The Anti-Photoaging Activity of Peptides from Pinctada martensii Meat
Abstract
1. Introduction
2. Results and Discussion
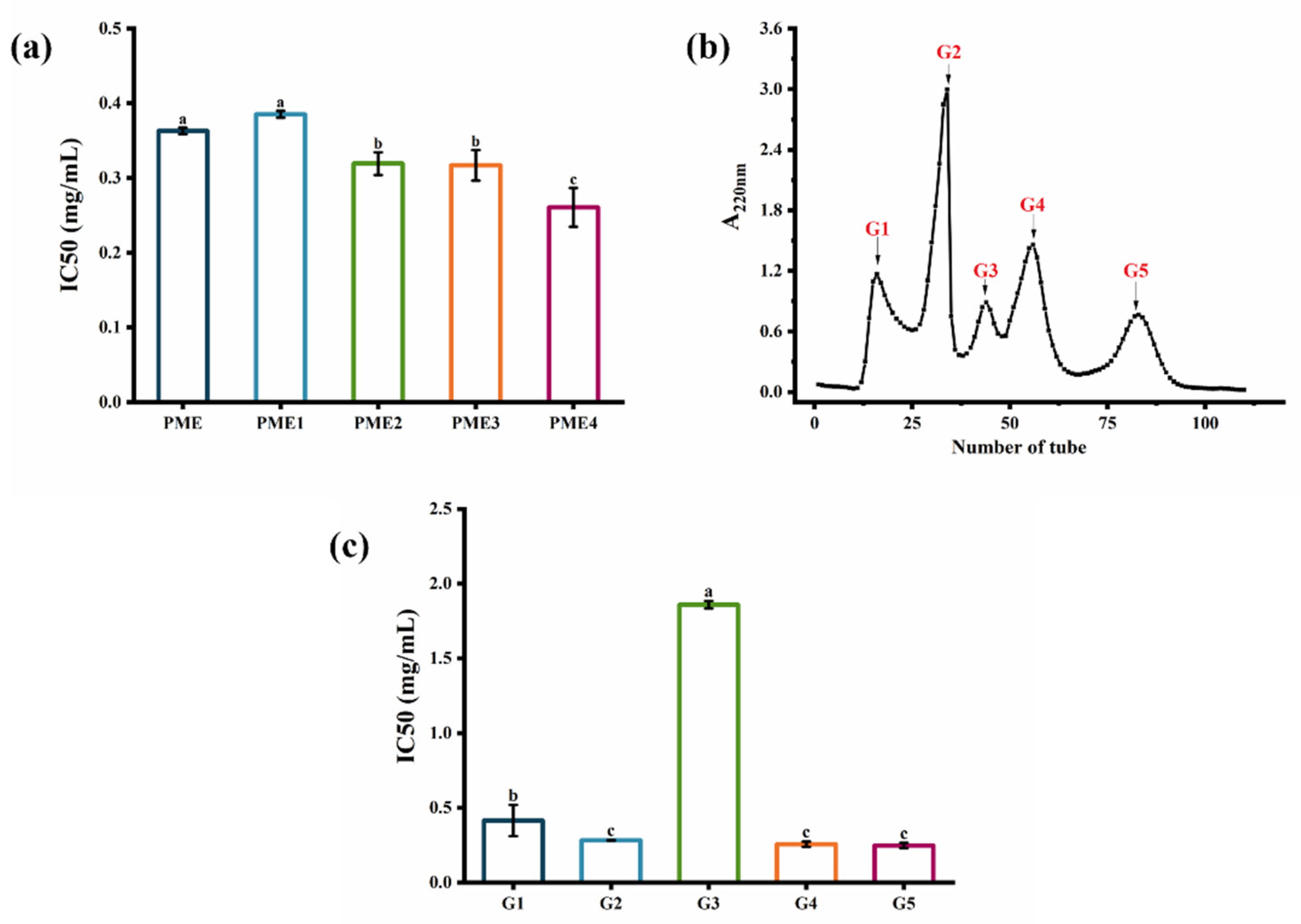
2.1. Anti-Photoaging Activity of Pinctada martensii Meat Hydrolysates (PME)
2.2. Screening of Fractions Separated by Ultrafiltration and Sephadex G-25 Gel Filtration Chromatography
2.3. Anti-Photoaging Mechanism of the Fraction G2
2.3.1. Effects of the Fraction G2 on Cell Viability, the Contents of MMP-1 and MMP-3
2.3.2. Effects of the Fraction G2 on the Expression of p38, JNK, ERK, MMP-1, and MMP-3
2.4. Identification of Peptides in the Fraction G2
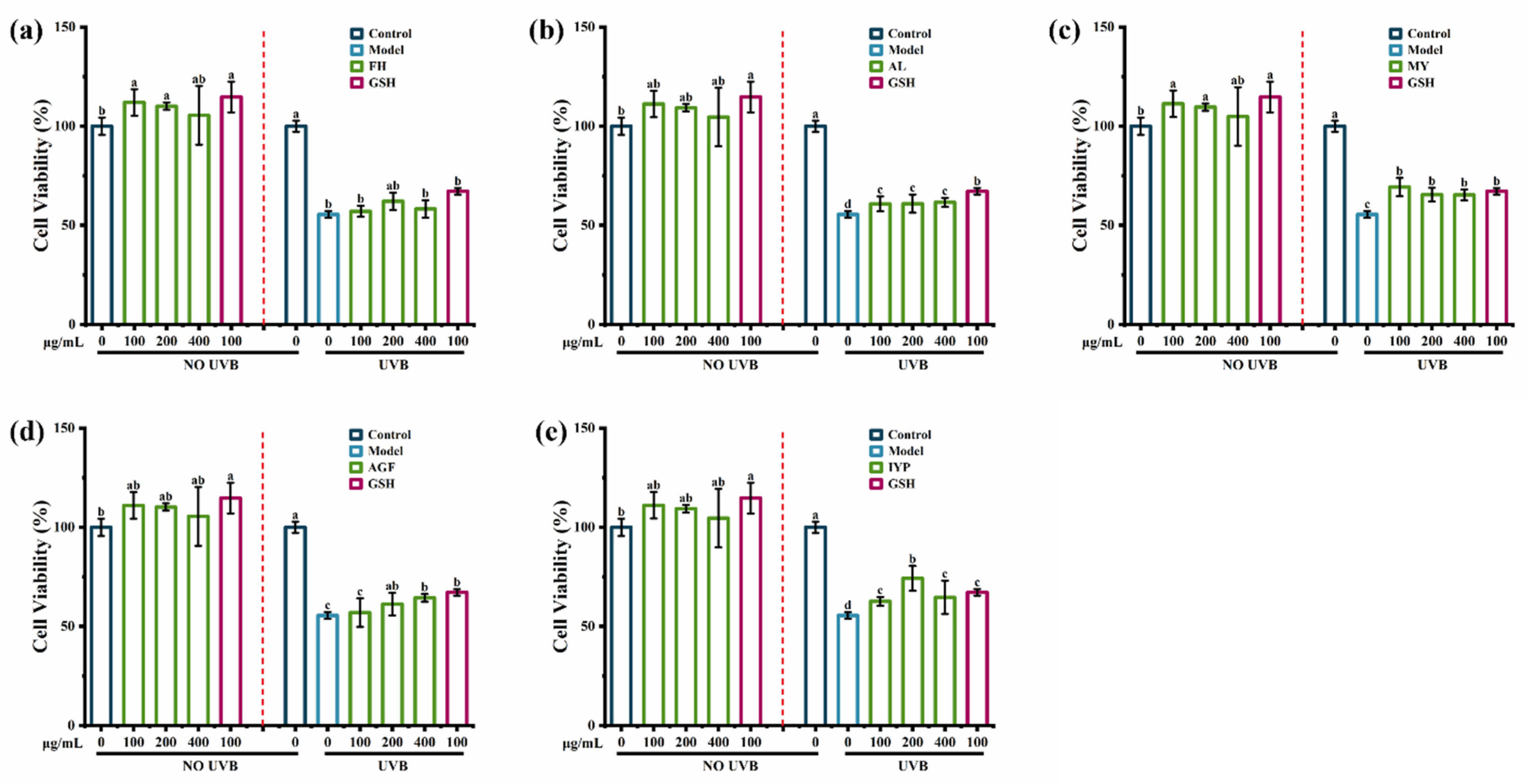
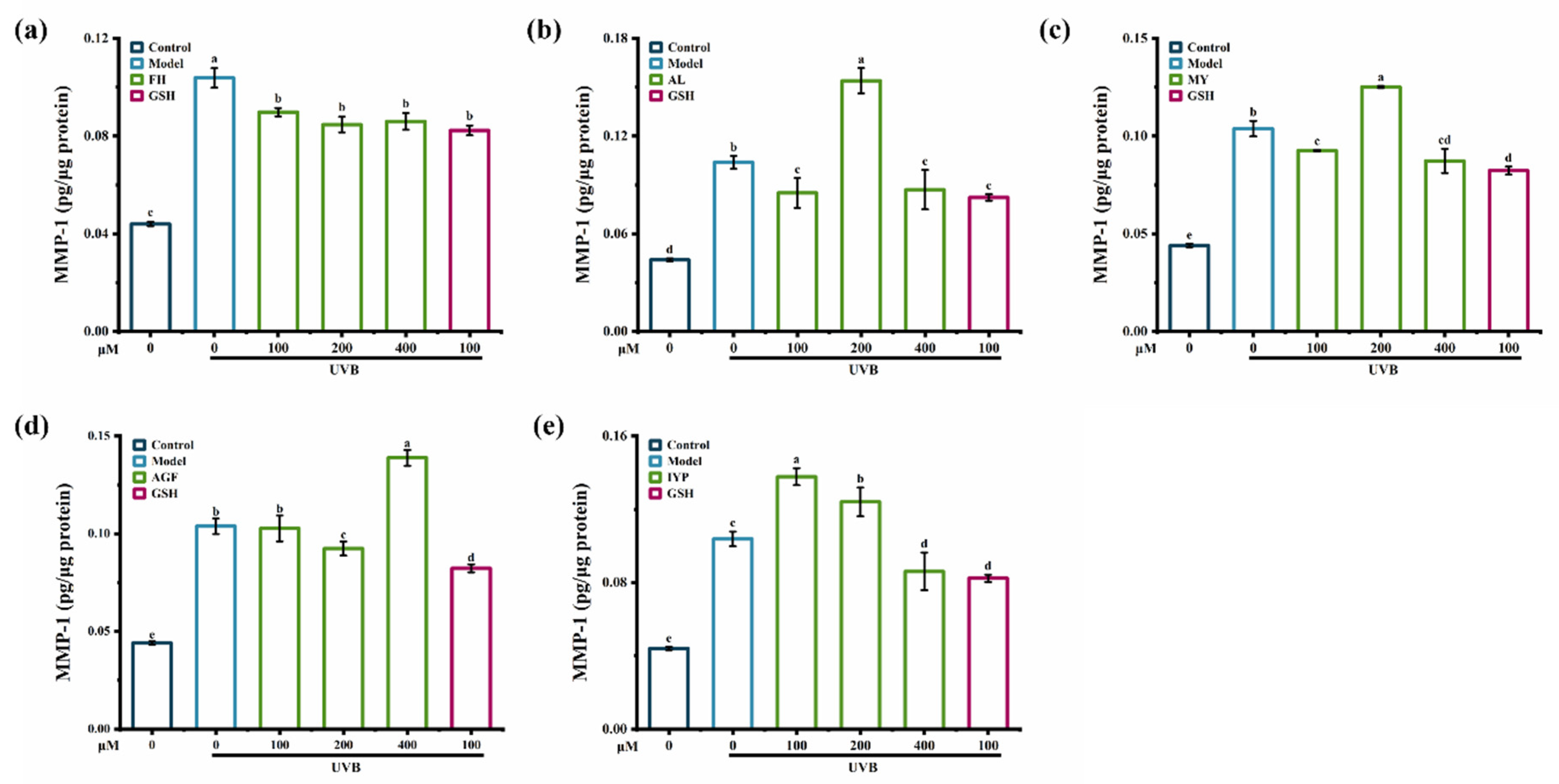
2.5. Anti-Photoaging Activity of Synthesized Peptides
2.5.1. Effects of Synthesized Peptides on Cell Viability
2.5.2. Effects of Synthesized Peptides on the Content of MMP-1 and MMP-3
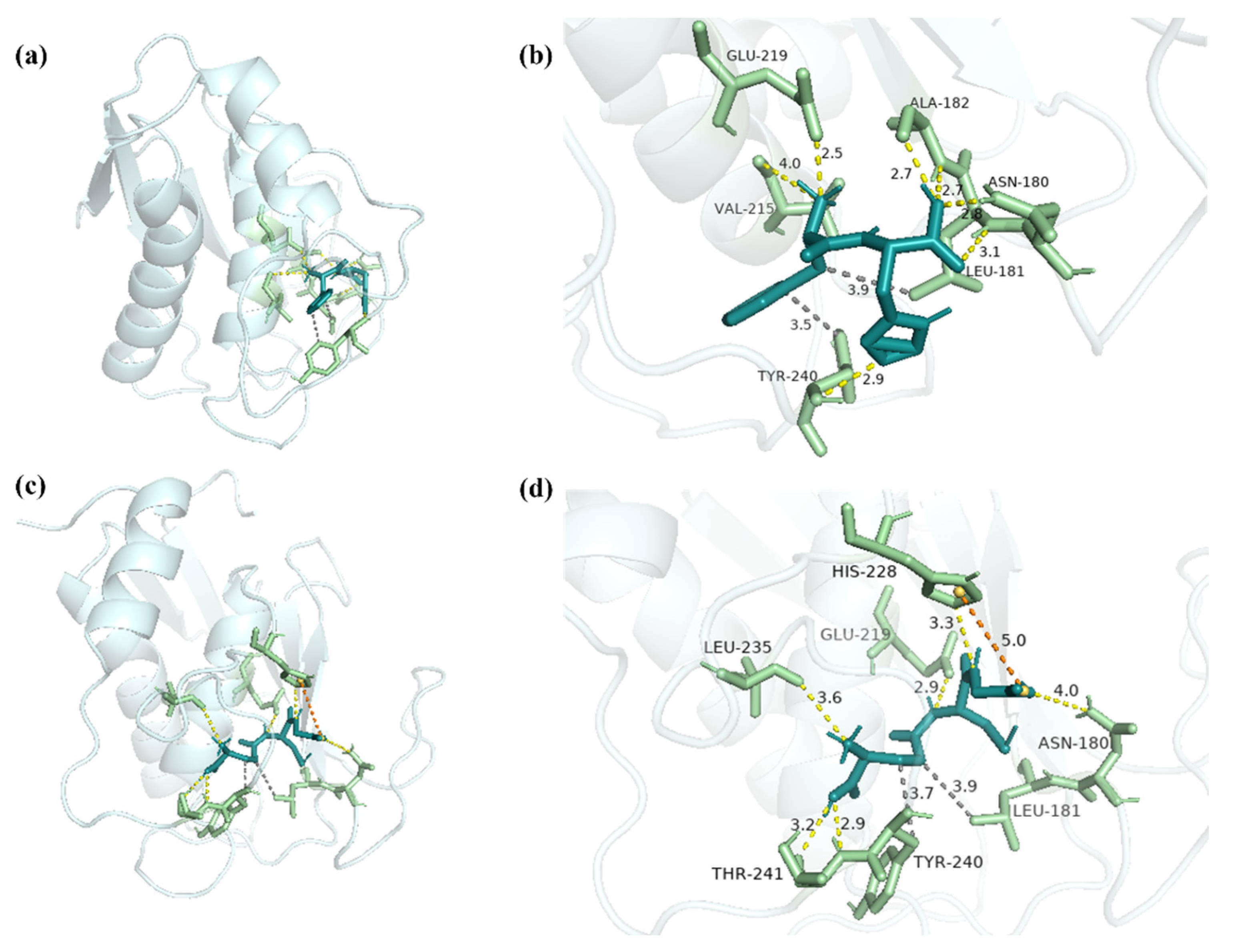
2.6. Molecular Docking Analysis of FH with MMP-1 and MMP-3
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Enzymatic Hydrolysis of Pinctada Martensii Meat
3.3. Separation and Purification of PME
3.4. Elastase Inhibition Activity Assay
3.5. Cell Culture and UVB Irradiation
3.6. Cell Viability Assay
3.7. Measurement of the Contents of MMP-1 and MMP-3
3.8. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) Analysis
3.9. Identification of Peptides by UHPLC-MS/MS
3.10. Peptide Synthesis
3.11. Molecular Docking Analysis with FH
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Petruk, G.; Del Giudice, R.; Rigano, M.M.; Monti, D.M. Antioxidants from Plants Protect against Skin Photoaging. Oxid. Med. Cell. Longev. 2018, 2018, 1454936. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.W.; Hung, Y.C.; Lin, T.Y.; Fang, J.Y.; Yang, P.M.; Chen, M.H.; Pan, T.L. Comparison of the Biological Impact of UVA and UVB upon the Skin with Functional Proteomics and Immunohistochemistry. Antioxidants 2019, 8, 569. