Acetyltransferase P300 Regulates Glucose Metabolic Reprogramming through Catalyzing Succinylation in Lung Cancer
Abstract
:1. Introduction
2. Results
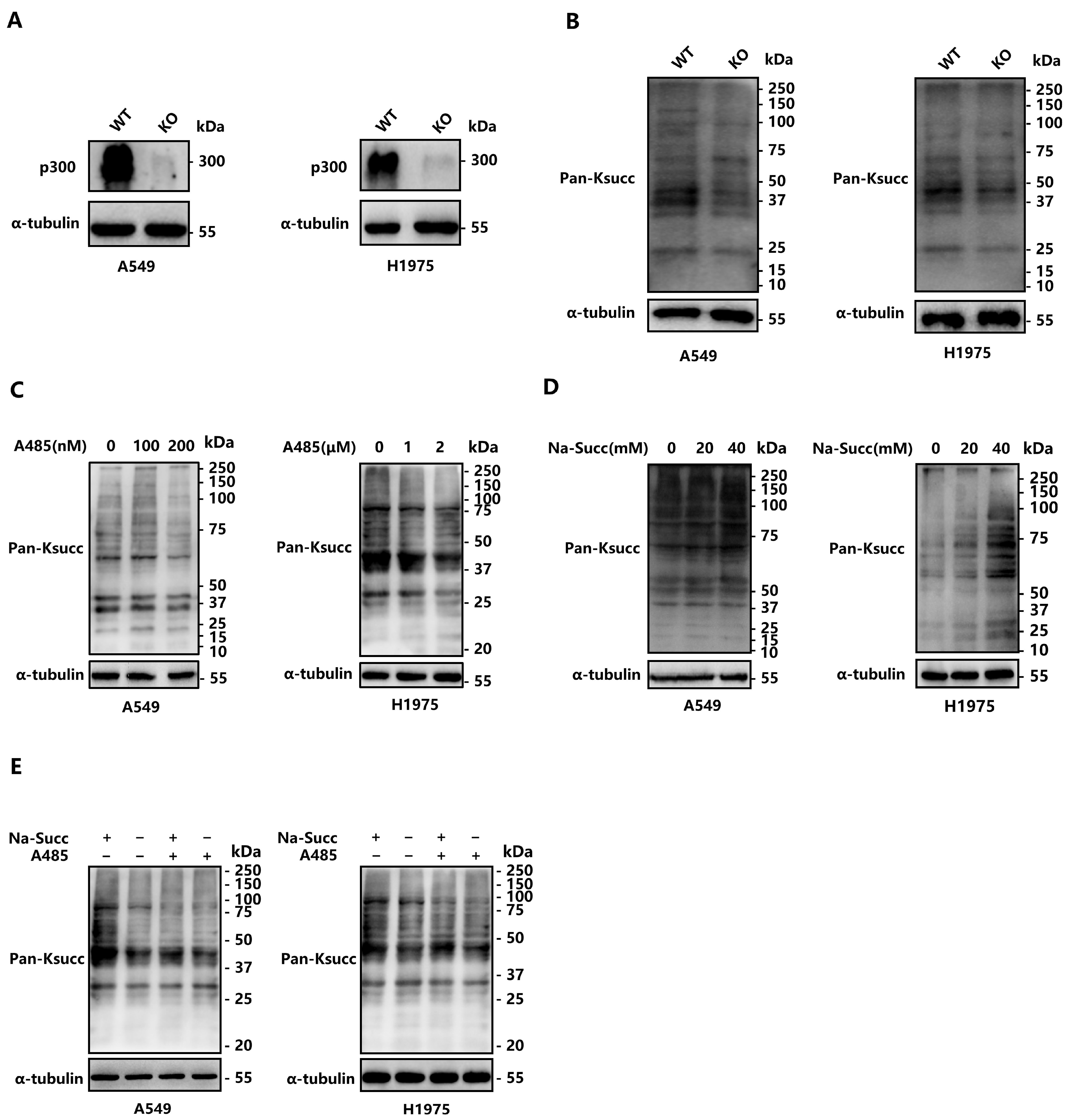
2.1. p300 Catalyzes Lysine Succinylation in LUAD Cells
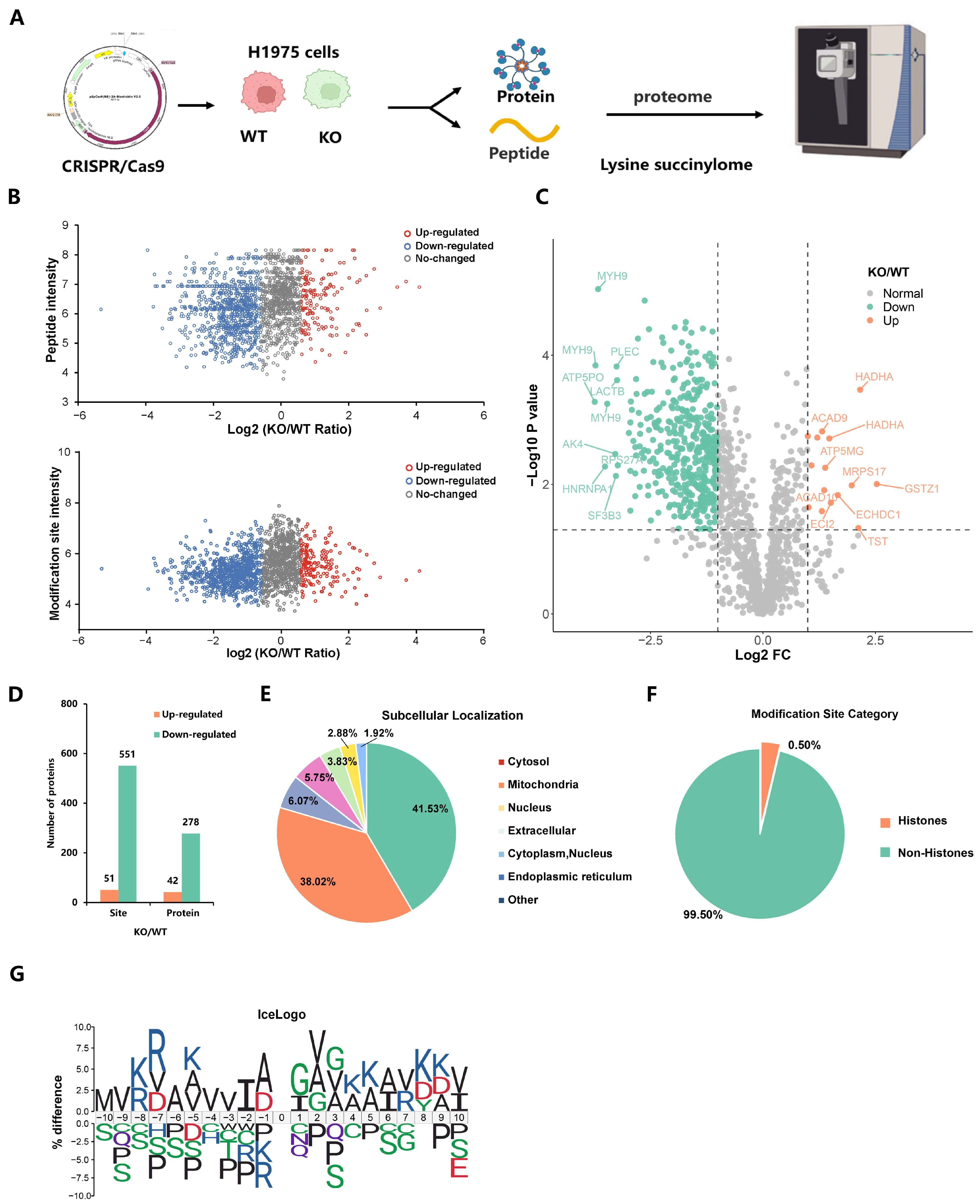
2.2. Deletion of EP300 Decreases Ksucc Levels on Various Proteins
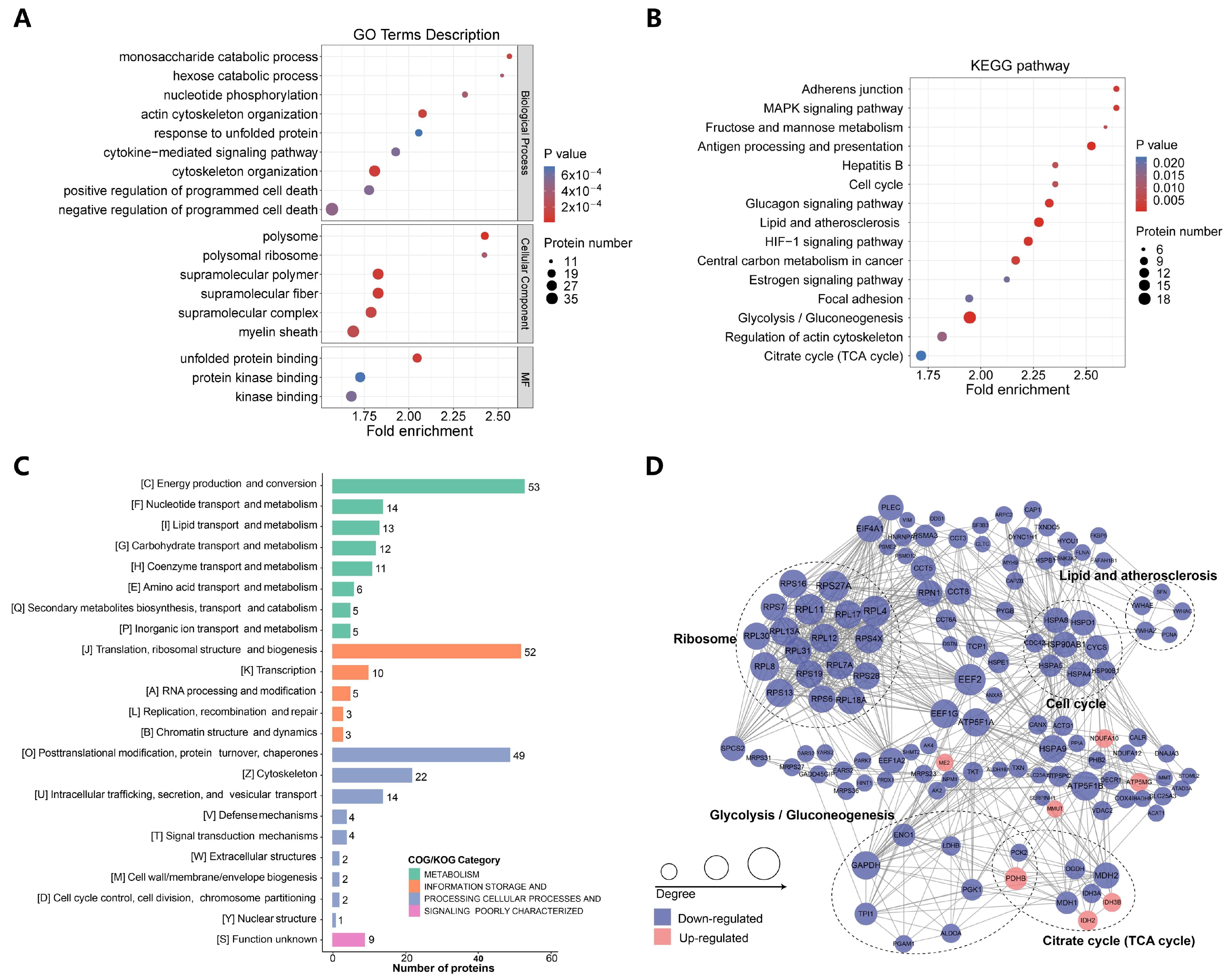
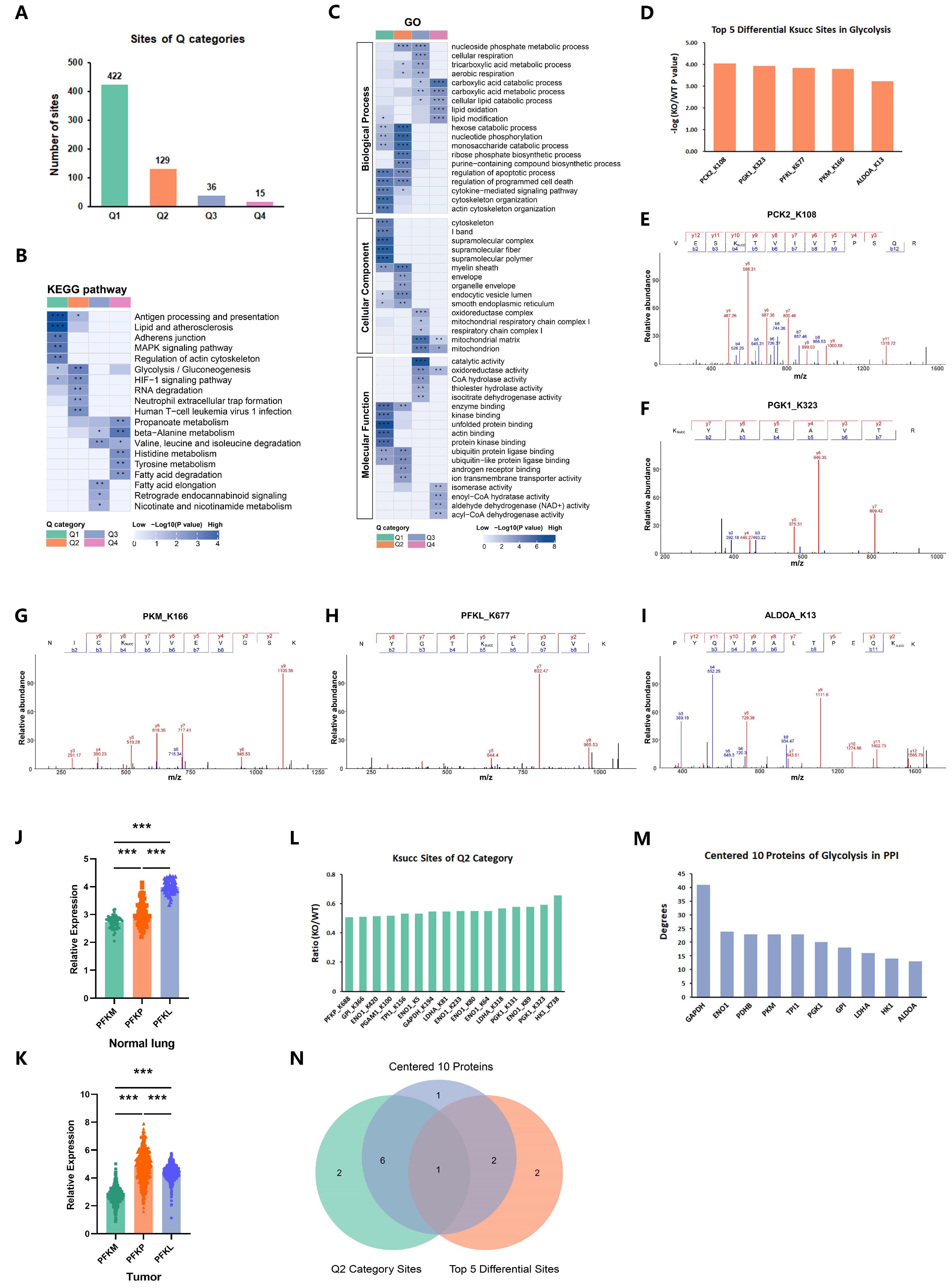
2.3. p300-Mediated Ksucc Modulates Tumor Carbohydrate Metabolism
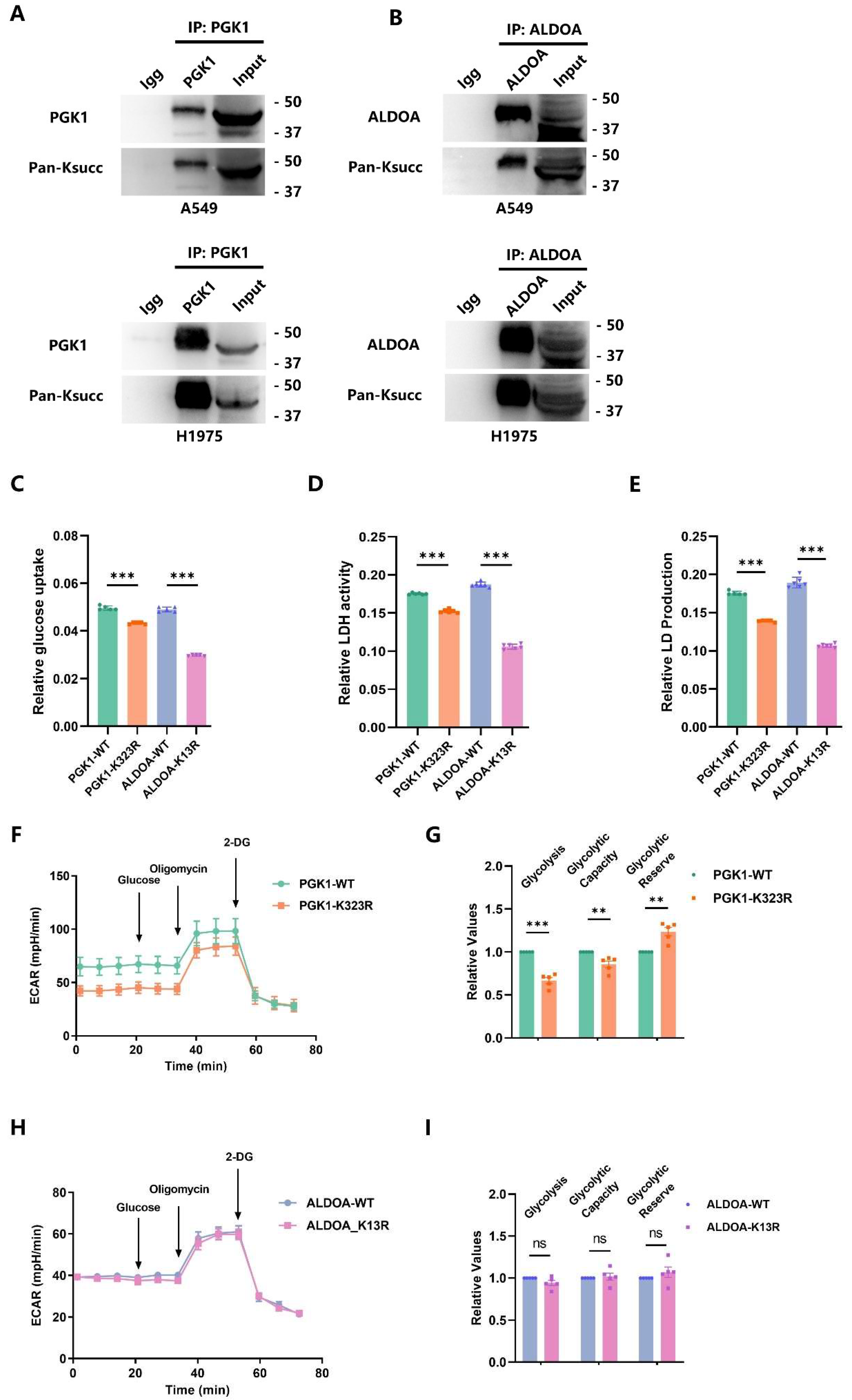
2.4. p300 Succinylates Key Glycolytic Enzymes
2.5. p300-Mediated PGK1-K323succ Enhances Tumor Glycolysis
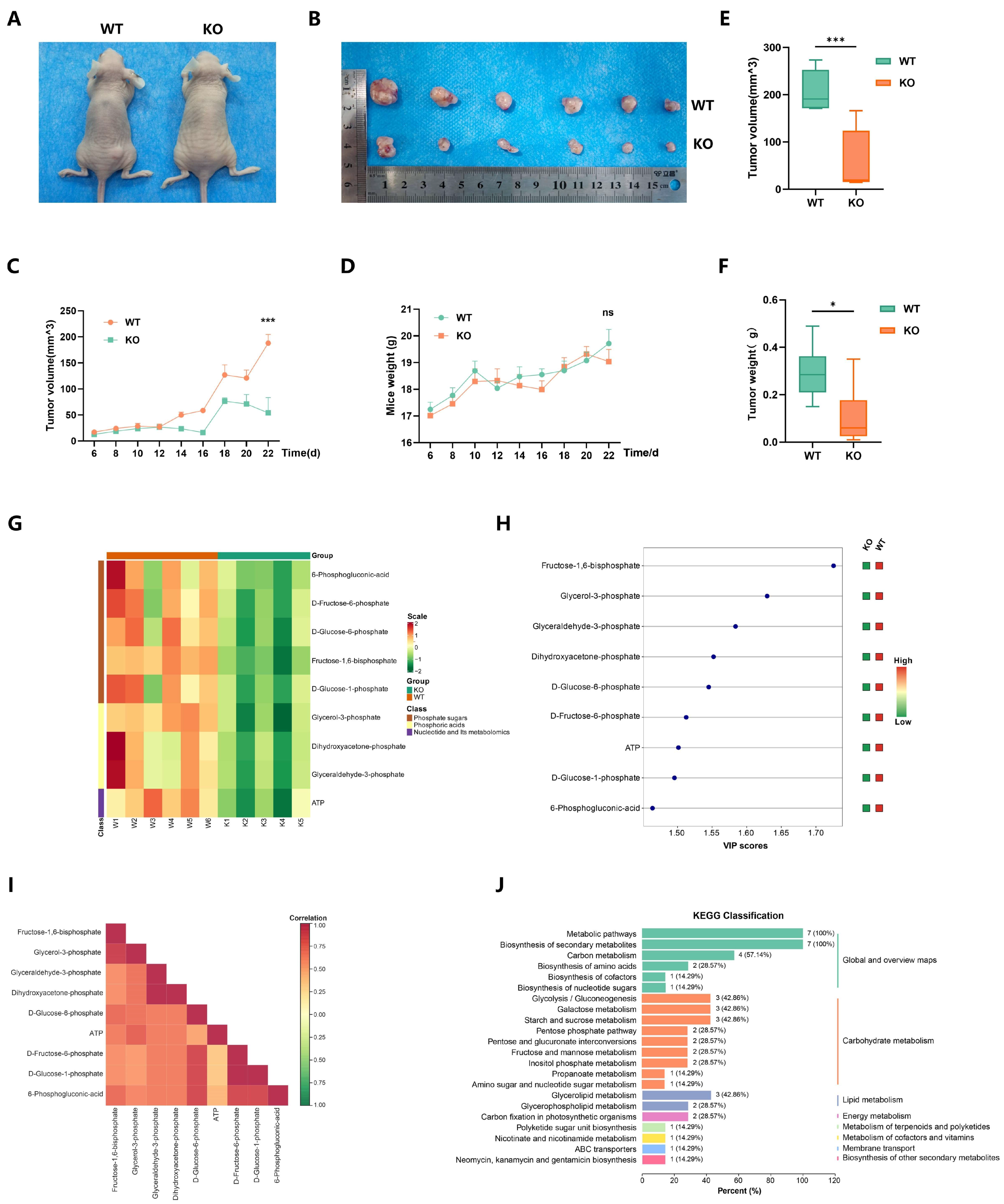
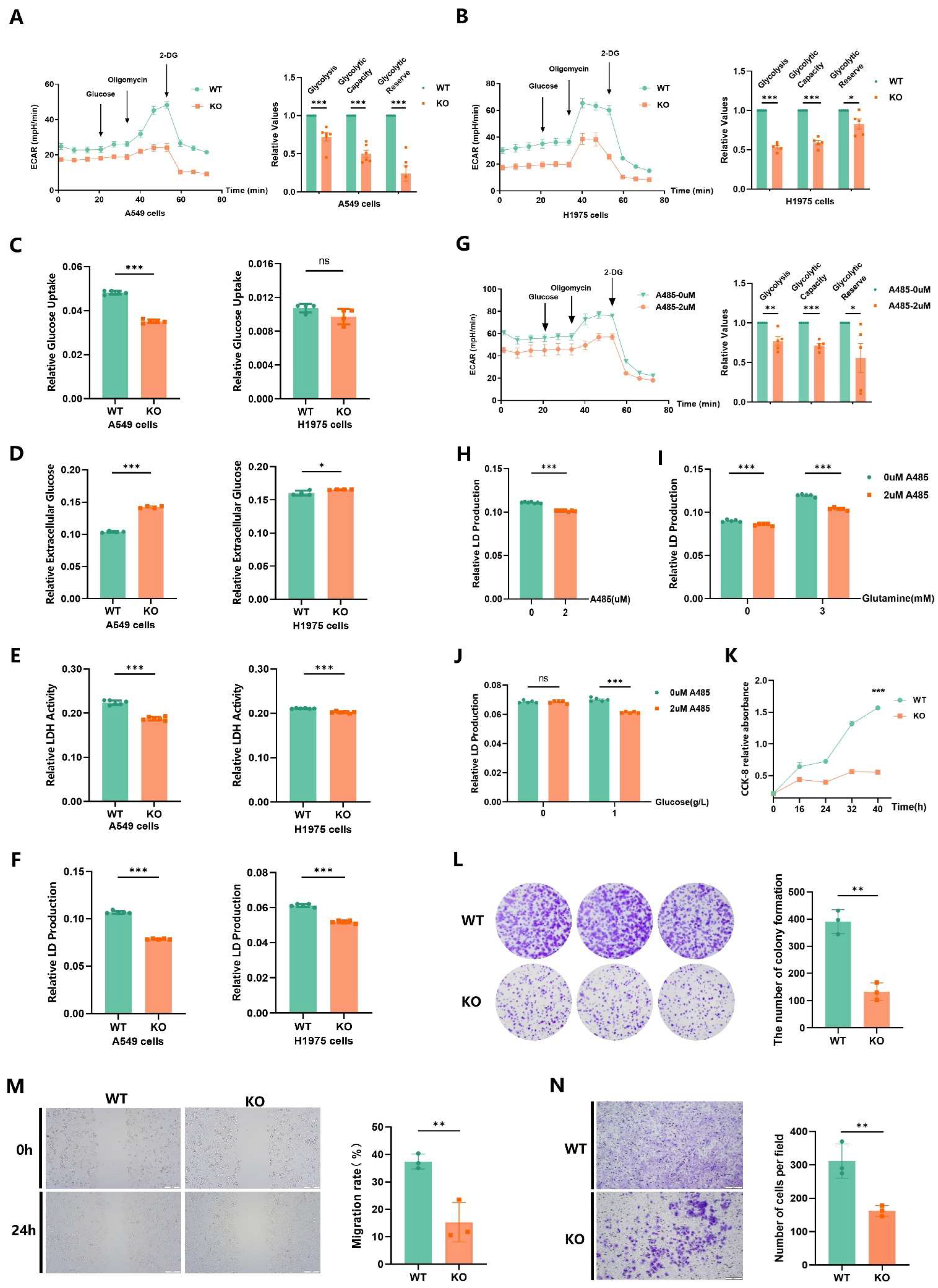
2.6. EP300 Knockout Inhibits Cell Energy Metabolism, Particularly Carbohydrate Metabolism In Vivo
2.7. EP300 Deficiency Impairs Glycolysis and Malignant Phenotypes of LUAD Cells
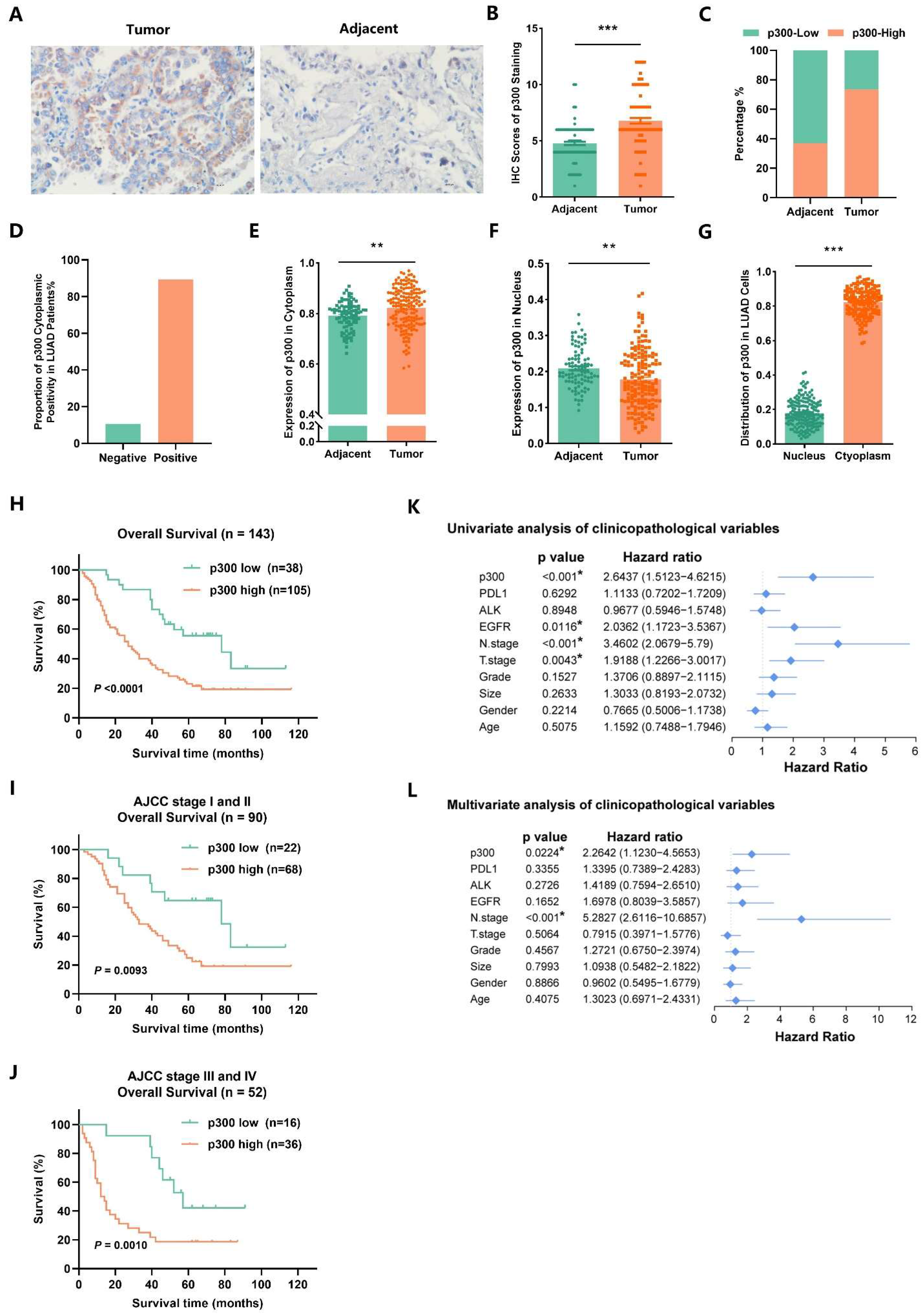
2.8. EP300 Coincides with Advanced Tumor Stage and Poor Prognosis of LUAD Individuals
3. Discussion
4. Materials and Methods
4.1. Key Resources Table
4.2. Cell Culture
4.3. Cell Lines
4.4. Western Blot and Antibodies
4.5. Quantitative Succinylome Analysis
4.5.1. Protein Extraction and Trypsin Digestion
4.5.2. Affinity Enrichment
4.5.3. LC-MS/MS Analysis
4.5.4. Database Search
4.5.5. Quantitative Analysis
4.5.6. Bioinformatics Methods
4.6. Targeted Metabolomics
4.7. Tests of Glucose Uptake, LDH Activity, and Lactic Acid Export
4.8. Seahorse Analysis
4.9. Proliferation, Migration, and Invasion Assays
4.10. LUAD Nude Mouse Models
4.11. IP and Co-IP
4.12. Immunohistochemistry Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Leiter, A.; Veluswamy, R.R.; Wisnivesky, J.P. The global burden of lung cancer: Current status and future trends. Nat. Rev. Clin. Oncol. 2023, 20, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Wang, L.; Jiang, M.; Zhao, D.; Wang, Y.; Zhang, F.; Li, J.; Zhang, X. A Review About Pembrolizumab in First-Line Treatment of Advanced NSCLC: Focus on KEYNOTE Studies. Cancer Manag. Res. 2020, 12, 6493–6509. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.; Dolde, C.; Lewis, B.C.; Wu, C.-S.; Dang, G.; Jungmann, R.A.; Dalla-Favera, R.; Dang, C.V. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 1997, 94, 6658–6663. [Google Scholar] [CrossRef]
- Guo, D.; Tong, Y.; Jiang, X.; Meng, Y.; Jiang, H.; Du, L.; Wu, Q.; Li, S.; Luo, S.; Li, M.; et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metab. 2022, 34, 1312–1324.e6. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ge, Y.; Dong, J.; Wang, H.; Zhao, T.; Wang, X.; Liu, J.; Gao, S.; Shi, L.; Yang, S.; et al. BZW1 Facilitates Glycolysis and Promotes Tumor Growth in Pancreatic Ductal Adenocarcinoma Through Potentiating eIF2α Phosphorylation. Gastroenterology 2022, 162, 1256–1271.e14. [Google Scholar] [CrossRef]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef]
