1. Introduction
The brain, which is the human body’s most important structural element, contains 50–100 trillion neurons [
1]. It is also known as the human body’s core section. Furthermore, it is known as the “processor” or “kernel” of the nervous system, and it plays the most important and critical role in the nervous system [
2,
3]. To the best of our knowledge, diagnosing brain disease is too difficult and complex due to the presence of the skull around it [
4].
Utilizing technology to evaluate individuals with the aim of identifying, tracking, and treating medical issues is known as medical imaging. In medical imaging, magnetic resonance imaging (MRI) is a precise and noninvasive technique that can be used to diagnose a variety of disorders. In the last few decades, many scholars have proposed various state-of-the-art methods for brain MRI classification, and most of them focused on various modules of the MRI systems.
A latest convolutional neural network-based MRI method, data expansion, and image processing were proposed by [
5] to recognize brain MRI images in various diseases. They compared the significance of their approach with pre-trained VGG-16 in the presence of transfer learning using a small dataset. Another deep learning-based method for detecting brain tumors in MRI images was created by [
6]. This approach was divided into three phases: in the first phase, the CNN-based classifiers were implemented; while in the second phase, a region-based CNN was utilized on the output of the first phase; finally, in the third phase, the boundary of the brain tumor was focused and segmented by the Chan-Vese energy function followed by the edge detection method. On the other hand, an integrated approach was designed by [
7], combining a mathematical morphological operator and an OASIS operator. In the first step, they extracted the largest connected area, such as the brain. After this, the unsupervised framework was employed to extract the various axial slices of brain. The main contribution of this paper was to identify the brains automatically, which was evaluated through five matrices using a publicly available dataset. Similarly, the glioma disease was analyzed by [
8], where they utilized the Gaussian Naïve Bayes technique. In their approach, they employed the grow cut method followed by 3D features on MRI images. Then, they statistically analyzed the corresponding values through Spearman and Mann–Whitney U tests and achieved better results than the standard MRI dataset. The authors of [
9] proposed an integrated approach for the detection of the tumor on brain MRI images. This approach is the combination of two well-known methods, morphological edge detection followed by fuzzy methods, respectively. In this method, the authors located the tumor through edge detection methods, while their performances were enhanced by a fuzzy algorithm, and showed the best recognition rate on a brain MRI dataset.
The authors [
10] recently developed a piece of work that used deep learning and transfer learning to classify different MRI pictures of brain tumors. They showed acceptable results on a small public dataset of brain tumor MRI images. Likewise, an artificial neural network (ANN) based approach was developed by [
11] that efficiently classified normal and abnormal MRI images. In this approach, they utilized a median filter in the pre-processing step in order to diminish the noise from MRI images. For the feature extraction, they employed the wavelet transform to extract the best features from the enhanced images. Then the dimensions are reduced by employing color moments. Finally, for the classification of normal and abnormal MRI images, the fast-forward ANN has been utilized. They utilized only 70 images for their corresponding experiments. In contrast, a cutting-edge system with three fundamental modules was created by [
12] for the identification of brain tumors. They utilized histogram equalization in the pre-processing module in order to enhance the contrast of the brain MRI images. While, in the feature extraction module, they used principal component analysis followed by independent analysis to extract prominent features. Finally, they utilized an integrated classifier that was based on Naïve Bayes recurrent neural networks. They claimed better performance using publicly available brain MRI datasets.
Additionally, [
13] established a reliable method for the recognition and segmentation of the brain tumor in MRI images. To distinguish between a wide range of tumor tissues in normal and abnormal MRI images and segment the tumor area accordingly, they used Berkeley’s wavelet transform followed by a deep learning classifier. Most of the afore-mentioned approaches showed better performances and acceptable results on small brain MRI images. However, their performances and classification accuracies are accordingly decreased on large brain MRI datasets.
