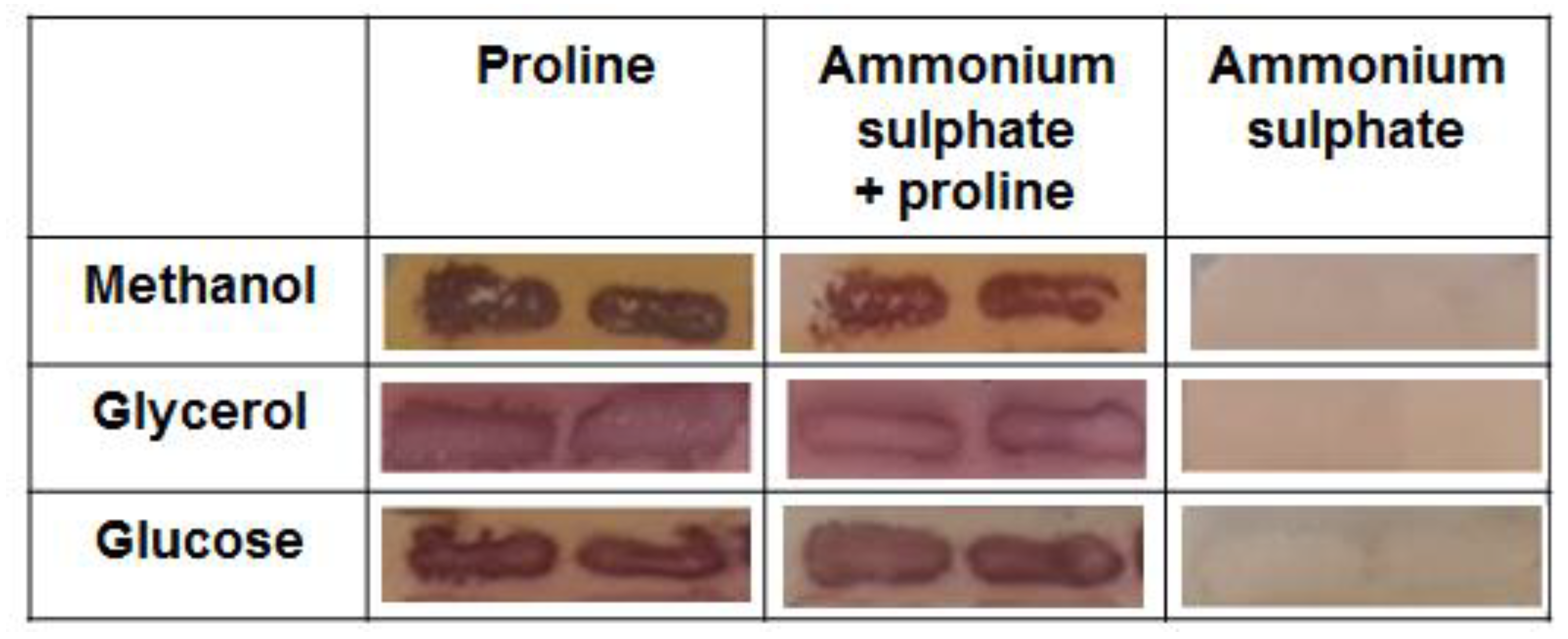
3.3. The Analysis of the Mitochondrial Activity in K. phaffii Cells
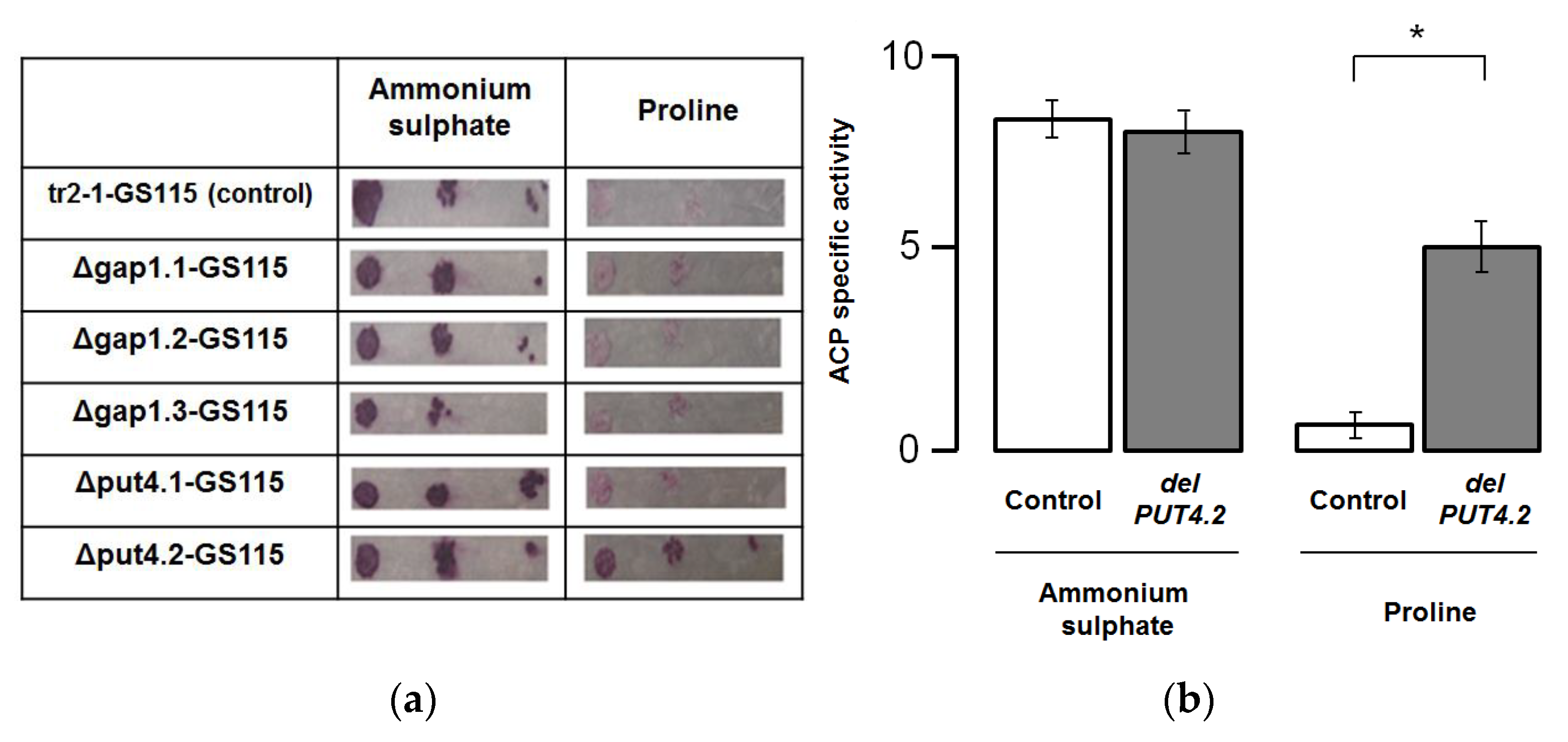
3.4. The Analysis of the Phenotypes of K. phaffii Strains Carrying the Deletions in Permease Genes
3.5. The Analysis of the Effects of the Deletions in Permease Genes on the AOX1 Promoter Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317. [Google Scholar] [CrossRef] [PubMed]
- Cregg, J.M.; Madden, K.R.; Barringer, K.J.; Thill, G.P.; Stillman, A.C. Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. Mol. Cell. Biol. 1989, 9, 1316–1323. [Google Scholar] [PubMed]
- Tschopp, J.F.; Brust, P.F.; Cregg, J.M.; Stillman, C.A.; Gingeras, T.R. Expression of thelacZgene from two methanol-regulated promoters inPichia pastoris. Nucleic Acids Res. 1987, 15, 3859–3876. [Google Scholar] [CrossRef] [PubMed]
- SEllis, S.B.; Brust, P.F.; Koutz, P.J.; Waters, A.F.; Harpold, M.M.; Gingeras, T.R. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. Mol. Cell. Biol. 1985, 5, 1111–1121. [Google Scholar]
- Kalender, Ö.; Çalık, P. Transcriptional regulatory proteins in central carbon metabolism of Pichia pastoris and Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2020, 104, 7273–7311. [Google Scholar] [CrossRef]
- Lin-Cereghino, G.P.; Godfrey, L.; de la Cruz, B.J.; Johnson, S.; Khuongsathiene, S.; Tolstorukov, I.; Yan, M.; Lin-Cereghino, J.; Veenhuis, M.; Subramani, S.; et al. Mxr1p, a Key Regulator of the Methanol Utilization Pathway and Peroxisomal Genes in Pichia pastoris. Mol. Cell. Biol. 2006, 26, 883–897. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Q.; Wang, J.; Bai, P.; Shi, L.; Shen, W.; Zhou, M.; Zhou, X.; Zhang, Y.; Cai, M. Mit1 Transcription Factor Mediates Methanol Signaling and Regulates the Alcohol Oxidase 1 (AOX1) Promoter in Pichia pastoris. J. Biol. Chem. 2016, 291, 6245–6261. [Google Scholar] [CrossRef]
- Sahu, U.; Rao, K.K.; Rangarajan, P.N. Trm1p, a Zn(II)2Cys6-type transcription factor, is essential for the transcriptional activation of genes of methanol utilization pathway, in Pichia pastoris. Biochem. Biophys. Res. Commun. 2014, 451, 158–164. [Google Scholar] [CrossRef]
- Shi, L.; Wang, X.; Wang, J.; Zhang, P.; Qi, F.; Cai, M.; Zhang, Y.; Zhou, X. Transcriptome analysis of Δmig1Δmig2 mutant reveals their roles in methanol catabolism, peroxisome biogenesis and autophagy in methylotrophic yeast Pichia pastoris. Genes Genom. 2018, 40, 399–412. [Google Scholar] [CrossRef]