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef]
- Meng, X.; Mujahid, H.; Zhang, Y.; Peng, X.; Redoña, E.D.; Wang, C.; Peng, Z. Comprehensive Analysis of the Lysine Succinylome and Protein Co-modifications in Developing Rice Seeds. Mol. Cell. Proteom. 2019, 18, 2359–2372. [Google Scholar] [CrossRef]
- Yang, Y.; Tapias, V.; Acosta, D.; Xu, H.; Chen, H.; Bhawal, R.; Anderson, E.T.; Ivanova, E.; Lin, H.; Sagdullaev, B.T.; et al. Altered succinylation of mitochondrial proteins, APP and tau in Alzheimer’s disease. Nat. Commun. 2022, 13, 1–19. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Boylston, J.A.; Sun, J.; Chen, Y.; Gucek, M.; Sack, M.N.; Murphy, E. Characterization of the cardiac succinylome and its role in ischemia–reperfusion injury. J. Mol. Cell. Cardiol. 2015, 88, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Y.; Fang, L.; Zhao, J.; Deng, S. The involvement of high succinylation modification in the development of prostate cancer. Front. Oncol. 2022, 12, 1034605. [Google Scholar] [CrossRef] [PubMed]
- Colak, G.; Xie, Z.; Zhu, A.Y.; Dai, L.; Lu, Z.; Zhang, Y.; Wan, X.; Chen, Y.; Cha, Y.H.; Lin, H.; et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteom. 2013, 12, 3509–3520. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Yu, H.; Liu, X.; Wang, T.; Yao, Y.; Zhou, Q.; Zheng, X.; Tan, F. Systematic identification of the lysine lactylation in the protozoan parasite Toxoplasma gondii. Parasites Vectors 2022, 15, 180. [Google Scholar] [CrossRef]
- Kurmi, K.; Hitosugi, S.; Wiese, E.K.; Boakye-Agyeman, F.; Gonsalves, W.I.; Lou, Z.; Karnitz, L.M.; Goetz, M.P.; Hitosugi, T. Carnitine Palmitoyltransferase 1A Has a Lysine Succinyltransferase Activity. Cell Rep. 2018, 22, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yuan, Y.; Yuan, H.; Wang, J.; Yun, H.; Geng, Y.; Zhao, M.; Li, L.; Weng, Y.; Liu, Z.; et al. Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Rep. 2021, 22, e50967. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Y.R.; Liu, K.; Yin, Z.; Liu, R.; Xia, Y.; Tan, L.; Yang, P.; Lee, J.-H.; Li, X.-J.; et al. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 2017, 552, 273–277. [Google Scholar] [CrossRef]
- Gibson, G.E.; Xu, H.; Chen, H.; Chen, W.; Denton, T.T.; Zhang, S. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. J. Neurochem. 2015, 134, 86–96. [Google Scholar] [CrossRef]