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Chen, X.; Li, X.; Chang, S.; Zhao, M.; You, L. Current trends in the anti-photoaging activities and mechanisms of dietary non-starch polysaccharides from natural resources. Crit. Rev. Food Sci. Nutr. 2021, 62, 9021–9035. [Google Scholar] [CrossRef]
- Fanjul-Fernandez, M.; Folgueras, A.R.; Cabrera, S.; Lopez-Otin, C. Matrix metalloproteinases: Evolution, gene regulation and functional analysis in mouse models. Biochim. Biophys. Acta 2010, 1803, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Kandi, S.; Baskaran Stephen, I.; Bing-Huei, C. Recent developments on production, purification and biological activity of marine peptides. Food Res. Int. 2021, 147, 110468. [Google Scholar]
- Bashir, K.M.I.; Sohn, J.H.; Kim, J.S.; Choi, J.S. Identification and characterization of novel antioxidant peptides from mackerel (Scomber japonicus) muscle protein hydrolysates. Food Chem. 2020, 323, 126809. [Google Scholar] [CrossRef]
- Sangtanoo, P.; Srimongkol, P.; Saisavoey, T.; Reamtong, O.; Karnchanatat, A. Anti-inflammatory action of two novel peptides derived from peanut worms (Sipunculus nudus) in lipopolysaccharide-induced RAW264.7 macrophages. Food Funct. 2020, 11, 552–560. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, Y.; Li, L. Relationship between primary structure or spatial conformation and functional activity of antioxidant peptides from Pinctada fucata. Food Chem. 2018, 264, 108–117. [Google Scholar] [CrossRef]
- Hao, R.; Du, X.; Yang, C.; Deng, Y.; Zheng, Z.; Wang, Q. Integrated application of transcriptomics and metabolomics provides insights into unsynchronized growth in pearl oyster Pinctada fucata martensii. Sci. Total Environ. 2019, 666, 46–56. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, F.; Qin, X.; Yang, X.; Zhang, C.; Wan, Z.; Lin, H. Investigation of the In Vivo, In Vitro, and In Silico Wound Healing Potential of Pinctada martensii Purified Peptides. Mar. Drugs 2022, 20, 417. [Google Scholar] [CrossRef]