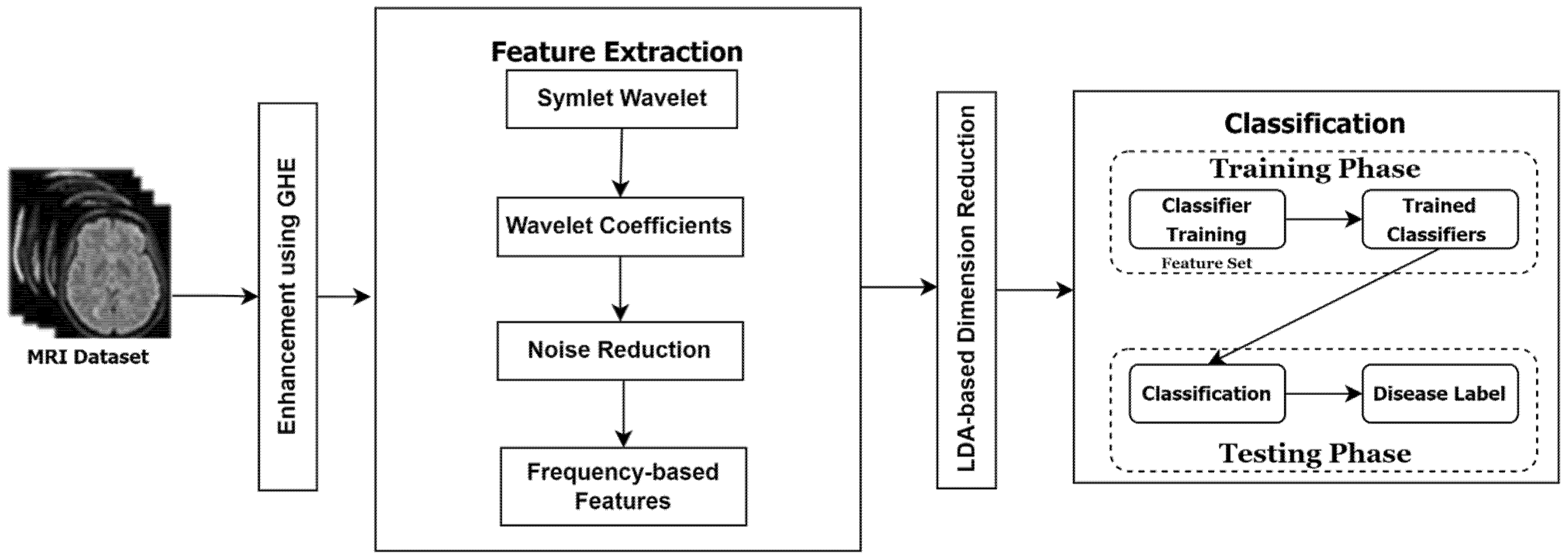
As a result, this study suggests a precise and effective system for the classification of brain diseases using MRI images. In this approach, the following contributions have been made.
In the preprocessing step, the MRI images have been enhanced through existing well-known techniques like global histogram equalization.
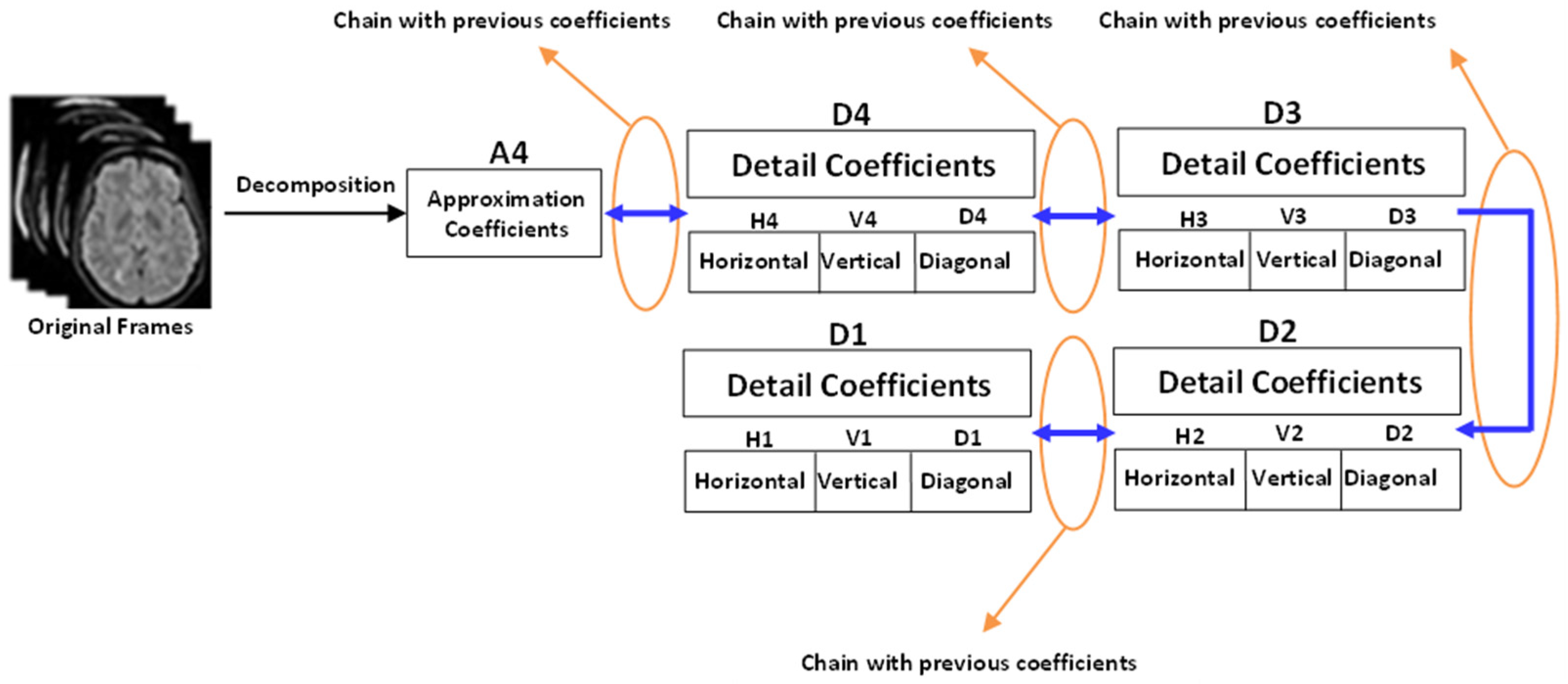
Then, for feature extraction, an accurate and robust technique is proposed that is based on symlet wavelet transform. This technique yields better classification outcomes because it can handle the orthogonal, biorthogonal, and reverse biorthogonal features of gray scale images. Our tests support the frequency-based supposition. The wavelet coefficients’ statistical reliance was assessed for each frame of grayscale MRI data. A gray scale frame’s joint probability is calculated by collecting geometrically aligned MRI images for each wavelet coefficient. In order to determine the wavelet coefficients obtained from these distributions, the mutual information between the two MRI images is used to calculate the statistical dependence’s intensity.
Following the extraction of the best feature, a linear discriminant analysis (LDA) was used to minimize the feature space’s dimensions.