- Xu, S.; Fan, L.; Jeon, H.-Y.; Zhang, F.; Cui, X.; Mickle, M.B.; Peng, G.; Hussain, A.; Fazli, L.; Gleave, M.E.; et al. p300-Mediated Acetylation of Histone Demethylase JMJD1A Prevents Its Degradation by Ubiquitin Ligase STUB1 and Enhances Its Activity in Prostate Cancer. Cancer Res. 2020, 80, 3074–3087. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Yang, Y.-W.; Chen, F.; Yu, L.; Shen, S.-H.; Li, K.; Cui, B.; Lv, X.-X.; Zhang, C.; Yang, C.; et al. TRIB3 reduces CD8+T cell infiltration and induces immune evasion by repressing the STAT1-CXCL10 axis in colorectal cancer. Sci. Transl. Med. 2022, 14, eabf0992. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Pinello, C.E.; Luo, J.; Li, D.; Wang, Y.; Zhao, L.Y.; Jahn, S.C.; Saldanha, S.A.; Planck, J.; Geary, K.R.; et al. Small-molecule inhibitors of acetyltransferase p300 identified by high-throughput screening are potent anticancer agents. Mol. Cancer Ther. 2013, 12, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Gajer, J.M.; Furdas, S.D.; Gründer, A.; Gothwal, M.; Heinicke, U.; Keller, K.; Colland, F.; Fulda, S.; Pahl, H.L.; Fichtner, I.; et al. Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo. Oncogenesis 2015, 4, e137. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Wang, K.; Zhao, Y.; Ma, Q.; Chen, Z.; Huang, W. Effects of the Acetyltransferase p300 on Tumour Regulation from the Novel Perspective of Posttranslational Protein Modification. Biomolecules 2023, 13, 417. [Google Scholar] [CrossRef]
- Welti, J.; Sharp, A.; Brooks, N.; Yuan, W.; McNair, C.; Chand, S.N.; Pal, A.; Figueiredo, I.; Riisnaes, R.; Gurel, B.; et al. Targeting the p300/CBP Axis in Lethal Prostate Cancer. Cancer Discov. 2021, 11, 1118–1137. [Google Scholar] [CrossRef]
- Gopalakrishna Iyer, N.; Chin, S.F.; Ozdag, H.; Daigo, Y.; Hu, D.E.; Cariati, M.; Brindle, K.; Aparicio, S.; Caldas, C. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc. Natl. Acad. Sci. USA 2004, 101, 7386–7391. [Google Scholar] [CrossRef]
- Zhao, L.-J.; Loewenstein, P.M.; Green, M. The adenovirus E1A oncoprotein N-terminal transcriptional repression domain enhances p300 autoacetylation and inhibits histone H3 Lys18 acetylation. Genes Cancer 2015, 6, 30–37. [Google Scholar] [CrossRef]
- Liu, C.; Pan, Y.; Wang, X.; Lu, J.; Huang, B.; Li, X. Activation of RASSF2A by p300 induces late apoptosis through histone hyperacetylation. Cell Biol. Int. 2010, 34, 1133–1139. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Okan, N.A.; Bales, E.; Nascimento, L.; Cole, P.A.; Medrano, E.E. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002, 62, 6231–6239. [Google Scholar]