- Kumar, N.V.; Rangarajan, P. Catabolite repression of phosphoenolpyruvate carboxykinase by a zinc finger protein under biotin- and pyruvate carboxylase-deficient conditions in Pichia pastoris. Microbiology 2011, 157, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cai, M.; Shi, L.; Wang, Q.; Zhu, J.; Wang, J.; Zhou, M.; Zhou, X.; Zhang, Y. PpNrg1 is a transcriptional repressor for glucose and glycerol repression of AOX1 promoter in methylotrophic yeast Pichia pastoris. Biotechnol. Lett. 2016, 38, 291–298. [Google Scholar] [CrossRef]
- Parua, P.; Ryan, P.M.; Trang, K.; Young, E.T. Pichia pastoris 14-3-3 regulates transcriptional activity of the methanol inducible transcription factor Mxr1 by direct interaction. Mol. Microbiol. 2012, 85, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Kong, C.; Xue, Y.; Liu, Y.; Cai, M.; Zhang, Y.; Jiang, T.; Zhou, X.; Zhou, M. Kinase Screening in Pichia pastoris Identified Promising Targets Involved in Cell Growth and Alcohol Oxidase 1 Promoter (PAOX1) Regulation. PLoS ONE 2016, 11, e0167766. [Google Scholar] [CrossRef]
- Zhan, C.; Bai, Z.; Wang, S.; Sun, Y.; Dai, X.; Liu, X.; Harvey, L.; McNeil, B.; Yang, Y. The Pichia pastoris transmembrane protein GT1 is a glycerol transporter and relieves the repression of glycerol on AOX1 expression. FEMS Yeast Res. 2016, 16, 1–10. [Google Scholar] [CrossRef][Green Version]
- Zhang, P.; Zhang, W.; Zhou, X.; Bai, P.; Cregg, J.M.; Zhang, Y. Catabolite Repression of Aox in Pichia pastoris Is Dependent on Hexose Transporter PpHxt1 and Pexophagy. Appl. Environ. Microbiol. 2010, 76, 6108–6118. [Google Scholar] [CrossRef] [PubMed]
- Rumiantsev, A.M.; Padkina, M.V.; Sambuk, E.V. Effect of nitrogen source on gene expression of first steps of methanol utilization pathway in Pichia pastoris. Russ. J. Genet. 2013, 49, 394–400. [Google Scholar] [CrossRef]
- Rumjantsev, A.; Bondareva, O.V.; Padkina, M.V.; Sambuk, E.V. Effect of Nitrogen Source and Inorganic Phosphate Concentration on Methanol Utilization and PEXGenes Expression in Pichia pastoris. Sci. World J. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Sahu, U.; Rangarajan, P.N. Methanol Expression Regulator 1 (Mxr1p) Is Essential for the Utilization of Amino Acids as the Sole Source of Carbon by the Methylotrophic Yeast, Pichia pastoris. J. Biol. Chem. 2016, 291, 20588–20601. [Google Scholar] [CrossRef]