Following the selection of the best features, the model is trained using logistic regression, which uses the coefficient values to determine which characteristics (i.e., which pixels) are crucial in deciding which class a sample belongs to. The per-class probability for each sample may be computed using the coefficient values, and the conditional probability for each class can be computed using this method. In general, the class with the highest probability might be found to acquire the predicted label.
In order to assess the performance of the proposed approach, a comprehensive set of experiments was performed using the brain MRI dataset, which has 24 various kinds of brain diseases.
For this assessment, a comprehensive dataset is collected from Harvard Medical School [
14] and Open Access Series of Imaging Studies (OASIS) [
15], which has total 24 various kinds of diseases such as Fatal stroke (FS), Motor neuron disease (MN), Glioma (GL), Vascular dementia (VD), Cavernous angioma (CA), Hypertensive encephalopathy (HY), Cerebral calcinosis (CC), Metastatic adenocarcinoma (MA), Chronic subdural hematoma (CS), Multiple embolic infarctions (MI), AIDS dementia (AD), Cerebral toxoplasmosis (CT), Meningioma (M), Pick’s disease (PD), Sarcoma (SR), Alzheimer’s disease (AL), Creutzfeld-Jakob disease (CJ), Metastatic bronchogenic carcinoma (MB), Alzheimer’s disease with visual agnosia (AV), Multiple sclerosis (MS), Lyme encephalopathy (LE), Herpes encephalitis (HE), Cerebral haemorrhage (CH), Huntington’s disease (HD), and normal brain (NB). The proposed technique achieved better performance on this comprehensive MRI dataset.