- Huang, H.; Tang, S.; Ji, M.; Tang, Z.; Shimada, M.; Liu, X.; Qi, S.; Locasale, J.W.; Roeder, R.G.; Zhao, Y.; et al. p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis. Mol. Cell 2018, 70, 663–678.e6. [Google Scholar] [CrossRef] [PubMed]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550, 128–132. [Google Scholar] [CrossRef]
- Wang, G.; Xu, Z.; Wang, C.; Yao, F.; Li, J.; Chen, C.; Sun, S. Differential phosphofructokinase-1 isoenzyme patterns associated with glycolytic efficiency in human breast cancer and paracancer tissues. Oncol. Lett. 2013, 6, 1701–1706. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martínez, C.; Aragón, J.J. Analysis of phosphofructokinase subunits and isozymes in ascites tumor cells and its original tissue, murine mammary gland. FEBS Lett. 1997, 409, 86–90. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Icard, P.; Shulman, S.; Farhat, D.; Steyaert, J.M.; Alifano, M.; Lincet, H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updates 2018, 38, 1–11. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Giatromanolaki, A. Warburg effect, lactate dehydrogenase, and radio/chemo-therapy efficacy. Int. J. Radiat. Biol. 2019, 95, 408–426. [Google Scholar] [CrossRef]
- Vaupel, P.; Multhoff, G. The Warburg Effect: Historical Dogma Versus Current Rationale. Adv. Exp. Med. Biol. 2021, 1269, 169–177. [Google Scholar]
- Contat, C.; Ancey, P.B.; Zangger, N.; Sabatino, S.; Pascual, J.; Escrig, S.; Jensen, L.; Goepfert, C.; Lanz, B.; Lepore, M.; et al. Combined deletion of Glut1 and Glut3 impairs lung adenocarcinoma growth. eLife 2020, 9, e53618. [Google Scholar] [CrossRef]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.; Skinner, M.E.; et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Stacpoole, P.W. Therapeutic Targeting of the Pyruvate Dehydrogenase Complex/Pyruvate Dehydrogenase Kinase (PDC/PDK) Axis in Cancer. JNCI J. Natl. Cancer Inst. 2017, 109, djx071. [Google Scholar] [CrossRef] [PubMed]
- Xiangyun, Y.; Xiaomin, N.; Linping, G.; Yunhua, X.; Ziming, L.; Yongfeng, Y.; Zhiwei, C.; Shun, L. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget 2017, 8, 6984–6993. [Google Scholar] [CrossRef] [PubMed]
- Hirschey, M.D.; Zhao, Y. Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Mol. Cell. Proteom. 2015, 14, 2308–2315. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef]
- Weinert, B.T.; Narita, T.; Satpathy, S.; Srinivasan, B.; Hansen, B.K.; Schölz, C.; Hamilton, W.B.; Zucconi, B.E.; Wang, W.W.; Liu, W.R.; et al. Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 2018, 174, 231–244.e12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sprung, R.; Tang, Y.; Ball, H.; Sangras, B.; Kim, S.C.; Falck, J.R.; Peng, J.; Gu, W.; Zhao, Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteom. 2007, 6, 812–819. [Google Scholar] [CrossRef]