- Liu, P.; Lan, X.; Yaseen, M.; Wu, S.; Feng, X.; Zhou, L.; Sun, J.; Liao, A.; Liao, D.; Sun, L. Purification, Characterization and Evaluation of Inhibitory Mechanism of ACE Inhibitory Peptides from Pearl Oyster (Pinctada fucata martensii) Meat Protein Hydrolysate. Mar. Drugs 2019, 17, 463. [Google Scholar] [CrossRef] [PubMed]
- Yanyan, W.; Qian, T.; Laihao, L.; Muhammad Naseem, K.; Xianqing, Y.; Zhongmin, Z.; Xiao, H.; Shengjun, C. Inhibitory effect of antioxidant peptides derived from Pinctada fucata protein on ultraviolet-induced photoaging in mice. J. Funct. Foods 2013, 5, 527–538. [Google Scholar]
- Liu, Y.; Zheng, L.; Xu, J.C.; Sun-Waterhouse, D.; Sun, B.G.; Su, G.W.; Zhao, M.M. Identification of novel peptides with high stability against in vitro hydrolysis from bovine elastin hydrolysates and evaluation of their elastase inhibitory activity. Int. J. Food Sci. Technol. 2020, 55, 99–108. [Google Scholar] [CrossRef]
- Aguilar-Toala, J.E.; Liceaga, A.M. Identification of chia seed (Salvia hispanica L.) peptides with enzyme inhibition activity towards skin-aging enzymes. Amino Acids 2020, 52, 1149–1159. [Google Scholar] [CrossRef]
- Peng, Z.; Gao, J.; Su, W.; Cao, W.; Zhu, G.; Qin, X.; Zhang, C.; Qi, Y. Purification and Identification of Peptides from Oyster (Crassostrea hongkongensis) Protein Enzymatic Hydrolysates and Their Anti-Skin Photoaging Effects on UVB-Irradiated HaCaT Cells. Mar. Drugs 2022, 20, 749. [Google Scholar] [CrossRef]
- Garg, C.; Sharma, H.; Garg, M. Skin photo-protection with phytochemicals against photo-oxidative stress, photo-carcinogenesis, signal transduction pathways and extracellular matrix remodeling—An overview. Ageing Res. Rev. 2020, 62, 101127. [Google Scholar] [CrossRef]