- Rumyantsev, A.M.; Soloviev, G.A.; Slepchenkov, A.V.; Sambuk, E.V. Effects of Deletions in Pichia pastoris RTG Genes on Phenotype and AOX1 Expression. Adv. Microbiol. 2018, 8, 439–450. [Google Scholar] [CrossRef][Green Version]
- Mukai, Y.; Kamei, Y.; Liu, X.; Jiang, S.; Sugimoto, Y.; Nanyan, N.S.M.; Watanabe, D.; Takagi, H. Proline metabolism regulates replicative lifespan in the yeast Saccharomyces cerevisiae. Microb. Cell 2019, 6, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H. Proline as a stress protectant in yeast: Physiological functions, metabolic regulations, and biotechnological applications. Appl. Microbiol. Biotechnol. 2008, 81, 211–223. [Google Scholar] [CrossRef]
- Marzluf, G.A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 1997, 61, 17–32. [Google Scholar] [PubMed]
- Andréasson, C.; Neve, E.P.A.; Ljungdahl, P. Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast 2004, 21, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Grenson, M.; Hou, C.; Crabeel, M. Multiplicity of the Amino Acid Permeases in Saccharomyces cerevisiae IV. Evidence for a General Amino Acid Permease. J. Bacteriol. 1970, 103, 770–777. [Google Scholar] [CrossRef]
- Zhu, X.; Garrett, J.; Schreve, J.; Michaeli, T. GNP1, the high-affinity glutamine permease of S. cerevisiae. Curr. Genet. 1996, 30, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Iraqui, I.; Vissers, S.; Bernard, F.; de Craene, J.-O.; Boles, E.; Urrestarazu, A.; AndréB. Amino Acid Signaling in Saccharomyces cerevisiae: A Permease-Like Sensor of External Amino Acids and F-Box Protein Grr1p Are Required for Transcriptional Induction of the AGP1 Gene, Which Encodes a Broad-Specificity Amino Acid Permease. Mol. Cell. Biol. 1999, 19, 989–1001. [Google Scholar] [CrossRef]
- Vandenbol, M.; Jauniaux, J.-C.; Grenson, M. Nucleotide sequence of the Saccharomyces cerevisiae PUT4 proline-permease-encoding gene: Similarities between CAN1, HIP1 and PUT4 permeases. Gene 1989, 83, 153–159. [Google Scholar] [CrossRef]
- Brandriss, M.C.; Magasanik, B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: Enzyme induction by proline. J. Bacteriol. 1979, 140, 498–503. [Google Scholar] [CrossRef]
- Wang, S.S.; Brandriss, M.C. Proline utilization in Saccharomyces cerevisiae: Analysis of the cloned PUT1 gene. Mol. Cell. Biol. 1986, 6, 2638–2645. [Google Scholar]
- Grenson, M.; Dubois, E.; Piotrowska, M.; Drillien, R.; Aigle, M. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Mol. Genet. Genom. 1974, 128, 73–85. [Google Scholar] [CrossRef]