- Sabari, B.R.; Tang, Z.; Huang, H.; Yong-Gonzalez, V.; Molina, H.; Kong, H.E.; Dai, L.; Shimada, M.; Cross, J.R.; Zhao, Y.; et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell. 2015, 58, 203–215. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, H.; Gao, P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 2022, 13, 877–919. [Google Scholar] [CrossRef]
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarska, Z.; Ortega, E.; Goudarzi, A.; Huang, H.; Kim, S.; Márquez, J.A.; Zhao, Y.; Khochbin, S.; Panne, D. Structure of p300 in complex with acyl-CoA variants. Nat. Chem. Biol. 2017, 13, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Ponugoti, B.; Kim, D.-H.; Xiao, Z.; Smith, Z.; Miao, J.; Zang, M.; Wu, S.-Y.; Chiang, C.-M.; Veenstra, T.D.; Kemper, J.K. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 2010, 285, 33959–33970. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Liu, Q.; Zhang, L.; Zhu, Q.; Wang, S.; Zhu, K.; Deng, R.; Liu, Y.; Yuan, G.; Wang, X.; et al. Selective inhibition of CBP/p300 HAT by A-485 results in suppression of lipogenesis and hepatic gluconeogenesis. Cell Death Dis. 2020, 11, 745. [Google Scholar] [CrossRef]
- Cai, L.Y.; Chen, S.J.; Xiao, S.H.; Sun, Q.J.; Ding, C.H.; Zheng, B.N.; Zhu, X.Y.; Liu, S.Q.; Yang, F.; Yang, Y.X.; et al. Targeting p300/CBP Attenuates Hepatocellular Carcinoma Progression through Epigenetic Regulation of Metabolism. Cancer Res. 2021, 81, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Liang, Y.; Kim, C.; Jaganathan, A.; Ji, D.; Han, X.; Yang, X.; Jia, Y.; Gu, R.; Wang, C.; et al. Structural mechanism of BRD4-NUT and p300 bipartite interaction in propagating aberrant gene transcription in chromatin in NUT carcinoma. Nat. Commun. 2023, 14, 378. [Google Scholar] [CrossRef]
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B.; et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013, 18, 920–933. [Google Scholar] [CrossRef]
- Casey, J.R.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11, 50–61. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, G.; Chu, H.; Wang, X.; Xiong, L.; Cai, G.; Liu, R.; Gao, H.; Tao, B.; Li, W.; et al. Macrophage-Associated PGK1 Phosphorylation Promotes Aerobic Glycolysis and Tumorigenesis. Mol. Cell 2018, 71, 201–215.e7. [Google Scholar] [CrossRef]
- Hu, H.; Zhu, W.; Qin, J.; Chen, M.; Gong, L.; Li, L.; Liu, X.; Tao, Y.; Yin, H.; Zhou, H.; et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 2017, 65, 515–528. [Google Scholar] [CrossRef]