- Gu, Y.; Han, J.; Jiang, C.; Zhang, Y. Biomarkers, oxidative stress and autophagy in skin aging. Ageing Res. Rev. 2020, 59, 101036. [Google Scholar] [CrossRef]
- Chen, J.; Liang, P.; Xiao, Z.; Chen, M.-F.; Gong, F.; Li, C.; Zhou, C.; Hong, P.; Jung, W.-K.; Qian, Z.-J. Antiphotoaging effect of boiled abalone residual peptide ATPGDEG on UVB-induced keratinocyte HaCaT cells. Food Nutr. Res. 2019, 63, 3508:1–3508:13. [Google Scholar] [CrossRef]
- Zheng, Z.; Xiao, Z.; He, Y.-L.; Tang, Y.; Li, L.; Zhou, C.; Hong, P.; Luo, H.; Qian, Z.-J. Heptapeptide Isolated from Isochrysis zhanjiangensis Exhibited Anti-Photoaging Potential via MAPK/AP-1/MMP Pathway and Anti-Apoptosis in UVB-Irradiated HaCaT Cells. Mar. Drugs 2021, 19, 626. [Google Scholar] [CrossRef]
- Liu, Y.; Su, G.; Zhou, F.; Zhang, J.; Zheng, L.; Zhao, M. Protective Effect of Bovine Elastin Peptides against Photoaging in Mice and Identification of Novel Antiphotoaging Peptides. J. Agric. Food Chem. 2018, 66, 10760–10768. [Google Scholar] [CrossRef]
- Van Laethem, A.; Van Kelst, S.; Lippens, S.; Declercq, W.; Vandenabeele, P.; Janssens, S.; Vandenheede, J.R.; Garmyn, M.; Agostinis, P. Activation of p38 MAPK is required for Bax translocation to mitochondria, cytochrome c release and apoptosis induced by UVB irradiation in human keratinocytes. FASEB J. 2004, 18, 1946–1948. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Liang, P.; Chen, J.; Chen, M.F.; Gong, F.; Li, C.; Zhou, C.; Hong, P.; Yang, P.; Qian, Z.J. A Peptide YGDEY from Tilapia Gelatin Hydrolysates Inhibits UVB-mediated Skin Photoaging by Regulating MMP-1 and MMP-9 Expression in HaCaT Cells. Photochem. Photobiol. 2019, 95, 1424–1432. [Google Scholar] [CrossRef] [PubMed]
- Jiaohan, L.; Hu, H.; Yan, F.; Tingting, Y.; Bafang, L. Identification of MMP-1 inhibitory peptides from cod skin gelatin hydrolysates and the inhibition mechanism by MAPK signaling pathway. J. Funct. Foods 2017, 33, 251–260. [Google Scholar]