- Bach, B.; Meudec, E.; Lepoutre, J.-P.; Rossignol, T.; Blondin, B.; Dequin, S.; Camarasa, C. New Insights into γ-Aminobutyric Acid Catabolism: Evidence for γ-Hydroxybutyric Acid and Polyhydroxybutyrate Synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2009, 75, 4231–4239. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Wu, S.; Letchworth, G.J. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 2004, 36, 152–154. [Google Scholar] [CrossRef]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1988. [Google Scholar]
- Samsonova, M.G.; Padkina, M.V.; Krasnopevtseva, N.G.; A Kozhin, S.; Smirnov, M.N. Genetico-biochemical study of acid phosphatases in Saccharomyces cerevisiae yeast. V. Genetic control of regulation of acid phosphatase II synthesis. Genetika 1975, 11, 104–10415. [Google Scholar]
- Padkina, M.V.; Krasnopevtseva, N.G.; Petrashen, M.G. Genetic and biochemical studies of acid phosphatases of Saccharomyces cerevisiae. Properties of acid phosphatases from different strains (Russian). Genetika 1974, 10, 100–110. [Google Scholar]
- Sánchez, N.S.; Königsberg, M. Using yeast to easily determine mitochondrial functionality with 1-(4,5-dimethylthiazol-2-yl)-3,5-diphenyltetrazolium bromide (MTT) assay. Biochem. Mol. Biol. Educ. 2006, 34, 209–212. [Google Scholar] [CrossRef]
- Vogl, T.; Sturmberger, L.; Kickenweiz, T.; Wasmayer, R.; Schmid, C.; Hatzl, A.-M.; Gerstmann, M.A.; Pitzer, J.; Wagner, M.; Thallinger, G.G.; et al. A Toolbox of Diverse Promoters Related to Methanol Utilization: Functionally Verified Parts for Heterologous Pathway Expression in Pichia pastoris. ACS Synth. Biol. 2016, 5, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Leggett, R.M.; Ramirez-Gonzalez, R.H.; Clavijo, B.J.; Ewaite, D.; Davey, R.P. Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front. Genet. 2013, 4, 288. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Fazius, F.; Shelest, E.; Gebhardt, P.; Brock, M. The fungal α-aminoadipate pathway for lysine biosynthesis requires two enzymes of the aconitase family for the isomerization of homocitrate to homoisocitrate. Mol. Microbiol. 2012, 86, 1508–1530. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Kaneko, I.; Yamada, N.; Sakuraba, Y.; Kamenosono, M.; Tutumi, S. Suppression of Mitochondrial Succinate Dehydrogenase, a Primary Target of β-Amyloid, and Its Derivative Racemized at Ser Residue. J. Neurochem. 1995, 65, 2585–2593. [Google Scholar] [CrossRef]