- Yu, T.; Zhao, Y.; Hu, Z.; Li, J.; Chu, D.; Zhang, J.; Li, Z.; Chen, B.; Zhang, X.; Pan, H.; et al. MetaLnc9 Facilitates Lung Cancer Metastasis via a PGK1-Activated AKT/mTOR Pathway. Cancer Res. 2017, 77, 5782–5794. [Google Scholar] [CrossRef] [PubMed]
- Yokomizo, C.; Yamaguchi, K.; Itoh, Y.; Nishimura, T.; Umemura, A.; Minami, M.; Yasui, K.; Mitsuyoshi, H.; Fujii, H.; Tochiki, N.; et al. High expression of p300 in HCC predicts shortened overall survival in association with enhanced epithelial mesenchymal transition of HCC cells. Cancer Lett. 2011, 310, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.-S.; Cai, M.-Y.; Chen, J.-W.; Guan, X.-Y.; Kung, H.-F.; Zeng, Y.-X.; Xie, D. High Expression of p300 in Human Breast Cancer Correlates with Tumor Recurrence and Predicts Adverse Prognosis. Chin. J. Cancer Res. 2011, 23, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Gu, M.L.; Meng, F.S.; Jiao, W.R.; Zhou, X.X.; Yao, H.P.; Ji, F. Histone acetyltransferase p300/CBP inhibitor C646 blocks the survival and invasion pathways of gastric cancer cell lines. Int. J. Oncol. 2017, 51, 1860–1868. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; He, D.; Cheng, L.; Huang, C.; Zhang, Y.; Rao, X.; Kong, Y.; Li, C.; Zhang, Z.; Liu, J.; et al. p300/CBP inhibition enhances the efficacy of programmed death-ligand 1 blockade treatment in prostate cancer. Oncogene 2020, 39, 3939–3951. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Liu, C.; Zhang, L.; Lv, D.; Weng, Y.; Cheng, Z.; Chen, X.; Zhan, J. Isonicotinylation is a histone mark induced by the anti-tuberculosis first-line drug isoniazid. Nat. Commun. 2021, 12, 5548. [Google Scholar] [CrossRef]
- Deng, Y.; Xie, M.; Li, Q.; Xu, X.; Ou, W.; Zhang, Y.; Xiao, H.; Yu, H.; Zheng, Y.; Liang, Y.; et al. Targeting Mitochondria-Inflammation Circuit by β-Hydroxybutyrate Mitigates HFpEF. Circ. Res. 2021, 128, 232–245. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Dimmer, E.C.; Huntley, R.P.; Alam-Faruque, Y.; Sawford, T.; O’Donovan, C.; Martin, M.J.; Bely, B.; Browne, P.; Mun Chan, W.; Eberhardt, R.; et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2012, 40, D565–D570. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Cheng, A.; Grant, C.E.; Noble, W.S.; Bailey, T.L. MoMo: Discovery of statistically significant post-translational modification motifs. Bioinformatics 2019, 35, 2774–2782. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.W.; Kim, H.C.; Kim, S.H.; Park, Y.A.; Cho, Y.B.; Yun, S.H.; Lee, W.Y.; Chun, H.-K. Prognostic impact of p300 expression in patients with colorectal cancer. J. Surg. Oncol. 2013, 108, 374–377. [Google Scholar] [CrossRef] [PubMed]