- Chongyang, L.; Yu, F.; Hongjie, D.; Qiang, W.; Ruichang, G.; Yuhao, Z. Recent progress in preventive effect of collagen peptides on photoaging skin and action mechanism. Food Sci. Hum. Wellness 2022, 11, 218–229. [Google Scholar]
- Bang, J.S.; Jin, Y.J.; Choung, S.-Y. Low molecular polypeptide from oyster hydrolysate recovers photoaging in SKH-1 hairless mice. Toxicol. Appl. Pharmacol. 2020, 386, 114844. [Google Scholar] [CrossRef]
- Park, S.K.; Van Hien, P.; Van Luong, H.; Yan, S.-W.; Byun, S.Y. Peptide Hydrolysates from Astragalus membranaceus Bunge Inhibit the Expression of Matrix Metalloproteinases in Human Dermal Fibroblasts. KSBB J. 2014, 29, 380–384. [Google Scholar] [CrossRef][Green Version]
- Ye, Y.; You, L.; Deng, Q.; Li, X.; Zhao, M. Preparation, structure identification and the anti-photoaging activity of peptide fraction OP-Ia from Ostrea rivularis. Rsc Adv. 2019, 9, 44–51. [Google Scholar] [CrossRef]
- Mo, Q.; Li, S.; You, S.; Wang, D.; Zhang, J.; Li, M.; Wang, C. Puerarin Reduces Oxidative Damage and Photoaging Caused by UVA Radiation in Human Fibroblasts by Regulating Nrf2 and MAPK Signaling Pathways. Nutrients 2022, 14, 4724. [Google Scholar] [CrossRef]
- Ahmed, T.; Sun, X.H.; Udenigwe, C.C. Role of structural properties of bioactive peptides in their stability during simulated gastrointestinal digestion: A systematic review. Trends Food Sci. Technol. 2022, 120, 265–273. [Google Scholar] [CrossRef]
- Xu, D.; Wang, W.; Liao, J.; Liao, L.; Li, C.; Zhao, M. Walnut protein hydrolysates, rich with peptide fragments of WSREEQEREE and ADIYTEEAGR ameliorate UV-induced photoaging through inhibition of the NF-kappa B/MMP-1 signaling pathway in female rats. Food Funct. 2020, 11, 10601–10616. [Google Scholar] [CrossRef]