- Regenberg, B.; Hansen, J. GAP1, a novel selection and counter-selection marker for multiple gene disruptions in Saccharomyces cerevisiae. Yeast 2000, 16, 1111–1119. [Google Scholar] [CrossRef]
- Shashkova, S.; Welkenhuysen, N.; Hohmann, S. Molecular communication: Crosstalk between the Snf1 and other signaling pathways. FEMS Yeast Res. 2015, 15, fov026. [Google Scholar] [CrossRef]
- Bernauer, L.; Radkohl, A.; Lehmayer, L.G.K.; Emmerstorfer-Augustin, A. Komagataella phaffii as Emerging Model Organism in Fundamental Research. Front. Microbiol. 2021, 11, 607028. [Google Scholar] [CrossRef] [PubMed]
- Ter Schure, E.G.; van Riel, N.A.W.; Verrips, C.T. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2000, 24, 67–83. [Google Scholar] [CrossRef] [PubMed]


| Strain | Genotype | Sourse |
|---|---|---|
| GS115 | his4 | Thermo Fisher Scientific, Waltham, MA, USA |
| X-33 | wt | Thermo Fisher Scientific, Waltham, MA, USA |
| tr2-1-GS115 | phox PAOX1-PHO5 HIS4 | 17, 18 |
| P1AP-GS115 | PPUT1-PHO5 HIS4 | This study |
| Δgap1.1-GS115 | Δkpgap1.1 ZeoR PAOX1-PHO5 HIS4 | This study |
| Δgap1.2-GS115 | Δkpgap1.2 ZeoR PAOX1-PHO5 HIS4 | This study |
| Δgap1.3-GS115 | Δkpgap1.3 ZeoR PAOX1-PHO5 HIS4 | This study |
| Δput4.1-GS115 | Δkpput4.1 ZeoR PAOX1-PHO5 HIS4 | This study |
| Δput4.2-GS115 | Δkpput4.2 ZeoR PAOX1-PHO5 HIS4 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rumyantsev, A.; Sidorin, A.; Volkov, A.; Al Shanaa, O.; Sambuk, E.; Padkina, M. Transcriptome Analysis Unveils the Effects of Proline on Gene Expression in the Yeast Komagataella phaffii. Microorganisms 2022, 10, 67. https://doi.org/10.3390/microorganisms10010067
Rumyantsev A, Sidorin A, Volkov A, Al Shanaa O, Sambuk E, Padkina M. Transcriptome Analysis Unveils the Effects of Proline on Gene Expression in the Yeast Komagataella phaffii. Microorganisms. 2022; 10(1):67. https://doi.org/10.3390/microorganisms10010067
Chicago/Turabian StyleRumyantsev, Andrey, Anton Sidorin, Artemii Volkov, Ousama Al Shanaa, Elena Sambuk, and Marina Padkina. 2022. "Transcriptome Analysis Unveils the Effects of Proline on Gene Expression in the Yeast Komagataella phaffii" Microorganisms 10, no. 1: 67. https://doi.org/10.3390/microorganisms10010067
APA StyleRumyantsev, A., Sidorin, A., Volkov, A., Al Shanaa, O., Sambuk, E., & Padkina, M. (2022). Transcriptome Analysis Unveils the Effects of Proline on Gene Expression in the Yeast Komagataella phaffii. Microorganisms, 10(1), 67. https://doi.org/10.3390/microorganisms10010067