The entire paper is organized as follows:
Section 2 describes the existing MRI systems along with their respective disadvantages.
Section 3 presents the proposed approach, while, the experimental setup is described in
Section 4. Based on the experimental setup, the results are shown in
Section 5. Finally,
Section 6 summarizes the proposed approach along with future directions.
2. Literature Review
In the past couple of years, lots of efficient and accurate studies have been done for the classification of numerous types of brain ailments using MRI images. Most of these studies showed the best performances on a small dataset of brain MRI. However, their performances degraded accordingly on larger testing datasets. Therefore, a robust and accurate framework has been designed that showed good classification results on a large brain MRI dataset.
A novel method has been proposed by [
16] that is based on statistical features coupled with various machine learning techniques. They claimed the best performance on a small MRI dataset. However, computational-wise, this approach is much more expensive. A state-of-the-art framework has been designed by [
17], which classified the Alzheimer disease using MRI images. In this framework, the corresponding MRI image has been enhanced in the preprocessing step, while the brain tissues are segmented in the post-processing step. Then several deep learning techniques (convolutional neural network) are employed to classify the corresponding disease. However, the convolutional neural network has an overfitting problem [
18]. Also, this approach has been tested and validated on a small dataset. Similarly, an accurate and robust method was proposed by [
19]. They utilized stepwise linear discriminant analysis (SWLDA) for feature extraction and support vector machines for classification on a large brain MRI dataset. They achieved the best performance using the MRI dataset. However, SWLDA is a linear method that might be employed in a small subspace of binary classification problems [
20].
On the other hand, an integrated approach was designed by [
21], where the authors integrated a feature-based classifier and an image-based classifier for brain tumor classification. Further, their proposed architecture was based on deep neural networks and deep convolutional networks. They achieved a comparable classification rate. However, a huge number of training images and the carefully constructed deep networks required for this approach [
22]. Similarly, a state-of-the-art framework was designed by [
23] in order to classify brain MRI along with gender and age. They utilized deep neural network, convolutional network, LeNet, AlexNet, ResNet, and SVM to classify abnormal and normal MRIs accurately. However, they showed better performance on a small dataset, and most of the experiments were in a static environment. Likewise, the authors of [
24] proposed an efficient brain image classification system using an MRI dataset. In their system, they extracted the features by shape and textual method, such as region based active contour, and showed good performance. However, the major limitation of the region-based method is its’ sensitivity to the initialization, and because of this, the region of interest does not segment properly [
25].
A cutting-edge method for classifying various brain illnesses using MRI images was reported by Nayak et al. [
26]. They utilized convolutional neural network-based dense EfficientNet coupled with min-mix normalization for categorization, and they showed better performance using the MRI dataset. However, this approach employs a huge number of operations, which make the model computationally slower [
27]. Similarly, an integrated framework was designed by [
28], where the authors employed a semantic segmentation network coupled with GoogleNet and a convolutional neural network (CNN) for brain tumor classification using MRI and CT images. They achieved better results using a small dataset of brain MRI and CT. However, in GoogleNet, the connected layers cannot manage various input image sizes [
29].
A fully automated brain tumor segmentation approach was developed by [
30] that was based on support vector machines and CNN. Moreover, the segmentation was done through the details of various techniques such as structural, morphological, and relaxometry. However, the methodologies utilized in this framework have comparatively lower significance with larger amounts of input MRI images [
31]. Because it is a challenging task for these methods to accurately detect the abnormalities in the brain MRI images [
31]. Moreover, a modified CNN based model was developed by [
32] for the analysis of brain tumors. The authors employed CNN along with parametric optimization techniques such as the sunflower optimization algorithm (SFOA), the forensic-based investigation algorithm (FBIA), and the material generation algorithm (MGA). They claimed the highest accuracy of classification using the MRI dataset. However, SFOA is very sensitive to initializing and premature convergence [
33]. Moreover, in MGA, the predictions are made based on single-slice inputs, hypothetically restraining the information available to the network [
34].