- Iosageanu, A.; Ilie, D.; Craciunescu, O.; Seciu-Grama, A.-M.; Oancea, A.; Zarnescu, O.; Moraru, I.; Oancea, F. Effect of Fish Bone Bioactive Peptides on Oxidative, Inflammatory and Pigmentation Processes Triggered by UVB Irradiation in Skin Cells. Molecules 2021, 26, 2691. [Google Scholar] [CrossRef] [PubMed]
- Chaoting, W.; Jixian, Z.; Haihui, Z.; Yuqing, D.; Haile, M. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar]
- Zhang, C.; Lv, J.; Qin, X.; Peng, Z.; Lin, H. Novel Antioxidant Peptides from Crassostrea Hongkongensis Improve Photo-Oxidation in UV-Induced HaCaT Cells. Marine Drugs 2022, 20, 100. [Google Scholar] [CrossRef]
- Carvalho, B.G.; Raniero, L.J.; Martin, A.A.; Favero, P.P. Phenylalanine ab initio models for the simulation of skin natural moisturizing factor. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 106, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Liu, B.; Zhuang, Y. Effects of rambutan (Nephelium lappaceum) peel phenolics and Leu-Ser-Gly-Tyr-Gly-Pro on hairless mice skin photoaging induced by ultraviolet irradiation. Food Chem. Toxicol. 2019, 129, 30–37. [Google Scholar] [CrossRef]
- Yathisha, U.G.; Bhat, I.; Karunasagar, I.; Mamatha, B.S. Antihypertensive activity of fish protein hydrolysates and its peptides. Crit. Rev. Food Sci. Nutr. 2019, 59, 2363–2374. [Google Scholar]
- Mohankumar, T.; Chandramohan, V.; Lalithamba, H.S.; Jayaraj, R.L.; Kumaradhas, P.; Sivanandam, M.; Hunday, G.; Vijayakumar, R.; Balakrishnan, R.; Manimaran, D. Design and molecular dynamic investigations of 7, 8-dihydroxyflavone derivatives as potential neuroprotective agents against alpha-synuclein. Sci. Rep. 2020, 10, 599. [Google Scholar] [CrossRef]
- Connelly, P.R.; Snyder, P.W.; Zhang, Y.; McClain, B.; Quinn, B.P.; Johnston, S.; Medek, A.; Tanoury, J.; Griffith, J.; Walters, W.P. The potency–insolubility conundrum in pharmaceuticals: Mechanism and solution for hepatitis C protease inhibitors. Biophys. Chem. 2015, 196, 100–108. [Google Scholar] [CrossRef]