| Clinicopathological Variables | EP300 Expression | χ2 | p | |
|---|---|---|---|---|
| High (n = 105) | Low (n = 38) | |||
| Gender | ||||
| Male | 65 | 21 | 0.513 | 0.474 |
| Female | 40 | 17 | ||
| Age (year) | ||||
| <60 | 65 | 21 | 0.513 | 0.474 |
| ≥60 | 40 | 17 | ||
| PDL1 expression | ||||
| Positive | 45 | 10 | 1.495 | 0.221 |
| Negative | 50 | 19 | ||
| EGFR expression | ||||
| Positive | 17 | 1 | 4.345 | 0.037 * |
| Negative | 72 | 29 | ||
| ALK expression | ||||
| Positive | 21 | 9 | 0.528 | 0.468 |
| Negative | 72 | 22 | ||
| Tumor size (cm) | ||||
| <5 | 76 | 27 | 0.024 | 0.876 |
| ≥5 | 29 | 11 | ||
| Tumor differentiation | ||||
| I–II | 68 | 22 | 0.673 | 0.412 |
| III–IV | 36 | 16 | ||
| T stage | ||||
| T1–T2 | 72 | 30 | 3.901 | 0.048 * |
| T3–T4 | 33 | 5 | ||
| N stage | ||||
| N0–N1 | 67 | 26 | 0.079 | 0.779 |
| N2–N3 | 18 | 8 | ||
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-p300 antibody | Santa Cruz, Santa Cruz, CA, USA | sc-48343 |
| Mouse monoclonal anti-α-tubulin antibody | Proteintech, Rosemont, IL, USA | 66031-1-lg |
| Mouse monoclonal anti-succinyllysine antibody | PTM Bio, Hangzhou, China | PTM-419 |
| Rabbit polyclonal anti-ALDOA antibody | Proteintech, Rosemont, IL, USA | 11217-1-AP |
| Rabbit polyclonal anti-PGK1 antibody | Proteintech, Rosemont, IL, USA | 17811-1-AP |
| Rabbit polyclonal anti-PKM antibody | Proteintech, Rosemont, IL, USA | 25659-1-AP |
| Rabbit recombinant anti-FLAG antibody | Proteintech, Rosemont, IL,USA | 80010-1-RR |
| Goat Anti-Rabbit IgG (H+L) secondary antibody, HRP | Invitrogen, Carlsbad, CA, USA | 31460 |
| Goat Anti-Mouse IgG (H+L) secondary antibody, HRP | Invitrogen, Carlsbad, CA, USA | 31430 |
| Critical Commercial Assays | ||
| Pierce Crosslink Immunoprecipitation Kit | Thermo Fisher Scientific, Waltham, MA, USA | 26147 |
| Pierce Co-Immunoprecipitation Kit | Thermo Fisher Scientific, Waltham, MA, USA | 26149 |
| Pierce BCA protein assay kit | Thermo Fisher Scientific, Waltham, MA, USA | 23225 |
| Glucose assay kit | Jianchengbio, Nanjing, China | F006-1-1 |
| Lactate dehydrogenase assay kit | Jianchengbio, Nanjing, China | A020-1-2 |
| Lactic acid assay kit | Jianchengbio, Nanjing, China | A019-2-1 |
| Seahorse XF RPMI Assay Medium Pack | Agilent, Santa Clara, CA, USA | 103681-100 |
| Seahorse XF Glycolysis Stress Test Kit | Agilent, Santa Clara, CA, USA | 103020-100 |
| General Two-Step Test Kit | ZSGB Bio, Beijing, China | PV-9000 |
| Plasmids | ||
| EP300 CRISPR/Cas9 KO plasmid | Sangon Biotech, Shanghai, China | This paper |
| pLV3-CMV-PGK1-3×FLAG-CopGFP | MiaoLing bio, Wuhan, China | This paper |