An integrated framework was proposed by [
35], which was based on the VGG19 feature extractor along with a progressive growing generative adversarial network (PGGAN) augmentation model for brain tumor classification using MRI images. They achieved good classification results on a publicly available MRI dataset. However, this approach cannot generate high-resolution images via the PGGAN model [
36]. Moreover, this approach might not generate new examples with objects in the desired condition [
37]. Another state-of-the-art scheme was proposed by [
38], which contained some steps such as preprocessing, segmentation, feature extraction, and classification. The image was enhanced via a Wiener filter followed by edge detection. The tumor was segmented by a mean shift clustering algorithm. The features were extracted from the segmented tumor through the gray level co-occurrence matrix (GLCM), and the classification was done by support vector machines. However, the GLCM method is robust to Gaussian noise, and the extracted features are based on the difference between the corresponding pixels, but the magnitude of the difference was not taken into account [
39].
A state-of-the-art fused method was developed by [
40] that was based on gray level co-occurrence matrix (GLCM), spatial grey level dependence matrix (SGLDM), and Harris hawks optimization (HHO) techniques followed by support vector machines for brain tumor detection. However, this approach depends on the manual selection of the region of interest, due to which the corresponding results in the dependence of parameter values on the extracted region might not be selected [
40].
As a result, in this work, a solid framework was created for the classification of various brain illnesses using an MRI dataset. A symlet wavelet-based feature extraction method was designed and is used in the proposed framework to extract the key features from brain MRI images. Furthermore, the dimensions of the feature space are reduced by LDA, and the classification is done through logistic regression. The proposed approach achieved the best classification results using MRI images compared to the existing publications.
6. Conclusions
In medical imaging, magnetic resonance imaging (MRI) is a precise and noninvasive technique that can be used to diagnose a variety of disorders. Various algorithms for brain MRI categorization have been developed by a number of researchers. On small MRI datasets, the majority of these algorithms did well and had higher identification rates. When dealing with larger MRI datasets, however, their performance degrades. As a result, the objective is to create a quick and precise classification system that can sustain a high identification rate across a sizable MRI dataset. As a result, in this study, a well-known enhancement method called global histogram equalization (GHE) is used to reduce undesirable information in MRI images. Furthermore, a reliable and accurate feature extraction technique is suggested for extracting and selecting the most prominent feature from an MRI picture. The suggested feature extraction method for grayscale photos is a compactly supported wavelet that has the greatest number of vanishing moments and the least amount of asymmetry for a given support width. Our study supports the frequency-based hypothesis. The statistical dependence of the wavelet coefficients is assessed for all grayscale MRI pictures. A gray scale frame’s joint probability is calculated by collecting geometrically aligned MRI pictures for each wavelet coefficient. Using mutual information for the wavelet coefficients derived using these distributions, the degree of statistical dependence between the two MRI images is evaluated. Furthermore, the linear discriminant analysis is used after extracting the features to choose the best features and lower the dimensions of the feature space, which may improve the performance of the recommended method for generating feature vectors. Finally, logistic regression is used to classify the brain illnesses. A huge dataset from Harvard Medical School and the OASIS is utilized, which comprises a total of 24 distinct types of brain disorders, to assess and test the suggested method.
In the proposed approach, the optimum set of features is extracted from the MRI images that are important for improving the accuracy. Subsequently, the rate of convergence is also one of the main factors improving the accuracy of this research; however, the number of features in this approach is not too high to reduce the computational complexity. Therefore, in the future, the proposed approach will be enclosed using MRI datasets in various healthcare domains. Moreover, the proposed approach is robust and efficient, which might be useful for real-time diagnostic applications in the future. Therefore, the proposed method might play a significant role in helping the radiologists and physicians with the initial diagnosis of the brain diseases using MRI.