- de Almeida, L.G.N.; Thode, H.; Eslambolchi, Y.; Chopra, S.; Young, D.; Gill, S.; Devel, L.; Dufour, A. Matrix Metalloproteinases: From Molecular Mechanisms to Physiology, Pathophysiology, and Pharmacology. Pharmacol. Rev. 2022, 74, 712–768. [Google Scholar] [CrossRef]
- Liping, S.; Qiuming, L.; Jian, F.; Xiao, L.; Yongliang, Z. Purification and Characterization of Peptides Inhibiting MMP-1 Activity with C Terminate of Gly-Leu from Simulated Gastrointestinal Digestion Hydrolysates of Tilapia (Oreochromis niloticus) Skin Gelatin. J. Agric. Food Chem. 2018, 66, 593–601. [Google Scholar] [CrossRef]
- Lee, K.E.; Bharadwaj, S.; Yadava, U.; Kang, S.G. Computational and In Vitro Investigation of (-)-Epicatechin and Proanthocyanidin B2 as Inhibitors of Human Matrix Metalloproteinase 1. Biomolecules 2020, 10, 1379. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Yang, S.; Liu, Y.; Zhou, C.; Hong, P.; Sun, S.; Qian, Z.J. A novel glyceroglycolipid from brown algae Ishige okamurae improve photoaging and counteract inflammation in UVB-induced HaCaT cells. Chem. Biol. Interact. 2022, 351, 109737. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, G.; Kaliamurthi, S.; Thiruganasambandam, R. Molecular docking studies of rutin on matrix metalloproteinase. Insights Biomed. 2016, 1, 1–5. [Google Scholar]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assuncao, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carriere, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]






| Purified Fractions | Protein Recovery Rates (%) |
|---|---|
| G1 | 21.47 ± 0.89 a |
| G2 | 14.01 ± 1.86 b |
| G3 | 21.15 ± 0.22 a |
| G4 | 2.30 ± 0.20 c |
| G5 | 1.68 ± 0.30 c |
| Sequence | PeptideRanker Score | Length | Mass (Da) | ToxinPred |
|---|---|---|---|---|
| Phe-His (FH) | 0.95283 | 2 | 303.14 | Non-Toxin |
| Ala-Leu (AL) | 0.4378 | 2 | 203.13 | Non-Toxin |
| Met-Tyr (MY) | 0.84347 | 2 | 392.21 | Non-Toxin |