| pLV3-CMV-PKM-3×FLAG-CopGFP | MiaoLing bio, Wuhan, China | This paper |
| pLV3-CMV-ALDOA-3×FLAG-CopGFP | MiaoLing bio, Wuhan, China | This paper |
| pLV3-CMV-PGK1-K323R-3×FLAG-CopGFP | MiaoLing bio, Wuhan, China | This paper |
| pLV3-CMV-PKM-K166R-3×FLAG-CopGFP | MiaoLing bio, Wuhan, China | This paper |
| pLV3-CMV-ALDOA-K13R-3×FLAG-CopGFP | MiaoLing bio, Wuhan, China | This paper |
| Biological Samples | ||
| Human LUAD tissue microarray | Outdo Biotech, Shanghai, China | HLugA180Su04, HLugA180Su08 |
| Cell Lines | ||
| A549 | ATCC, Gaithersburg, MD, USA | CCL-185 |
| H1975 | ATCC, Gaithersburg, MD, USA | CRL-5908 |
| Organisms | ||
| BALB/c nude mice | HFK Bioscience, Beijing, China | BALB/c nude mice |
| Reagents | ||
| Lipofectamine 2000 | Invitrogen, Carlsbad, CA, USA | 11668019 |
| Blasticidin | Invitrogen, Carlsbad, CA, USA | Ant-bl-1 |
| Ampicillin sodium | Solarbio, Beijing, China | A8180 |
| CCK-8 | Engreen, Beijing, China | EC008 |
| Hematoxylin stain solution, Gill I | Solarbio, Beijing, China | G4492 |
| Matrigel gel | R&D Systems, Minneapolis, MN, USA | 3432-010-01 |
| Software and Algorithms | ||
| Prism 8.0 | GraphPad, San Diego, CA, USA | N/A |
| SPSS 25.0 | IBMCorp, New York, NY, USA | N/A |
| ImageJ 1.51 | National Institutes of Health, Bethesda, MD, USA | N/A |
| Image Lab 6.1 | BIO-RAD, Hercules, CA, USA | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.; Zeng, Q.; Wang, K.; Qian, M.; Li, J.; Wang, H.; Zhang, H.; Jiang, J.; Chen, Z.; Huang, W. Acetyltransferase P300 Regulates Glucose Metabolic Reprogramming through Catalyzing Succinylation in Lung Cancer. Int. J. Mol. Sci. 2024, 25, 1057. https://doi.org/10.3390/ijms25021057
Ma Q, Zeng Q, Wang K, Qian M, Li J, Wang H, Zhang H, Jiang J, Chen Z, Huang W. Acetyltransferase P300 Regulates Glucose Metabolic Reprogramming through Catalyzing Succinylation in Lung Cancer. International Journal of Molecular Sciences. 2024; 25(2):1057. https://doi.org/10.3390/ijms25021057
Chicago/Turabian StyleMa, Qingzhi, Qingmei Zeng, Kun Wang, Meirui Qian, Jingzhuo Li, Hao Wang, Huijie Zhang, Jianli Jiang, Zhinan Chen, and Wan Huang. 2024. "Acetyltransferase P300 Regulates Glucose Metabolic Reprogramming through Catalyzing Succinylation in Lung Cancer" International Journal of Molecular Sciences 25, no. 2: 1057. https://doi.org/10.3390/ijms25021057
APA StyleMa, Q., Zeng, Q., Wang, K., Qian, M., Li, J., Wang, H., Zhang, H., Jiang, J., Chen, Z., & Huang, W. (2024). Acetyltransferase P300 Regulates Glucose Metabolic Reprogramming through Catalyzing Succinylation in Lung Cancer. International Journal of Molecular Sciences, 25(2), 1057. https://doi.org/10.3390/ijms25021057