| Ala-Gly-Phe (AGF) | 0.9568 | 3 | 294.14 | Non-Toxin |
| Ile-Tyr-Pro (IYP) | 0.57726 | 3 | 303.14 | Non-Toxin |
| FH | GSH | |
|---|---|---|
| Hydrophobic interaction | Leu181, Tyr240 | Leu181, Tyr240 |
| Hydrogen bonds | Asn180, Leu181, Ala182, VAL215, Glu219, Tyr240 | Asn180, Glu219, His228, Leu235, Thr241 |
| Salt bridges | - | His228 |
| π-stacking | - | - |
| FH | GSH | |
|---|---|---|
| Hydrophobic interaction | Leu164, Val198, His201 | - |
| Hydrogen bonds | Asn162, Leu164, Ala165, Pro221 | Asn162, Leu164, Ala165, Glu202, Tyr223 |
| Salt bridges | - | - |
| π-stacking | His201 | - |
| Gene | Forward Primer (5′-3′) | Reverse Primer (3′-5′) |
|---|---|---|
| MMP-1 | GATGTGGAGTGCCTGATGTG | TGCTTGACCCTCAGAGACCT |
| MMP-3 | CACTCACAGACCTGACTCGG | GAGTCAGGGGGAGGTCCATA |
| p38 | ATGCCAAGCCATGAGGCAA | GCATCTTCTCCAGCAAGTCG |
| ERK | GCCGAAGCACCATTCAAGTT | CCTCTGAGCCCTTGTCCTGA |
| JNK | CAGCCCTCTCCTTTAGGTGC | GCTGCTGCTTCTAGACTGCT |
| GAPDH | TCCACTGGCGTCTTCACCACCAT | GGAGGCATTGCTGATGATCTTGAGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, M.; Qiu, H.; Zhou, J.; Yang, C.; Chen, Y.; You, L. The Anti-Photoaging Activity of Peptides from Pinctada martensii Meat. Mar. Drugs 2022, 20, 770. https://doi.org/10.3390/md20120770
Wei M, Qiu H, Zhou J, Yang C, Chen Y, You L. The Anti-Photoaging Activity of Peptides from Pinctada martensii Meat. Marine Drugs. 2022; 20(12):770. https://doi.org/10.3390/md20120770
Chicago/Turabian StyleWei, Mengfen, Huamai Qiu, Jie Zhou, Chenghao Yang, Yifan Chen, and Lijun You. 2022. "The Anti-Photoaging Activity of Peptides from Pinctada martensii Meat" Marine Drugs 20, no. 12: 770. https://doi.org/10.3390/md20120770
APA StyleWei, M., Qiu, H., Zhou, J., Yang, C., Chen, Y., & You, L. (2022). The Anti-Photoaging Activity of Peptides from Pinctada martensii Meat. Marine Drugs, 20(12), 770. https://doi.org/10.3390/md20120770








